Published online Mar 21, 2014. doi: 10.3748/wjg.v20.i11.2927
Revised: January 6, 2014
Accepted: January 19, 2014
Published online: March 21, 2014
Hepatitis C virus (HCV) is a serious public health problem affecting 170 million carriers worldwide. It is a leading cause of chronic hepatitis, cirrhosis, and liver cancer and is the primary cause for liver transplantation worldwide. HCV genotype 6 (HCV-6) is restricted to South China, South-East Asia, and it is also occasionally found in migrant patients from endemic countries. HCV-6 has considerable genetic diversity with 23 subtypes (a to w). Although direct sequencing followed by phylogenetic analysis is the gold standard for HCV-6 genotyping and subtyping, there are also now rapid genotyping tests available such as the reverse hybridization line probe assay (INNO-LiPA II; Innogenetics, Zwijnaarde, Belgium). HCV-6 patients present with similar clinical manifestations as patients infected with other genotypes. Based on current evidence, the optimal treatment duration of HCV-6 with pegylated interferon/ribavirin should be 48 wk, although a shortened treatment duration of 24 wk could be sufficient in patients with low pretreatment viral load who achieve rapid virological response. In addition, the development of direct-acting antiviral agents is ongoing, and they give high response rate when combined with standard therapy. Herein, we review the epidemiology, classification, diagnosis and treatment as it pertain to HCV-6.
Core tip: Hepatitis C virus (HCV) genotype 6 is restricted to South China, South-East Asia, and it is occasionally found in migrant patients from endemic countries. Treatment response rates are lower than those of genotype 3 but higher than those of genotype 1. Based on current evidence, the optimal treatment duration of HCV-6 should be 48 wk. Shortened treatment duration of 24 wk could be sufficient in patients with low pretreatment viral load who achieve rapid virological response. The development of direct-acting antiviral agents is ongoing, and they give high response rate when combined with standard therapy. We review the epidemiology, classification, diagnosis and treatment.
- Citation: Thong VD, Akkarathamrongsin S, Poovorawan K, Tangkijvanich P, Poovorawan Y. Hepatitis C virus genotype 6: Virology, epidemiology, genetic variation and clinical implication. World J Gastroenterol 2014; 20(11): 2927-2940
- URL: https://www.wjgnet.com/1007-9327/full/v20/i11/2927.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i11.2927
Hepatitis C virus (HCV) infection is an important worldwide public health problem. Most HCV cases become chronic hepatitis C (CHC), which may advance to liver fibrosis, cirrhosis, and hepatocellular carcinoma. The global prevalence of HCV infection is estimated at more than 170 million people[1-3], and some studies estimate that mortality related to HCV infection (death from liver failure or hepatocellular carcinoma) will continue to increases over the next two decades[4].
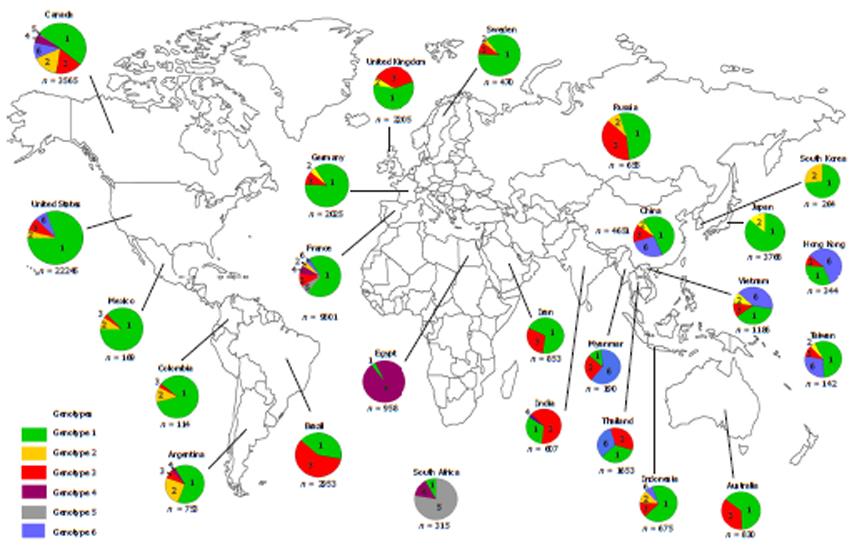
Hepatitis C virus is a member of the Flaviviridae family and belongs to the genus Hepacivirus. HCV is classified into six major genotypes (1-6) and subdivided into various subtypes named in alphabetical order from a to z. Currently, sequencing of HCV isolates has identified more than 83 subtypes from the six genotypes[5]. The genomes among HCV genotypes differ from each other by approximately 30%-35% while the genomes among the subtypes differ by 15%-20%. The prevalence of HCV genotypes varies geographically: HCV-1 is found worldwide including developed regions such as North America and Europe. HCV-2 has high prevalence in Central and West Africa as well as some western countries, while HCV-3 is predominantly found in the Far Eastern countries and the Indian subcontinent[6]. Meanwhile, HCV genotypes 4, 5 and 6 are endemic to specific geographical areas: HCV-4 is mainly found in Egypt and Sub-Saharan Africa, HCV-5 in South Africa[7], and HCV-6 in South China and South-East Asian countries[8-10] (Figure 1)[10-34].
HCV-6 is highly diverse with 23 subtypes currently known[8,9,30]. This genetic diversity may be the result of a long period of viral circulation[14]. In addition, repeated viral exposure through activities such as intravenous drug use leads to more than one viral strain circulating in the host, which in turn increases the chance of viral recombination events among circulating HCV genotypes and strains. The high variation and accumulation of HCV-6 in Southeast Asia also supports the idea that this area may be the origin and worldwide distribution center of this genotype.
Various factors determine treatment response. In the case of host factors, age, sex, race, fibrosis and steatosis level all have important influences on treatment outcome[35-37], while the most important viral factors for predicting response to IFN-based therapy are genotype and viral load at baseline. In the past 10 years, the standard treatment of HCV patients has been a combination of pegylated interferon (PEG-IFN) and ribavirin (RBV) with a 24 to 48 wk regimen depending on the viral genotype of each infected individual. Thus, HCV genotype is considered one of the most robust independent predictors for sustained virological response (SVR). Most clinical studies on PEG-IFN/RBV efficacy have been based on common genotypes such as genotype 1, 2 and 3, while scant clinical data has been generated concerning genotype 6. However, due to this genotype’s extreme genetic diversity, it may be important to study it in a controlled, clinical setting in order to gauge the standard therapy’s efficacy[5].
Currently, the few available studies suggest that treatment with the longer 48-wk regimen may lead to a higher rate of SVR. On the other hand though, treatment with the 24-wk regimen may also lead to a similar SVR rate in subgroups of patients, as in the case of patients with genotypes 2 and 3[38]. In 2011, newer agents known as direct-acting antivirals (DAA) were approved for use in conjunction with PEG-IFN and RBV for the treatment of HCV-1. However, the efficacy and safety of DAA in the treatment of the HCV-6 patients still needs to be assessed.
This study aimed to review the virology, epidemiology, genetic variation and clinical implication of HCV genotype 6. All data were retrieved and selected from related HCV-6 topics from PubMed database.
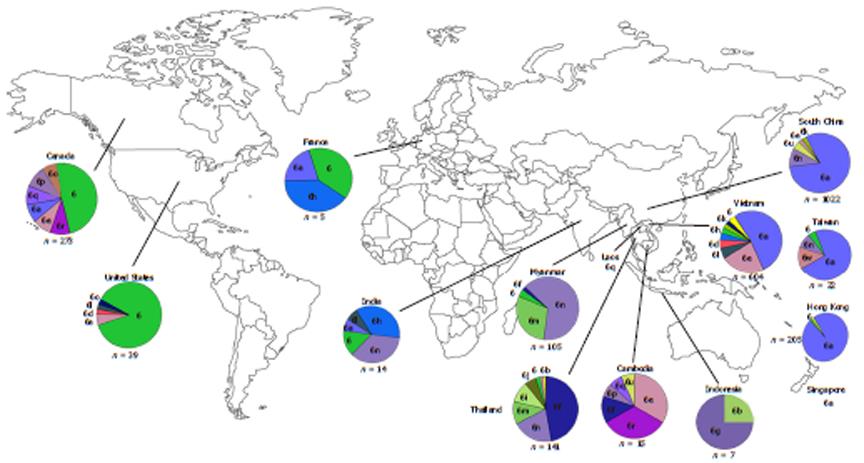
Within endemic countries, HCV-6 shows variability in subtype prevalence. Vietnam has reported HCV-6 prevalence of 51%-54.4% in Ho Chi Minh City and 47.1% elsewhere with the most common subtypes being 6a followed by 6e and 6l. In total, 12 subtypes have thus far been identified in Vietnam (6a, 6c, 6e, 6f, 6h, 6k, 6l, 6n, 6o, 6p, 6r and 6t)[39-41]. Although there is limited information regarding HCV epidemiology in Laos and Cambodia, a few studies have reported a high proportion of HCV-6 in these countries also. Among Laos blood donors, 95.6% of the HCV RNA positive samples were classified as HCV-6 with various subtypes being found including 6b, 6h, 6k, 6l, 6n, 6o, 6q and unclassified subtypes[42,43]. A study of Cambodian migrants in Thailand reported that HCV seroprevalence in this group is similar to their guest country (2.3%). It was also found that HCV-6 is the most dominate genotype in Cambodian migrants with 52% of the HCV-RNA carriers testing positive for this genotype with the subtype breakdown as the following; 6e (20%), 6r (20%), 6f (8%) and 6p (4%)[42]. Similar to its neighboring countries, Thailand also has several endemic HCV-6 subtypes (6b, 6c, 6e, 6f, 6i, 6m and 6n) along with novel subtypes (6u and 6v), which are found in the North and Northeast[43]. However, unlike the Cambodian migrant population in Thailand, this genotype contributes a lower proportion (20.1%) of overall infection in comparison with HCV-3 and HCV-1[32]. In Myanmar, HCV-6 prevalence has gone from being the third most prevalent HCV genotype in 2004 to the first after analysis of a large number of blood samples from 2007. HCV-6 is especially prevalent in the Northern part of Myanmar with subtypes 6f, 6n and 6m predominating[44-47]. Despite is ubiquitous presence in the above-mentioned countries, however, HCV-6 is only rarely reported in other proximal countries. For example, only three samples of subtype 6g (previously designated as genotype 1a) have been reported in Indonesia since 1996[48], while only a single recent report of subtype 6n has been found in Kuala Lumpur, Malaysia, and notably, this patient was co-infected with HIV-1 and had a history of IVDU[49].
HCV-6 can also be found outside of the immediate South East Asian region in countries such as South China, Hong Kong and Taiwan (Figure 2)[28,40,43,46-48,50-55].
Intravenous drug abuse is the transmission route suspected of being most responsible for the high frequency of this genotype in certain parts of Asia and the factor driving a continuous discovery of new subtypes. In China, HCV is frequently found in the South in patients with HIV-1 co-infection and IDU history[56]. For example, prevalence of HCV-6 in chronic hepatitis cases is 12.9% to 14.2% while being 28.2% to 51.5% in IDUs. In addition, similar to Vietnam, 6a is the major subtype in IDUs from all study groups[17,57,58]. A similar trend of HCV-6 infection can also be observed in Hong Kong. More than half of infected IVDU have this genotype (53.2% to 58.5%), which is much higher than the prevalence in the general population (23.6%), and subtype 6a is the most common subtype[52,59]. In Taiwan, there has been no report of HCV-6 until 2010, but since then there is a growing prevalence of HCV-6 and subtype 6a along with novel subtypes being reported in prisoners and IDUs[53].
There is now considerable evidence to support the hypothesis that HCV-6 originated in Southeast Asia. First, this genotype is mainly observed in countries such as Vietnam, Cambodia, Laos, Myanmar and Thailand with the prevalence of HCV-6 in these countries ranging from approximately 20% to more than 50% of all genotypes. Second, there are a great number of known and novel subtypes circulating within these populations[41]. Third, when HCV-6 is found outside of its endemic area, such as in countries like US, Canada and Australia, the virus is almost exclusively isolated from Asian immigrants[7,60]. Fourth, a study in Laos showed that HCV-6 is highly divergent from other genotypes, and that it has distinct genetic differences from other strains, which suggests that there may yet be unclassified subtypes existing in this area. Thus, the accumulation of such genetic heterogeneity suggests that this genotype has circulated, adapted and evolved in this area for a long period of time (Table 1)[39,46,61-64].
| Country | Ref. | Genotype 6 prevalence |
| Myanmar | Lwin et al[46] | 1333 (49) |
| Vietnam (Hanoi, Vietnam) | Pham et al[39] | 238 (47.1) |
| Thailand | ||
| (blood donors throughout country) | Kanistanon et al[61] | NR (18) |
| (blood donors in northern Thailand provinces) | Jutavigittum et al[62] | 326 (31) |
| (blood donors from central Thailand) | Akkarathamrongsin et al[63] | 375 (30) |
| Hong Kong, China (Blood donors) | Leung et al[64] | 910 (27) |
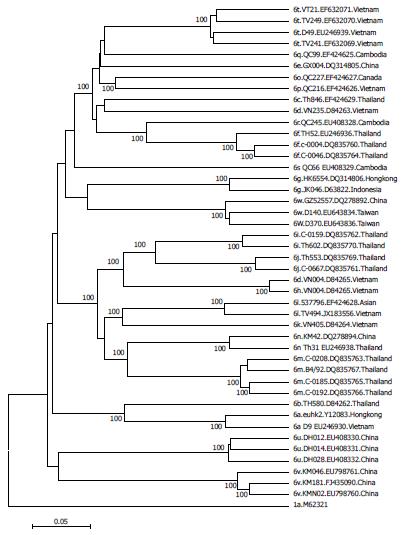
Evolutionary analysis of HCV-6 subtypes hypothesizes that these subtypes evolved from a common ancestor more than 1000 years ago, and that some subtypes may have maintained their endemicity via local epidemics during the 20th century initiated and propagated by modern medicine, blood transfusion and IVDU[42]. However, each of the subtypes seems generally restricted to different locations such as subtype 6d in Vietnam, 6f in Thailand, 6g in Indonesia and 6r in Cambodia. In addition, strains isolated from the same country tend to cluster together in a HCV-6 phylogenetic tree (Figure 3)[42,47].
HCV-6 has spread through other East Asian countries by multiple transmission routes with one of the most effective routes being IDU. In the general population, HCV-1 is still the predominant genotype across Asia. However, HCV-6 - and especially subtype 6a - is the dominant genotype among IVDUs[53,58,59]. An example of this is the phylogeographic and coalescent analysis of HCV-6’s spread through China. Analysis shows that subtype 6a and some other Chinese strains may have originated in Vietnam and spread to neighboring Guangxi and Yunnan provinces[65,66]. Finally, since Guangdong is a major gateway to China, this province may have been the origin of subtype 6a dispersion throughout other regions of the country[58].
Similar to other RNA viruses, HCV has a high tendency for genetic heterogeneity due to a lack of proof reading activity by its viral RNA polymerase. This genetic drift results in an accumulation of viral mutations, which will then be selected forby forces of environmental pressure. Thus, only the fittest strains survive and become the major circulating viral population. Of course, however, this process of fixation requires a relatively long period of time. Meanwhile, viral recombination can result in novel strains overnight, although the drastic nature of recombination can be a danger to viral survival. So far, the evidence suggests that novel HCV strains mainly accumulate through genetic drift by collecting viral mutation instead of recombination, as there have only been two reported cases of HCV-6-specific recombination occurring[67,68]; recombinants RF_2i/6p and RF_2b/6w from a Vietnamese blood donor and an IDU in Taiwan, respectively[53,69].
Since HCV genotype is such an important consideration for predicting an effective treatment regimen, several different genotyping methods have been developed. These methods include direct nucleic acid sequencing[70], a reverse hybridization line probe assay (LiPA)[71], subtype-specific reverse transcription (RT)-PCR[72], DNA restriction fragment length polymorphism[73], heteroduplex mobility analysis[74], primer-specific and mispair extension analysis[75], melting curve analysis with fluorescence resonance energy transfer probes[76] and serologic genotyping (Figure 4)[5,77,78].
However, not every method is equal. The Asian Pacific Association for the Study of the Liver states that genotype discrimination based on primers from the 5’untranslated region (5’UTR) do not distinguish some of the HCV-6 subtypes prevalent in Southeast Asia, and that these subtypes should instead be classified as genotype 1 or 1b[79], available methods generally use distinct motifs found within the HCV genome for HCV genotype[5]. In addition, previous studies reported the mistyping of HCV-6 as genotype 1 by the INNO-LiPA I assay (Innogenetics, Zwijnaarde, Belgium)[80,81]. However, the INNO-LiPA HCV II (Innogenetics, Zwijnaarde, Belgium) genotyping assay seems to overcome the deficiencies demonstrated by the INNO-LiPA I assay, as it has shown remarkable improvement in genotyping accuracy and differentiation between HCV-1 and HCV-6 variants by using core sequencing data as well as 5’ UTR data[82,83].
Acute HCV infection is infrequently diagnosed and leads to chronic infection in about 80% of cases[84]. Clinical manifestations can occur, usually within 7 to 8 wk (range: 2-26 wk) after exposure to HCV, but the majority of persons have either no symptoms or only mild symptoms, and fulminant hepatic failure due to acute HCV infection is very rare. Although clinical features will be present in less than 25% of infected patients, symptoms of acute hepatitis include jaundice, malaise, nausea and right upper quadrant pain[85]. While the infection becomes chronic in most cases, chronic infection is either asymptomatic or has only mild nonspecific symptoms such as fatigue as long as cirrhosis and hepatocellular carcinoma are not present. Other clinical manifestations are possible, however, such as nausea, weakness, myalgia, arthralgia and weight loss[86]. Although there have been many papers describing HCV-6’s epidemiology, the clinical characteristics have not been well described in those studies. Nguyen et al[87] reported that patients with HCV-6 presented similar clinical manifestations as those with genotype 1 or 2/3. They also found that people with HCV genotype 1 and 6 had a somewhat higher baseline viral load than those with others genotypes. However, when comparing HCV-6 patients with patients infected with other genotypes, these differences were not statistically significant with regard to host factors (e.g., age, history of smoking, alcohol use, family history of CHC, hepatitis B, hepatocellular carcinoma and liver-related death), baseline laboratory values (e.g., ALT, total bilirubin, albumin, white blood cell count, platelet count), and liver histology[55,88]. However, steatosis is a chief modulator of clinical course of HCV infection[89].
The current standard-of-care for treatment of HCV-infected patients is a combination of PEG-IFN and RBV. Among the various viral and host factors, HCV genotype is one of the most important predictors of response to treatment and is used to guide the duration of treatment. Patients with HCV genotype 1 are typically treated for 48 wk, whereas patients with genotype 2 and 3 are treated for 24 wk[35,36,90]. Limited studies have suggested that the response rate of HCV-6 may be at an intermediate level between those of genotypes 3 and 1[87,91-93].
Virological response kinetics during therapy has emerged as an important prognostic factor of treatment outcome in patients with chronic HCV infection[94,95]. Absence of an early virological response (EVR) at week 12 during therapy is the best negative predictor for non-response to treatment. In contrast, rapid virological response (RVR; defined as undetectable HCV RNA at week 4) is regarded as the most important predictor for SVR (defined as undetectable HCV RNA at week 24 after the end of therapy) and has emerged as an important milestone to guide the appropriate duration of therapy. For example, in patients with genotype 1, an individualized approach to therapy designed according to early viral kinetics has been adopted to optimize therapeutic outcome in patients. Recent clinical trials have used RVR to identify those patients with low baseline viral load that may benefit from shortened treatment duration of 24 wk[96-98].
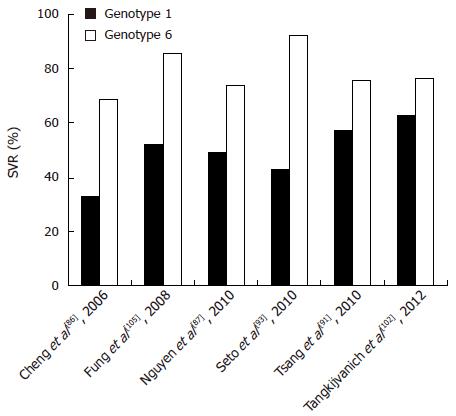
Currently, although the treatment outcome of patients with HCV-6 has so far not been exhaustively studied, a few studies exit which give hints as to what the standard course of care should be. Most previous studies have reported that genotype 6 behaves more similar to genotypes 2 and 3 (SVR rates of 76%-80%)[87,99,100] and thus responds better to therapy than genotype 1 (SVR rates of 46%-52%)[101]. For example, Nguyen et al[87] demonstrated that patients with HCV-6 had significantly better SVRs to PEG-IFN and RBV combination therapy than patients with genotype 1 (74% vs 49%). Furthermore, Tangkijvanich et al[102] reported that the SVR of HCV-6 is higher than that of genotype 1 but lower than that of genotype 3 (Figure 5).
Most prior studies of HCV-6 included patients treated for 48-52 wk[103-105]. A small study of Asian-American patients comparing a 48-wk to a shortened 24-wk regimen showed a significantly higher SVR rate in those treated by the 48-wk course (74% vs 49%)[38]. However, the limitation of the study was its retrospective design and the results were not analyzed with regard to an intention-to-treat method. A retrospective study conducted in China showed that the rate of SVR in 22 patients with HCV-6 treated for 24 wk was comparable to that of genotypes 2/3 (82% and 83%, respectively)[92]. In that study, the positive predictive values of RVR and EVR for HCV-6 were comparable with those for genotypes 2/3 (87% vs 91% and 86% vs 87%, respectively).
A randomized controlled trial of 60 patients with HCV-6 demonstrated that there was no significant difference in SVR rates in patients treated with 48-wk and 24-wk regimens (79% and 70%, respectively)[101]. In that study, RVR was a significant predictor of SVR in the 48-wk group and tending towards significance in the 24-wk group, although a sizeable number of patients did not have RVR measurement performed. Recently, Thu Thuy et al[106] conducted an open-label randomized trial in Vietnam, which aimed at assessing the rate of SVR in HCV-6 chronic HCV following 48 and 24 wk of PEG-IFN and RBV combination therapy. They demonstrated that RVR was achieved in the majority of HCV-6 patients and similar and high rates of SVR were noted following 48- and 24-wk therapy (71% vs 60%, respectively, P = 0.24).
The feasibility of a response-guided therapy by individualizing the duration of treatment according to viral kinetics in patients with genotype 6 was first investigated by Fired et al[107]. In that pilot study, more than 70% of patients with HCV-6 achieved RVR and received an abbreviated 24-wk regimen. Among them, the rate of relapse was approximately 10%, and nearly 90% of them eradicated the virus. These data were consistent with observations regarding treatment of HCV genotypes 1, 2, 3 and 4, which suggest that monitoring RVR might be useful to guide treatment duration for patients with HCV-6. In particular, therapy might be shortened to 24 wk in patients with low pretreatment viral load who achieve RVR, whereas a 48-wk course was appropriate for those who cleared the virus after week 4. The abbreviated regimen could offer advantages by reducing unnecessary medication exposure, which may make the treatment of HCV-6 more affordable and maximize the cost effectiveness of therapy. However, further prospective randomized trials are required to evaluate the response-guided strategy in a larger number of patients with HCV-6.
Although PEG-IFN represents the backbone of treatment, combination with RBV has been shown to help prevent relapse. Current guidelines recommend a weight-adjusted dose of RBV in combination with PEG-IFN for treating patients with genotype 1, while a flat, low dose of RBV (800 mg/d) is recommended for treating patients with genotype 3[95]. However, a weight-adjusted dose of RBV might be useful to enhance the response rate in patients with genotype 3 who do not achieve RVR and in those with RVR undergoing abbreviated therapy[108,109]. Currently, the optimal dose of RBV for treatment of patients with HCV-6 is unknown. In previous studies, daily weight-based or fixed doses of RBV had been used, rendering comparisons rather complicated. Nonetheless, recent prospective trials adopted a weight-based dosage of RBV (1000-1200 mg/d) for abbreviated treatment (24 wk), which might result in achieving SVR equivalent to that obtained with longer treatment duration (48 wk)[101]. These data might reflect the need of a weight-based dosage of RBV in patients with HCV-6 undergoing abbreviated therapy.
Treatment with PEG-IFN and RBV has been shown to be safe in patients with HCV-6, but treatment discontinuation or dose reduction due to side-effects is typical. Although HCV genotypes play a role in achieving SVR, there is no significant difference in the frequency or types of side effects experienced among patients of genotypes 1, 2, 3 or 6[87,104,105] taking PEG-IFN and RBV, although side effect profiles do appear to differ among patients of different ethnicities. For example, compared with Caucasians, Asian patients are more likely to decrease their RBV dose or discontinue the therapy due to anemia. In addition, Asian patients reported symptoms of depression, more commonly than Caucasian patients[110,111]. Other common side effects include flu-like symptoms (fever, fatigue, headache, malign, and loss of appetite), dyspepsia and some cases with rash, weight loss, arthralgia and alopecia[110]. However, these symptoms are often mild and tolerable and without the requirement for PEG-IFN and/or RBV dose modification.
As in studies of patients with other HCV genotypes, pretreatment predictors of response are useful for advising patients on their likelihood of SVR. Pretreatment host and viral characteristics affect early viral kinetics. Once treatment has been initiated, outcome depends on how fast HCV RNA becomes undetectable. Multivariate analyses have identified various predictors of response in HCV-6 such as youth (< 40-50 years)[100,105], low BMI, treatment adherence and RVR[35,100,105]. Among these predictors - and concordant with observations in other HCV genotypes - the importance of RVR (undetectable HCV RNA after 4 wk of treatment) in the prediction of SVR has been further substantiated in HCV-6, wherein the positive predictive value to achieve SVR in patients with RVR has been 83%-87%[87,105].
Recent studies have reported that one of the strongest baseline predictors of SVR in HCV genotype 1 are single-nucleotide polymorphisms (SNPs) on chromosome 19 in or near the interleukin-28B gene (IL28B, encoding interferon lambda-3). Following antiviral treatment, patients carrying the CC genotype of one of these predictive SNPs (rs12979860) have a twofold (95%CI: 1.8-2.3) greater rate of SVR than patients who carry the TT alleles (78% for the CC genotype, 38% for the TC genotype, and 26% for the TT genotype). Interestingly, the C allele frequency is much higher in white and Asian populations than in black populations[112]. More recently, a variant upstream of IFNL3 creating a novel gene, designated as IFNL4, has been discovered[113]. This region harbors a dinucleotide variant (ss469415590) that is found in two alternative forms (ΔG or TT alleles). The ss469415590 indel is more strongly associated with treatment response of HCV-1 infection in African-American individuals compared to rs12979860[113].
Data regarding the association of the SNPs with antiviral response in HCV-6 infected patients are still very limited. A recent study from Hong Kong showed that rs8099917, another IL28B polymorphism, was associated with response to PEG-IFN/RBV therapy in HCV-6 infected patients[114]. In that report, the favorable TT genotype of rs8099917, when compared to the unfavorable TG genotype, was significantly associated with an increased SVR rate (96.2% and 62.5%, respectively) and was the only clinical parameter that predicted SVR.
In 2011, direct-acting antivirals (telaprevir and boceprevir) were approved by the US. Food and Drug Administration for treatment of HCV genotype 1. They are first generation NS3-4A protease inhibitors (PI), targeting the protease enzyme that cleaves the HCV polyprotein thus inhibiting the replication process. The addition of a DAA to PEG-IFN/RBV has reduced treatment duration and side effects, and improve efficacy and cost[115]. Thus, the development of DAA represents a significant milestone in improving the efficacy of HCV treatment, especially in patients with HCV genotype 1.
Currently, clinical results of the use of DAAs for patients with HCV-6 are limited[116]. For example, monotherapy with TMC 435, a second-generation NS3/4A PI with pan-genotype antiviral activity, could induce a significant mean viremia decrease of -4.35 log10 UI/mL after 8 days in patients with HCV-6. In addition, five patients with HCV-6 were included in the ATOMIC study and treated with sofosbuvir (GS 7977), a NS5B polymerase inhibitor, plus PEG-IFN/RBV for 24 wk. The RVR rate at week 4 and the SVR rate 12 wk after the end of the treatment were both 100%.
Three percent of the world’s population is infected with HCV. Of that 3%, HCV-6 accounts for a disproportionately high burden high of prevalence in Southeast Asia and the surrounding areas as well as in infection drug users and people with thalassemia major. Previous literature suggest older tests may have misclassified HCV-6 as genotype 1, but newer line probe assays have shown impressive improvement in genotyping accuracy and differentiation between HCV genotype 1 and HCV-6 variants. Clinical characteristics and predictors of poor response are similar for patients with HCV-6 and other HCV genotypes. Current data suggests that the response rate of HCV-6 may be at an intermediate level between those of genotypes 3 and 1. Thus, the optimal treatment duration of HCV-6 should be 48 wk, although shortened treatment duration of 24 wk could be sufficient in patients with low pretreatment viral load who achieve RVR. In addition, there are currently conflicting data on the role of IL28B testing in predicting treatment response in patients with HCV-6.
Further studies will be required to arrive at a sensitively diagnostic method for HCV-6/subtypes, optimal treatment duration, and early predictors for treatment response and drugs (DAA) to achieve higher SVR rates in patients with HCV-6.
We would like to express our gratitude to the entire staff of the Gastroenterology unit department of Medicine, Center of Excellence in Clinical Virology, Department of Biochemistry, Faculty of Medicine, Chulalongkorn University and Hospital, Thai Red Cross Society. Finally, we would like to thank Brian Muchmore for reviewing our manuscript.
P- Reviewers: Castiella A, Chiu KW, Dang SS, Lonardo A S- Editor: Gou SX L- Editor: A E- Editor: Zhang DN
| 1. | Leary TP, Gutierrez RA, Muerhoff AS, Birkenmeyer LG, Desai SM, Dawson GJ. A chemiluminescent, magnetic particle-based immunoassay for the detection of hepatitis C virus core antigen in human serum or plasma. J Med Virol. 2006;78:1436-1440. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 45] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 2. | Moreno M, Pérez-Alvarez R, Rodrigo L, Pérez-López R, Suárez-Leiva P. Long-term evolution of serum and liver viral markers in patients treated for chronic hepatitis C and sustained response. J Viral Hepat. 2006;13:28-33. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 3. | Ramia S, Eid-Fares J. Distribution of hepatitis C virus genotypes in the Middle East. Int J Infect Dis. 2006;10:272-277. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 71] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 4. | Kim WR. The burden of hepatitis C in the United States. Hepatology. 2002;36:S30-S34. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 128] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 5. | Chao DT, Abe K, Nguyen MH. Systematic review: epidemiology of hepatitis C genotype 6 and its management. Aliment Pharmacol Ther. 2011;34:286-296. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 6. | Simmonds P. Genetic diversity and evolution of hepatitis C virus--15 years on. J Gen Virol. 2004;85:3173-3188. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 622] [Cited by in F6Publishing: 575] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 7. | Antaki N, Craxi A, Kamal S, Moucari R, Van der Merwe S, Haffar S, Gadano A, Zein N, Lai CL, Pawlotsky JM. The neglected hepatitis C virus genotypes 4, 5 and 6: an international consensus report. Liver Int. 2010;30:342-355. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 128] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 8. | Mellor J, Holmes EC, Jarvis LM, Yap PL, Simmonds P. Investigation of the pattern of hepatitis C virus sequence diversity in different geographical regions: implications for virus classification. The International HCV Collaborative Study Group. J Gen Virol. 1995;76:2493-2507. [PubMed] [Cited in This Article: ] |
| 9. | Simmonds P, Bukh J, Combet C, Deléage G, Enomoto N, Feinstone S, Halfon P, Inchauspé G, Kuiken C, Maertens G. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42:962-973. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1070] [Cited by in F6Publishing: 1058] [Article Influence: 55.7] [Reference Citation Analysis (0)] |
| 10. | Alter MJ. Epidemiology of hepatitis C virus infection. World J Gastroenterol. 2007;13:2436-2441. [PubMed] [Cited in This Article: ] |
| 11. | Armstrong GL, Wasley A, Simard EP, McQuillan GM, Kuhnert WL, Alter MJ. The prevalence of hepatitis C virus infection in the United States, 1999 through 2002. Ann Intern Med. 2006;144:705-714. [PubMed] [Cited in This Article: ] |
| 12. | Wiessing L, Guarita B, Giraudon I, Brummer-Korvenkontio H, Salminen M, Cowan SA. European monitoring of notifications of hepatitis C virus infection in the general population and among injecting drug users (IDUs) - the need to improve quality and comparability. Euro Surveill. 2008;13:pii: 18884. [PubMed] [Cited in This Article: ] |
| 13. | Amon JJ, Garfein RS, Ahdieh-Grant L, Armstrong GL, Ouellet LJ, Latka MH, Vlahov D, Strathdee SA, Hudson SM, Kerndt P. Prevalence of hepatitis C virus infection among injection drug users in the United States, 1994-2004. Clin Infect Dis. 2008;46:1852-1858. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 128] [Cited by in F6Publishing: 159] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 14. | Touzet S, Kraemer L, Colin C, Pradat P, Lanoir D, Bailly F, Coppola RC, Sauleda S, Thursz MR, Tillmann H. Epidemiology of hepatitis C virus infection in seven European Union countries: a critical analysis of the literature. HENCORE Group. (Hepatitis C European Network for Co-operative Research. Eur J Gastroenterol Hepatol. 2000;12:667-678. [PubMed] [Cited in This Article: ] |
| 15. | Burguete-García AI, Conde-González CJ, Jiménez-Méndez R, Juárez-Díaz Y, Meda-Monzón E, Torres-Poveda K, Madrid-Marina V. Hepatitis C seroprevalence and correlation between viral load and viral genotype among primary care clients in Mexico. Salud Publica Mex. 2011;53 Suppl 1:S7-12. [PubMed] [Cited in This Article: ] |
| 16. | Viazov S, Kuzin S, Paladi N, Tchernovetsky M, Isaeva E, Mazhul L, Vasychova F, Widell A, Roggendorf M. Hepatitis C virus genotypes in different regions of the former Soviet Union (Russia, Belarus, Moldova, and Uzbekistan). J Med Virol. 1997;53:36-40. [PubMed] [Cited in This Article: ] |
| 17. | Zhou Y, Wang X, Mao Q, Fan Y, Zhu Y, Zhang X, Lan L, Jiang L, Tan W. Changes in modes of hepatitis C infection acquisition and genotypes in southwest China. J Clin Virol. 2009;46:230-233. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 25] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 18. | Utama A, Budiarto BR, Monasari D, Octavia TI, Chandra IS, Gani RA, Hasan I, Sanityoso A, Miskad UA, Yusuf I. Hepatitis C virus genotype in blood donors and associated liver disease in Indonesia. Intervirology. 2008;51:410-416. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 15] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 19. | Samimi-Rad K, Nasiri Toosi M, Masoudi-Nejad A, Najafi A, Rahimnia R, Asgari F, Shabestari AN, Hassanpour G, Alavian SM, Asgari F. Molecular epidemiology of hepatitis C virus among injection drug users in Iran: a slight change in prevalence of HCV genotypes over time. Arch Virol. 2012;157:1959-1965. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 20. | Berg T, Hopf U, Stark K, Baumgarten R, Lobeck H, Schreier E. Distribution of hepatitis C virus genotypes in German patients with chronic hepatitis C: correlation with clinical and virological parameters. J Hepatol. 1997;26:484-491. [PubMed] [Cited in This Article: ] |
| 21. | Campiotto S, Pinho JR, Carrilho FJ, Da Silva LC, Souto FJ, Spinelli V, Pereira LM, Coelho HS, Silva AO, Fonseca JC. Geographic distribution of hepatitis C virus genotypes in Brazil. Braz J Med Biol Res. 2005;38:41-49. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 124] [Cited by in F6Publishing: 130] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 22. | Kamal SM. Hepatitis C genotype 4 therapy: increasing options and improving outcomes. Liver Int. 2009;29 Suppl 1:39-48. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 21] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 23. | Mangia A, Mottola L. What’s new in HCV genotype 2 treatment. Liver Int. 2012;32 Suppl 1:135-140. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 13] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 24. | Oh DJ, Park YM, Seo YI, Lee JS, Lee JY. Prevalence of hepatitis C virus infections and distribution of hepatitis C virus genotypes among Korean blood donors. Ann Lab Med. 2012;32:210-215. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 25. | Das BR, Kundu B, Khandapkar R, Sahni S. Geographical distribution of hepatitis C virus genotypes in India. Indian J Pathol Microbiol. 2002;45:323-328. [PubMed] [Cited in This Article: ] |
| 26. | Zarife MA, Silva LK, Silva MB, Lopes GB, Barreto ML, Teixeira Mda G, Dourado I, Reis MG. Prevalence of hepatitis C virus infection in north-eastern Brazil: a population-based study. Trans R Soc Trop Med Hyg. 2006;100:663-668. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 28] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 27. | Akkarathamrongsin S, Hacharoen P, Tangkijvanich P, Theamboonlers A, Tanaka Y, Mizokami M, Poovorawan Y. Molecular epidemiology and genetic history of hepatitis C virus subtype 3a infection in Thailand. Intervirology. 2013;56:284-294. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 23] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 28. | Narahari S, Juwle A, Basak S, Saranath D. Prevalence and geographic distribution of Hepatitis C Virus genotypes in Indian patient cohort. Infect Genet Evol. 2009;9:643-645. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 56] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 29. | Zali MR, Mayumi M, Raoufi M, Nowroozi A. Hepatitis C virus genotypes in the Islamic Republic of Iran: a preliminary study. East Mediterr Health J. 2000;6:372-377. [PubMed] [Cited in This Article: ] |
| 30. | Gededzha MP, Selabe SG, Kyaw T, Rakgole JN, Blackard JT, Mphahlele MJ. Introduction of new subtypes and variants of hepatitis C virus genotype 4 in South Africa. J Med Virol. 2012;84:601-607. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 18] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 31. | Chamberlain RW, Adams NJ, Taylor LA, Simmonds P, Elliott RM. The complete coding sequence of hepatitis C virus genotype 5a, the predominant genotype in South Africa. Biochem Biophys Res Commun. 1997;236:44-49. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 73] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 32. | Waheed Y, Shafi T, Safi SZ, Qadri I. Hepatitis C virus in Pakistan: a systematic review of prevalence, genotypes and risk factors. World J Gastroenterol. 2009;15:5647-5653. [PubMed] [Cited in This Article: ] |
| 33. | Manos MM, Shvachko VA, Murphy RC, Arduino JM, Shire NJ. Distribution of hepatitis C virus genotypes in a diverse US integrated health care population. J Med Virol. 2012;84:1744-1750. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 69] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 34. | Nguyen LH, Nguyen MH. Systematic review: Asian patients with chronic hepatitis C infection. Aliment Pharmacol Ther. 2013;37:921-936. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 94] [Cited by in F6Publishing: 85] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 35. | Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Gonçales FL, Häussinger D, Diago M, Carosi G, Dhumeaux D. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975-982. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4847] [Cited by in F6Publishing: 4689] [Article Influence: 213.1] [Reference Citation Analysis (0)] |
| 36. | Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, Goodman ZD, Koury K, Ling M, Albrecht JK. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958-965. [PubMed] [Cited in This Article: ] |
| 37. | Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T, Bochud M, Battegay M, Bernasconi E, Borovicka J. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338-1345, 1345.e1-7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 850] [Cited by in F6Publishing: 854] [Article Influence: 61.0] [Reference Citation Analysis (0)] |
| 38. | Nguyen MH, Trinh HN, Garcia R, Nguyen G, Lam KD, Keeffe EB. Higher rate of sustained virologic response in chronic hepatitis C genotype 6 treated with 48 weeks versus 24 weeks of peginterferon plus ribavirin. Am J Gastroenterol. 2008;103:1131-1135. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 39] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 39. | Pham DA, Leuangwutiwong P, Jittmittraphap A, Luplertlop N, Bach HK, Akkarathamrongsin S, Theamboonlers A, Poovorawan Y. High prevalence of Hepatitis C virus genotype 6 in Vietnam. Asian Pac J Allergy Immunol. 2009;27:153-160. [PubMed] [Cited in This Article: ] |
| 40. | Noppornpanth S, Lien TX, Poovorawan Y, Smits SL, Osterhaus AD, Haagmans BL. Identification of a naturally occurring recombinant genotype 2/6 hepatitis C virus. J Virol. 2006;80:7569-7577. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 115] [Cited by in F6Publishing: 122] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 41. | Pham VH, Nguyen HD, Ho PT, Banh DV, Pham HL, Pham PH, Lu L, Abe K. Very high prevalence of hepatitis C virus genotype 6 variants in southern Vietnam: large-scale survey based on sequence determination. Jpn J Infect Dis. 2011;64:537-539. [PubMed] [Cited in This Article: ] |
| 42. | Pybus OG, Barnes E, Taggart R, Lemey P, Markov PV, Rasachak B, Syhavong B, Phetsouvanah R, Sheridan I, Humphreys IS. Genetic history of hepatitis C virus in East Asia. J Virol. 2009;83:1071-1082. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 159] [Cited by in F6Publishing: 161] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 43. | Hübschen JM, Jutavijittum P, Thammavong T, Samountry B, Yousukh A, Toriyama K, Sausy A, Muller CP. High genetic diversity including potential new subtypes of hepatitis C virus genotype 6 in Lao People’s Democratic Republic. Clin Microbiol Infect. 2011;17:E30-E34. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 44. | Shinji T, Lwin AA, Gokan K, Obika M, Ryuko H, Khin M, Okada S, Koide N. Three type 6 hepatitis C virus subgroups among blood donors in the Yangon area of Myanmar are identified as subtypes 6m and 6n, and a novel subtype by sequence analysis of the core region. Acta Med Okayama. 2006;60:345-349. [PubMed] [Cited in This Article: ] |
| 45. | Shinji T, Kyaw YY, Gokan K, Tanaka Y, Ochi K, Kusano N, Mizushima T, Fujioka S, Shiraha H, Lwin AA. Analysis of HCV genotypes from blood donors shows three new HCV type 6 subgroups exist in Myanmar. Acta Med Okayama. 2004;58:135-142. [PubMed] [Cited in This Article: ] |
| 46. | Lwin AA, Shinji T, Khin M, Win N, Obika M, Okada S, Koide N. Hepatitis C virus genotype distribution in Myanmar: Predominance of genotype 6 and existence of new genotype 6 subtype. Hepatol Res. 2007;37:337-345. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 53] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 47. | Akkarathamrongsin S, Praianantathavorn K, Hacharoen N, Theamboonlers A, Tangkijvanich P, Poovorawan Y. Seroprevalence and genotype of hepatitis C virus among immigrant workers from Cambodia and Myanmar in Thailand. Intervirology. 2011;54:10-16. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 48. | Tokita H, Okamoto H, Iizuka H, Kishimoto J, Tsuda F, Lesmana LA, Miyakawa Y, Mayumi M. Hepatitis C virus variants from Jakarta, Indonesia classifiable into novel genotypes in the second (2e and 2f), tenth (10a) and eleventh (11a) genetic groups. J Gen Virol. 1996;77:293-301. [PubMed] [Cited in This Article: ] |
| 49. | Ng KT, Lee YM, Al-Darraji HA, Xia X, Takebe Y, Chan KG, Lu L, Mahadeva S, Kamarulzaman A, Tee KK. Genome sequence of the hepatitis C virus subtype 6n isolated from malaysia. Genome Announc. 2013;1:pii: e00168-12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 3] [Reference Citation Analysis (0)] |
| 50. | Yan Z, Fan K, Wang Y, Fan Y, Tan Z, Deng G. Changing pattern of clinical epidemiology on hepatitis C virus infection in southwest china. Hepat Mon. 2012;12:196-204. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 19] [Reference Citation Analysis (0)] |
| 51. | Apichartpiyakul C, Apichartpiyakul N, Urwijitaroon Y, Gray J, Natpratan C, Katayama Y, Fujii M, Hotta H. Seroprevalence and subtype distribution of hepatitis C virus among blood donors and intravenous drug users in northern/northeastern Thailand. Jpn J Infect Dis. 1999;52:121-123. [PubMed] [Cited in This Article: ] |
| 52. | Chan DP, Lee SS, Lee KC. The effects of widespread methadone treatment on the molecular epidemiology of hepatitis C virus infection among injection drug users in Hong Kong. J Med Virol. 2011;83:1187-1194. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 53. | Lee YM, Lin HJ, Chen YJ, Lee CM, Wang SF, Chang KY, Chen TL, Liu HF, Chen YM. Molecular epidemiology of HCV genotypes among injection drug users in Taiwan: Full-length sequences of two new subtype 6w strains and a recombinant form_2b6w. J Med Virol. 2010;82:57-68. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 64] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 54. | Chowdhury A, Santra A, Chaudhuri S, Dhali GK, Chaudhuri S, Maity SG, Naik TN, Bhattacharya SK, Mazumder DN. Hepatitis C virus infection in the general population: a community-based study in West Bengal, India. Hepatology. 2003;37:802-809. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 127] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 55. | Nguyen NH, Vutien P, Trinh HN, Garcia RT, Nguyen LH, Nguyen HA, Nguyen KK, Nguyen MH. Risk factors, genotype 6 prevalence, and clinical characteristics of chronic hepatitis C in Southeast Asian Americans. Hepatol Int. 2010;4:523-529. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 56. | Garten RJ, Lai S, Zhang J, Liu W, Chen J, Vlahov D, Yu XF. Rapid transmission of hepatitis C virus among young injecting heroin users in Southern China. Int J Epidemiol. 2004;33:182-188. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 125] [Cited by in F6Publishing: 138] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 57. | Garten RJ, Zhang J, Lai S, Liu W, Chen J, Yu XF. Coinfection with HIV and hepatitis C virus among injection drug users in southern China. Clin Infect Dis. 2005;41 Suppl 1:S18-S24. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 79] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 58. | Fu Y, Qin W, Cao H, Xu R, Tan Y, Lu T, Wang H, Tong W, Rong X, Li G. HCV 6a prevalence in Guangdong province had the origin from Vietnam and recent dissemination to other regions of China: phylogeographic analyses. PLoS One. 2012;7:e28006. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 57] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 59. | Zhou DX, Tang JW, Chu IM, Cheung JL, Tang NL, Tam JS, Chan PK. Hepatitis C virus genotype distribution among intravenous drug user and the general population in Hong Kong. J Med Virol. 2006;78:574-581. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 37] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 60. | Murphy DG, Willems B, Deschênes M, Hilzenrat N, Mousseau R, Sabbah S. Use of sequence analysis of the NS5B region for routine genotyping of hepatitis C virus with reference to C/E1 and 5’ untranslated region sequences. J Clin Microbiol. 2007;45:1102-1112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 208] [Cited by in F6Publishing: 212] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 61. | Kanistanon D, Neelamek M, Dharakul T, Songsivilai S. Genotypic distribution of hepatitis C virus in different regions of Thailand. J Clin Microbiol. 1997;35:1772-1776. [PubMed] [Cited in This Article: ] |
| 62. | Jutavijittum P, Jiviriyawat Y, Yousukh A, Pantip C, Maneekarn N, Toriyama K. Genotypic distribution of hepatitis C virus in voluntary blood donors of northern Thailand. Southeast Asian J Trop Med Public Health. 2009;40:471-479. [PubMed] [Cited in This Article: ] |
| 63. | Akkarathamrongsin S, Praianantathavorn K, Hacharoen N, Theamboonlers A, Tangkijvanich P, Tanaka Y, Mizokami M, Poovorawan Y. Geographic distribution of hepatitis C virus genotype 6 subtypes in Thailand. J Med Virol. 2010;82:257-262. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 47] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 64. | Leung N, Chu C, Tam JS. Viral hepatitis C in Hong Kong. Intervirology. 2006;49:23-27. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 31] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 65. | Xia X, Luo J, Bai J, Yu R. Epidemiology of hepatitis C virus infection among injection drug users in China: systematic review and meta-analysis. Public Health. 2008;122:990-1003. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 121] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 66. | Rong X, Lu L, Wang J, Xiong H, Huang J, Chen J, Huang K, Xu R, Wang M, Zhang X. Correlation of viral loads with HCV genotypes: higher levels of virus were revealed among blood donors infected with 6a strains. PLoS One. 2012;7:e52467. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 67. | Kalinina O, Norder H, Mukomolov S, Magnius LO. A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J Virol. 2002;76:4034-4043. [PubMed] [Cited in This Article: ] |
| 68. | Kurbanov F, Tanaka Y, Avazova D, Khan A, Sugauchi F, Kan N, Kurbanova-Khudayberganova D, Khikmatullaeva A, Musabaev E, Mizokami M. Detection of hepatitis C virus natural recombinant RF1_2k/1b strain among intravenous drug users in Uzbekistan. Hepatol Res. 2008;38:457-464. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 41] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 69. | Noppornpanth S, Sablon E, De Nys K, Truong XL, Brouwer J, Van Brussel M, Smits SL, Poovorawan Y, Osterhaus AD, Haagmans BL. Genotyping hepatitis C viruses from Southeast Asia by a novel line probe assay that simultaneously detects core and 5’ untranslated regions. J Clin Microbiol. 2006;44:3969-3974. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 41] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 70. | Simmonds P, Holmes EC, Cha TA, Chan SW, McOmish F, Irvine B, Beall E, Yap PL, Kolberg J, Urdea MS. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J Gen Virol. 1993;74:2391-2399. [PubMed] [Cited in This Article: ] |
| 71. | Stuyver L, Wyseur A, van Arnhem W, Hernandez F, Maertens G. Second-generation line probe assay for hepatitis C virus genotyping. J Clin Microbiol. 1996;34:2259-2266. [PubMed] [Cited in This Article: ] |
| 72. | Okamoto H, Sugiyama Y, Okada S, Kurai K, Akahane Y, Sugai Y, Tanaka T, Sato K, Tsuda F, Miyakawa Y. Typing hepatitis C virus by polymerase chain reaction with type-specific primers: application to clinical surveys and tracing infectious sources. J Gen Virol. 1992;73:673-679. [PubMed] [Cited in This Article: ] |
| 73. | Nakao T, Enomoto N, Takada N, Takada A, Date T. Typing of hepatitis C virus genomes by restriction fragment length polymorphism. J Gen Virol. 1991;72:2105-2112. [PubMed] [Cited in This Article: ] |
| 74. | White PA, Zhai X, Carter I, Zhao Y, Rawlinson WD. Simplified hepatitis C virus genotyping by heteroduplex mobility analysis. J Clin Microbiol. 2000;38:477-482. [PubMed] [Cited in This Article: ] |
| 75. | Hu YW, Balaskas E, Furione M, Yen PH, Kessler G, Scalia V, Chui L, Sher G. Comparison and application of a novel genotyping method, semiautomated primer-specific and mispair extension analysis, and four other genotyping assays for detection of hepatitis C virus mixed-genotype infections. J Clin Microbiol. 2000;38:2807-2813. [PubMed] [Cited in This Article: ] |
| 76. | Schröter M, Zöllner B, Schäfer P, Landt O, Lass U, Laufs R, Feucht HH. Genotyping of hepatitis C virus types 1, 2, 3, and 4 by a one-step LightCycler method using three different pairs of hybridization probes. J Clin Microbiol. 2002;40:2046-2050. [PubMed] [Cited in This Article: ] |
| 77. | Dixit V, Quan S, Martin P, Larson D, Brezina M, DiNello R, Sra K, Lau JY, Chien D, Kolberg J. Evaluation of a novel serotyping system for hepatitis C virus: strong correlation with standard genotyping methodologies. J Clin Microbiol. 1995;33:2978-2983. [PubMed] [Cited in This Article: ] |
| 78. | Cai Q, Zhao Z, Liu Y, Shao X, Gao Z. Comparison of three different HCV genotyping methods: core, NS5B sequence analysis and line probe assay. Int J Mol Med. 2013;31:347-352. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 61] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 79. | McCaughan GW, Omata M, Amarapurkar D, Bowden S, Chow WC, Chutaputti A, Dore G, Gane E, Guan R, Hamid SS. Asian Pacific Association for the Study of the Liver consensus statements on the diagnosis, management and treatment of hepatitis C virus infection. J Gastroenterol Hepatol. 2007;22:615-633. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 115] [Cited by in F6Publishing: 119] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 80. | Chinchai T, Noppornpanth S, Bedi K, Theamboonlers A, Poovorawan Y. 222 base pairs in NS5B region and the determination of hepatitis C virus genotype 6. Intervirology. 2006;49:224-229. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 81. | Bukh J, Purcell RH, Miller RH. Sequence analysis of the core gene of 14 hepatitis C virus genotypes. Proc Natl Acad Sci USA. 1994;91:8239-8243. [PubMed] [Cited in This Article: ] |
| 82. | Chinchai T, Labout J, Noppornpanth S, Theamboonlers A, Haagmans BL, Osterhaus AD, Poovorawan Y. Comparative study of different methods to genotype hepatitis C virus type 6 variants. J Virol Methods. 2003;109:195-201. [PubMed] [Cited in This Article: ] |
| 83. | Bouchardeau F, Cantaloube JF, Chevaliez S, Portal C, Razer A, Lefrère JJ, Pawlotsky JM, De Micco P, Laperche S. Improvement of hepatitis C virus (HCV) genotype determination with the new version of the INNO-LiPA HCV assay. J Clin Microbiol. 2007;45:1140-1145. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 94] [Cited by in F6Publishing: 100] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 84. | Farci P, Alter HJ, Shimoda A, Govindarajan S, Cheung LC, Melpolder JC, Sacher RA, Shih JW, Purcell RH. Hepatitis C virus-associated fulminant hepatic failure. N Engl J Med. 1996;335:631-634. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 123] [Cited by in F6Publishing: 123] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 85. | National Institutes of Health Consensus Development Conference Panel statement: management of hepatitis C. Hepatology. 1997;26:2S-10S. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 335] [Cited by in F6Publishing: 368] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 86. | Cheng JT, Hsien C, Sun HE, Tong MJ. The emerging importance of chronic hepatitis C infection in Asian Americans. Am J Gastroenterol. 2006;101:2737-2743. [PubMed] [Cited in This Article: ] |
| 87. | Nguyen NH, VuTien P, Garcia RT, Trinh H, Nguyen H, Nguyen K, Levitt B, Nguyen MH. Response to pegylated interferon and ribavirin in Asian American patients with chronic hepatitis C genotypes 1 vs 2/3 vs 6. J Viral Hepat. 2010;17:691-697. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 32] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 88. | Seong MH, Kil H, Kim JY, Lee SS, Jang ES, Kim JW, Jeong SH, Kim YS, Bae SH, Lee YJ. Clinical and epidemiological characteristics of Korean patients with hepatitis C virus genotype 6. Clin Mol Hepatol. 2013;19:45-50. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 89. | Adinolfi LE, Restivo L, Zampino R, Lonardo A, Loria P. Metabolic alterations and chronic hepatitis C: treatment strategies. Expert Opin Pharmacother. 2011;12:2215-2234. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 57] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 90. | Chevaliez S, Pawlotsky JM. Hepatitis C virus: virology, diagnosis and management of antiviral therapy. World J Gastroenterol. 2007;13:2461-2466. [PubMed] [Cited in This Article: ] |
| 91. | Tsang OT, Zee JS, Chan JM, Li RS, Kan YM, Li FT, Lo FH, Chow DA, Cheung KW, Chan KH. Chronic hepatitis C genotype 6 responds better to pegylated interferon and ribavirin combination therapy than genotype 1. J Gastroenterol Hepatol. 2010;25:766-771. [PubMed] [Cited in This Article: ] |
| 92. | Zhou YQ, Wang XH, Hong GH, Zhu Y, Zhang XQ, Hu YJ, Mao Q. Twenty-four weeks of pegylated interferon plus ribavirin effectively treat patients with HCV genotype 6a. J Viral Hepat. 2011;18:595-600. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 93. | Seto WK, Lai CL, Fung J, Hung I, Yuen J, Young J, Wong DK, Yuen MF. Natural history of chronic hepatitis C: genotype 1 versus genotype 6. J Hepatol. 2010;53:444-448. [PubMed] [Cited in This Article: ] |
| 94. | de Leuw P, Sarrazin C, Zeuzem S. How to use virological tools for the optimal management of chronic hepatitis C. Liver Int. 2011;31 Suppl 1:3-12. [PubMed] [Cited in This Article: ] |
| 95. | Ghany MG, Strader DB, Thomas DL, Seeff LB. Diagnosis, management, and treatment of hepatitis C: an update. Hepatology. 2009;49:1335-1374. [PubMed] [Cited in This Article: ] |
| 96. | Ferenci P, Laferl H, Scherzer TM, Gschwantler M, Maieron A, Brunner H, Stauber R, Bischof M, Bauer B, Datz C. Peginterferon alfa-2a and ribavirin for 24 weeks in hepatitis C type 1 and 4 patients with rapid virological response. Gastroenterology. 2008;135:451-458. [PubMed] [Cited in This Article: ] |
| 97. | Jensen DM, Morgan TR, Marcellin P, Pockros PJ, Reddy KR, Hadziyannis SJ, Ferenci P, Ackrill AM, Willems B. Early identification of HCV genotype 1 patients responding to 24 weeks peginterferon alpha-2a (40 kd)/ribavirin therapy. Hepatology. 2006;43:954-960. [PubMed] [Cited in This Article: ] |
| 98. | Zeuzem S, Buti M, Ferenci P, Sperl J, Horsmans Y, Cianciara J, Ibranyi E, Weiland O, Noviello S, Brass C. Efficacy of 24 weeks treatment with peginterferon alfa-2b plus ribavirin in patients with chronic hepatitis C infected with genotype 1 and low pretreatment viremia. J Hepatol. 2006;44:97-103. [PubMed] [Cited in This Article: ] |
| 99. | Nguyen MH, Keeffe EB. Prevalence and treatment of hepatitis C virus genotypes 4, 5, and 6. Clin Gastroenterol Hepatol. 2005;3:S97-S101. [PubMed] [Cited in This Article: ] |
| 100. | Yuen MF, Lai CL. Response to combined interferon and ribavirin is better in patients infected with hepatitis C virus genotype 6 than genotype 1 in Hong Kong. Intervirology. 2006;49:96-98. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 32] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 101. | Lam KD, Trinh HN, Do ST, Nguyen TT, Garcia RT, Nguyen T, Phan QQ, Nguyen HA, Nguyen KK, Nguyen LH. Randomized controlled trial of pegylated interferon-alfa 2a and ribavirin in treatment-naive chronic hepatitis C genotype 6. Hepatology. 2010;52:1573-1580. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 31] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 102. | Tangkijvanich P, Komolmit P, Mahachai V, Poovorawan K, Akkarathamrongsin S, Poovorawan Y. Response-guided therapy for patients with hepatitis C virus genotype 6 infection: a pilot study. J Viral Hepat. 2012;19:423-430. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 103. | Hui CK, Yuen MF, Sablon E, Chan AO, Wong BC, Lai CL. Interferon and ribavirin therapy for chronic hepatitis C virus genotype 6: a comparison with genotype 1. J Infect Dis. 2003;187:1071-1074. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 49] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 104. | Dev AT, McCaw R, Sundararajan V, Bowden S, Sievert W. Southeast Asian patients with chronic hepatitis C: the impact of novel genotypes and race on treatment outcome. Hepatology. 2002;36:1259-1265. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 79] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 105. | Fung J, Lai CL, Hung I, Young J, Cheng C, Wong D, Yuen MF. Chronic hepatitis C virus genotype 6 infection: response to pegylated interferon and ribavirin. J Infect Dis. 2008;198:808-812. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 57] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 106. | Thu Thuy PT, Bunchorntavakul C, Tan Dat H, Rajender Reddy K. A randomized trial of 48 versus 24 weeks of combination pegylated interferon and ribavirin therapy in genotype 6 chronic hepatitis C. J Hepatol. 2012;56:1012-1018. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 107. | Fried MW, Hadziyannis SJ, Shiffman ML, Messinger D, Zeuzem S. Rapid virological response is the most important predictor of sustained virological response across genotypes in patients with chronic hepatitis C virus infection. J Hepatol. 2011;55:69-75. [PubMed] [Cited in This Article: ] |
| 108. | Mangia A. Individualizing treatment duration in hepatitis C virus genotype 2/3-infected patients. Liver Int. 2011;31:36-41. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 9] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 109. | Martin P, Jensen DM. Ribavirin in the treatment of chronic hepatitis C. J Gastroenterol Hepatol. 2008;23:844-855. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 56] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 110. | Vutien P, Nguyen NH, Trinh HN, Li J, Garcia RT, Garcia G, Nguyen KK, Nguyen HA, Levitt BS, Keeffe EB. Similar treatment response to peginterferon and ribavirin in Asian and Caucasian patients with chronic hepatitis C. Am J Gastroenterol. 2010;105:1110-1115. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 19] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 111. | Fancher TL, Ton H, Le Meyer O, Ho T, Paterniti DA. Discussing depression with Vietnamese American patients. J Immigr Minor Health. 2010;12:263-266. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 112. | Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399-401. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2776] [Cited by in F6Publishing: 2666] [Article Influence: 177.7] [Reference Citation Analysis (0)] |
| 113. | Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H, Hergott D, Porter-Gill P, Mumy A, Kohaar I. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet. 2013;45:164-171. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 701] [Cited by in F6Publishing: 729] [Article Influence: 66.3] [Reference Citation Analysis (0)] |
| 114. | Seto WK, Tsang OT, Liu K, Chan JM, Wong DK, Fung J, Lai CL, Yuen MF. Role of IL28B and inosine triphosphatase polymorphisms in the treatment of chronic hepatitis C virus genotype 6 infection. J Viral Hepat. 2013;20:470-477. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 115. | Kiser JJ, Flexner C. Direct-acting antiviral agents for hepatitis C virus infection. Annu Rev Pharmacol Toxicol. 2013;53:427-449. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 68] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 116. | Wartelle-Bladou C, Le Folgoc G, Bourlière M, Lecomte L. Hepatitis C therapy in non-genotype 1 patients: the near future. J Viral Hepat. 2012;19:525-536. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |