Published online May 15, 2024. doi: 10.4251/wjgo.v16.i5.2123
Peer-review started: January 11, 2024
First decision: January 30, 2024
Revised: February 19, 2024
Accepted: March 13, 2024
Article in press: March 13, 2024
Published online: May 15, 2024
MicroRNAs (miRNAs) regulate gene expression and play a critical role in cancer physiology. However, there is still a limited understanding of the function and regulatory mechanism of miRNAs in gastric cancer (GC).
To investigate the role and molecular mechanism of miRNA-145-5p (miR145-5p) in the progression of GC.
Real-time polymerase chain reaction (RT-PCR) was used to detect miRNA expression in human GC tissues and cells. The ability of cancer cells to migrate and invade was assessed using wound-healing and transwell assays, respectively. Cell proliferation was measured using cell counting kit-8 and colony formation assays, and apoptosis was evaluated using flow cytometry. Expression of the epithelial-mesenchymal transition (EMT)-associated protein was determined by Western blot. Targets of miR-145-5p were predicated using bioinformatics analysis and verified using a dual-luciferase reporter system. Serpin family E member 1
GC tissues and cells had reduced miR-145-5p expression and SERPINE1 was identified as a direct target of this miRNA. Overexpression of miR-145-5p was associated with decreased GC cell proliferation, invasion, migration, and EMT, and these effects were reversed by forcing SERPINE1 expression. Kaplan-Meier plot analysis revealed that patients with higher SERPINE1 expression had a shorter survival rate than those with lower SERPINE1 expression. Nude mouse tumorigenesis experiments confirmed that miR-145-5p targets SERPINE1 to regulate extracellular signal-regulated kinase-1/2 (ERK1/2).
This study found that miR-145-5p inhibits tumor progression and is expressed in lower amounts in patients with GC. MiR-145-5p was found to affect GC cell proliferation, migration, and invasion by negatively regulating SERPINE1 levels and controlling the ERK1/2 pathway.
Core Tip: Abnormal microRNAs (miRNAs) expression is found in multiple diseases, including cancer, where it contributes to tumor progression. The exact role of miRNA-145-5p (miR-145-5p) in gastric cancer (GC) remains poorly understood. The current study assessed the molecular pathways used by miR-145-5p to regulate GC in tumor tissues, cell lines, and a nude mouse model. MiR-145-5p was shown to target serpin family E member 1, regulate the extracellular signal-regulated kinase-1/2 pathway, and directly impact GC progression, playing an important role in this disease. The findings identify the molecular mechanism of GC progression.
- Citation: Bai HX, Qiu XM, Xu CH, Guo JQ. MiRNA-145-5p inhibits gastric cancer progression via the serpin family E member 1- extracellular signal-regulated kinase-1/2 axis. World J Gastrointest Oncol 2024; 16(5): 2123-2140
- URL: https://www.wjgnet.com/1948-5204/full/v16/i5/2123.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i5.2123
Gastric cancer (GC) is a common malignancy of the digestive system that poses a substantial threat to human health. According to the GLOBOCAN 2018 estimates of worldwide cancer incidence and mortality, the incidence rate of GC ranked fifth (5.7%), and the mortality rate ranked third (8.3%)[1], and these rates are increasing annually[2,3]. GC has a simple histological classification, with 90% of cases originating from adenocarcinoma, however, there are also special types of this disease[4]. GC occurrence and prognosis are impacted by several factors, including alcohol consumption, smoking, obesity, gender, and diet[5]. The International Agency for Research on Cancer classified Helicobacter pylori as a first-class carcinogen of non-cardia GC[6]. Previous researchers have proposed that intestinal GC progresses as follows: Normal gastric mucosa → chronic inflammation → atrophic gastritis → intestinal metaplasia → precancerous lesions → GC (Correa mode)[7,8]. GC occurrence, progression, invasion, and metastasis are closely related to differential gene expression, gene mutations, and epigenetic changes[9]. In recent years, gastroscopy has aided the early diagnosis and treatment of GC. However, due to low screening rates and atypical early symptoms, late-stage diagnosis of GC patients remains high, resulting in the loss of surgical opportunities, high metastasis rates, and poor prognosis. While surgery, chemotherapy, and radiation therapy can prolong the time to GC recurrence and metastasis time, half of patients still experience recurrence and metastasis within 2 years after surgery and ultimately die[10]. Thus, an improved unders
MicroRNAs (miRNAs) are highly conserved single-stranded small endogenous non-coding RNAs, with a length of 20-25 nucleotides (nt), that are transcribed from miRNA genes. Although miRNAs are never translated into proteins, they play pivotal regulatory roles at the transcriptional and post-transcriptional levels by inhibiting gene expression[11]. Abnormal miRNA expression is associated with multiple diseases, including cancer, during which they modulate tumor progression and metastasis[12-15]. Intervening at the miRNA level is shown to alter tumor progression and offer new treatment opportunities, establishing their role as potential therapeutic targets[[16-18].
miRNA-145-5p (miR-145-5p), encoded by the MIR145 gene on chromosome 5, is a tumor suppressor miRNA in numerous human cancers, including bladder[19], prostate[20], breast[21], cervical[22], esophageal squamous cell[23], and colorectal[24]. Zhou et al[25] found that miR-145-5p could induce GC cell differentiation by directly targeting the KLF5 3'-UTR. However, the exact role of miR-145-5p in GC remains poorly understood. In this study, we used real-time polymerase chain reaction (RT-PCR), transwell assay, colony formation assay, Western blot, dual-luciferase reporter system, nude mice model, and many other methods to find that the expression of miR-145-5p was negatively related to GC progression by targeting serpin family E member 1 (SERPINE1) through signal-regulated kinase-1/2 (ERK1/2) pathway.
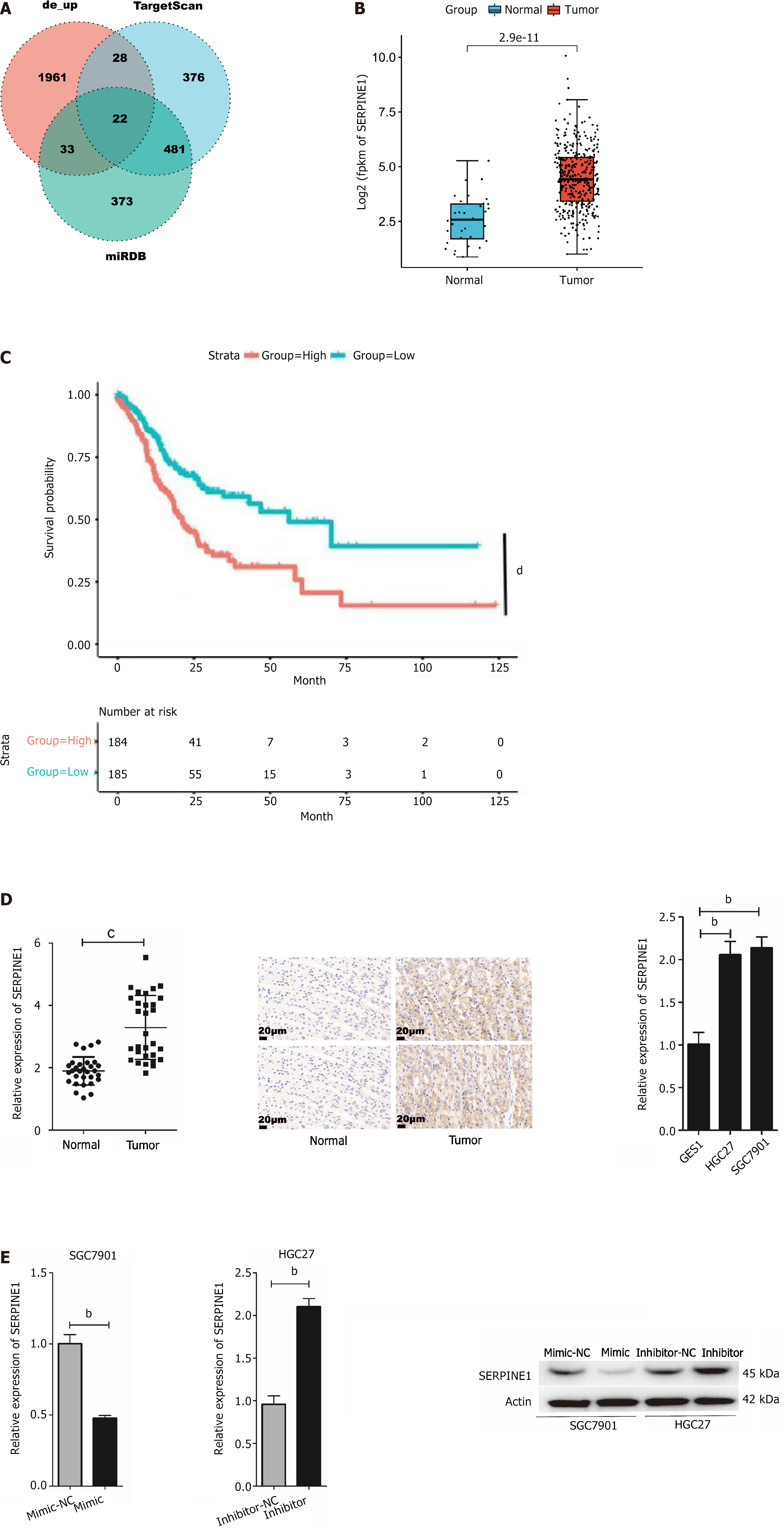
GC miRNA and mRNA expression data were acquired from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). The “edgeR” package was used to obtain the differentially expressed miRNAs and mRNAs and a boxplot was constructed and confirmed as the research object. Two databases (TargetScan and miRDB) were employed to predict the downstream target mRNAs of miR-145-5p. A Venn diagram was created to find the potential target mRNA.
The human GC cell lines (BGC-823, MGC-803, HGC-27, and SGC-7901) and the immortalized normal gastric mucosal cell line (GES-1) were purchased from the Cell Bank of the Chinese Academy of Sciences (CAS, China). All cell lines were cultured in RPMI-1640 medium (MACGENE, China) supplemented with 10% fetal bovine serum (EXcell Bio, China), 100 μg/mL streptomycin, and 100 U/mL Penicillin and incubated at 37 °C and 5% CO2. When the cells reached a confluency of 80%-90%, they were passaged and reseeded at an appropriate density.
For functional assays, miR-145-5p mimic (mimic), miR-145-5p inhibitor (inhibitor), NC for miR-145-5p mimic (mimic-NC), and NC for miR-145-5p inhibitor (inhibitor-NC) were synthesized and purified by Ribobio (China). Transfections were performed using Lipofectamine2000 (Invitrogen, United States) following the provided guidelines: When the cells reached the logarithmic growth phase, trypLE was used to digest the cells. The cells were then centrifuged, resuspended, and counted on a 6-well plate (with a fusion degree of about 80% during cell transfection. The mixed siRNA (100 nM) and an appropriate amount of Lipofectamine2000 dissolved in serum free Opti MEM I was added to the plate. After transfection, the culture medium was replaced after culturing the cells in a CO2 incubator at 37 °C for 4-6 h. The cells were incubated for 48 h before proceeding with additional experiments.
To verify the role of SERPINE1 in GC, the SGC-7901 and HGC-27 cells were transfected with lentivirus (sourced from Genomedicech), China. The cells were classified into two groups: Overexpressed SERPINE1 (OE) and control (OE-NC). Both the HGC27 and SGC7901 cells were cultivated overnight in 6-well plates. After reaching 40%-50% confluency, lentiviral infection was introduced. Stable transfectants were subsequently selected using 2 ug/mL puromycin screening. The expression of SERPINE1 in stably transfected cell lines was assessed using western blotting and q-PCR.
For the rescue experiment, HGC27 and SGC7901 cells were stably transfected with miR-145-5p-mimic (mimic) and miR-145-5p-mimic-NC (mimic-NC) and divided into four groups: Mimic-NC + OE-NC, mimic + OE-NC, mimic-NC + OE, and mimic + OE.
TRNzol reagent (Tiangen Biotech, China) was used for total RNA extraction and then reverse-transcribed into cDNA using a miRNA 1st Strand cDNA Synthesis Kit (Vazyme Biotech Co., Ltd, China). The real-time quantitative PCR (qRT-PCR) process was carried out using SYBR Green (Tiangen Biotech Co, Ltd, China) according to a specific thermal cycling protocol. U6 and actin served as endogenous controls, and the target miRNA and mRNA relative expression levels were calculated using the 2-ΔΔCt method. The primers used in this study were synthesized by Beijing Dingguo Changsheng Biotech. Co., Ltd (Table 1).
| Gene | Primer sequence |
| miR-145-5p | F: 5′- GCGTTTGGCAATGGTAGAACT -3′ |
| R: 5′- AGTGCAGGGTCCGAGGTATT -3′ | |
| U6 | F: 5′- CGGTCCAGTTTTCCCAGGA -3′ |
| R: 5′- AGTGCAGGGTCCGAGGTATT-3′ | |
| SERPINE1 | F: 5′- GCAAGGCACCTCTGAGAACT -3′ |
| R: 5′- GGGTGAGAAAACCACGTTGC -3′ | |
| Actin | F: 5′- ACACTGTGCCCATCTACG -3′ |
| R: 5′- TGTCACGCACGATTTCC -3′ |
Total protein was isolated from the cells using 1 × sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) Sample Loading Buffer (Beyotime Biotechnology, China). A BCA Protein Assay Kit (Beyotime Biotechnology, China) was used to determine the concentration of the protein samples, and the samples were quantified at different concentrations. The proteins were then boiled at 100°C for 10 min with 100 μL of loading buffer (Beyotime Biotechnology, China) and separated by SDS-PAGE at 120 V. After electrophoresis, the proteins were transferred onto PVDF membranes and blocked using 5% BSA/TBST for 60 min. the primary rabbit polyclonal antibodies, SERPINE1 (Proteintech, United States), α-smooth muscle actin (α-SMA) (Proteintech, United States), β-catenin (Proteintech, United States), vimentin (HuaBio, China), ERK1/2 (HuaBio, China), phosphorylation extracellular signal-regulated kinase1/2 (p-ERK1/2) (HuaBio, China) and actin (ZSGB-BIO, China), and the mouse monoclonal antibody E-cad (HuaBio, China), were added to the membranes and incubated overnight at 4 °C. Secondary goat anti-rabbit IgG or goat anti-mouse IgG (ZSGB-BIO, China) antibodies were then added and the membrane was incubated at room temperature for 1 h and then washed three times with TBST buffer for 10 min. The Immobilon Western Chemilum HRP Substrate (WBKLS0500, Millipore, United States) was used to visualize the protein bands, and Image Pro Plus 6.0 (Media Cybernetics, United States) software was employed to analyze the relative protein levels.
Tumor tissue from human or nude mice was fixed with 4% formaldehyde, dehydrated, embedded in paraffin, and cut into 4 μm thick tissue sections. After heating at 60 °C for 1 h, the slices were dewaxed and rehydrated with a concentration gradient of alcohol (100, 95, 90, 80, and 70%). The tissue sections were boiled in 10 mmol/L sodium citrate buffer (pH 6.0) at a high temperature to recover the antigens, and incubated for 20 min with 3% hydrogen peroxide to block endogenous peroxidase activity. BSA (5%) was added to the sections and sealed for 30 min. The slices were incubated overnight with primary anti-SERPINE1 (Proteintech, United States, 1:500) and anti-Ki67 (Proteintech, United States, 1:10000) antibodies at 4 °C. The tissue was then incubated with a secondary antibody and observed under a microscope. After the slices were slightly dried, the freshly prepared DAB chromogenic solution (TIANGEN, PA140212) was dripped onto the circle. The coloration time was controlled under the microscope, with brown color representing positive staining. The slices were then washed with tap water to stop the coloration. Harris hematoxylin (BOSTER, AR1108) was used to restain the samples for 5 min (the time was controlled by the degree of staining). The samples were then washed in tap water, differentiated with 1% hydrochloric acid alcohol for a few seconds, and rinsed in tap water. When the ammonia water was blue, the samples were rinsed in running water, dehydrated, and sealed. The images were observed and captured using a fluorescence microscope.
A Dual-Luciferase Reporter Gene Assay was conducted to predict the binding site of miR-145-5p and SERPINE1 3'-UTR. Wild-type and mutant SERPINE1 3'-UTR fragments were synthesized and cloned into the pmirGLO plasmid. The amplified sequences of the wild type 3'UTR of SERPINE1 (WT-3'UTR) were cloned into the pmirGLO vectors (Promega Corp., WI, United States) for the construction of luciferase Wt-SERPINE1 vectors and mutant vectors (luciferase Mut-SERPINE1) were synthesized by Sangon Biotech (China). MiR-145-5p mimic and NC were co-transfected into 293T cells with Wt-SERPINE1 or Mut-SERPINE1 (SCSP-502, National Collection of Authenticated Cell Cultures, China), respectively. After 48 h, Firefly and Renilla luciferase activities were measured using the Varioskan LUX Multi-function enzyme labeling instrument (Thermo Scientific, United States).
GC cell proliferation was measured using a cell counting kit-8 (CCK-8), Meilunbio, China. GC cells at a density of 5 × 103 cells/well were suspended in 100 μL and incubated in 96-well plates at 5% CO2 and 37 °C for 48 h. CCK-8 reagent (10 μL) was introduced into each well for a 4-h incubation. Absorbance values were recorded at 450 nm.
Cells were dispensed into 6-well plates at a density of 1000 cells/well to assess colony formation. After 10 d, the colonies were preserved using methanol and stained with 0.1% crystal violet alcohol solvent (G1014-50ML, Servicebio, China) for 15 min. Images of the stained colonies were obtained and their numbers were determined.
Flow cytometry was performed according to the Annexin V-FITC/PI apoptosis kit (Multi Sciences, China) instructions. To test the samples, 5 × 105 cells, including those in the culture supernatant, were collected by centrifuging with pre-cooled phosphate-buffered saline (PBS) at 4 °C and 1500 rpm for 5 min. Annexin V-FITC (5 μL) and 10 μL PI were added to each tube, mixed gently, and incubated in the dark at room temperature for 5 min. A CytoFLEX (BECKMAN, United States) was used to analyze the proportion of apoptotic cells in each sample. Annexin V-FITC was detected using the FITC detection channel (Ex = 488 nm; Em = 530 nm), and PI was detected using the PI detection channel (Ex = 535 nm; Em = 615 nm).
A 24-well Transwell BD Matrigel system (FN, Corning, Costar, China) with an 8 μm pore size was used to measure cellular invasion. Approximately 7.5 × 104 cells were added to the upper chamber pre-coated with a Matrigel matrix (Yes Service Biotech, China), while the lower chamber was filled with RPMI-1640 medium containing 10% FBS to act as an attractant. After 24 h of incubation at 37 °C, non-invading cells on the Matrigel membrane surface were removed. The invading cells were fixed and stained using 4% methanol and 0.1% crystal violet, and two random fields were used to conduct the cell counts under a microscope.
A wound-healing assay was performed to evaluate cellular migration. First, horizontal lines were drawn on the back of a 6-well plate using a marker pen and ruler, with each well crossed by at least five evenly spaced lines. Cells were resuscitated in RPMI-1640 medium containing 10% FBS, seeded on six-well plates at a density of 3 × 105 cells/well density, and incubated overnight. After cell reached 100% confluence, scratched cells with a 200 μL pipette tip and changed previous medium to serum-free medium. Following a 48-h culture, the cells were washed with PBS, and images of the scratch area in each well were taken at various time points. ImageJ software was used to measure the width of the scratch area and cell mobility was calculated to compare differences in tumor cell migration.
Fifteen male nude BALB/c mice, aged 4-6 wk and weighing 18-22 g, were purchased from the Experimental Animal Center of Shandong University. After 1 wk of adaptive feeding, 15 nude mice were randomly divided into three groups: the SERPINE1-NC group (NC) (n = 5), the SERPINE1-overexpression group (OE) (n = 5), and the SERPINE1-OE + PD98059 group (OE + PD98059) (n = 5). The logarithmic growth phase cells in each group were resuspended at 5 × 107/mL, mixed with matrix glue at 1:1, and 0.20 mL of the cell solution was subcutaneously injected into nude mice (0.5 × 107 cells/mouse) to establish a tumor implantation model, with the axillary site used as the injection site. After 18 d, the mice were sacrificed for analysis. The tumor was completely extracted, its mass and volume were calculated, and its length and width were determined to assess growth. Tumor volume was calculated using the formula ½ × length × width2. The average volume of the tumors in each group was used to create a growth curve.
All human and animal experiments were approved by the Clinical Research Ethics Committee of Liaocheng People's Hospital (Liaocheng, China) and following the ethical standards of the institutional and national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Written informed consent was obtained from all individual participants included in this study.
IBM SPSS Statistics 26 and GraphPad Prism 5 were used for all statistical analyses. Comparisons between two different groups across three independent experiments were performed using the student’s t-test. A one-way analysis of variance (ANOVA) was used for multiple group comparisons. The data are presented as the means ± SD. P < 0.05 was considered statistically significant.
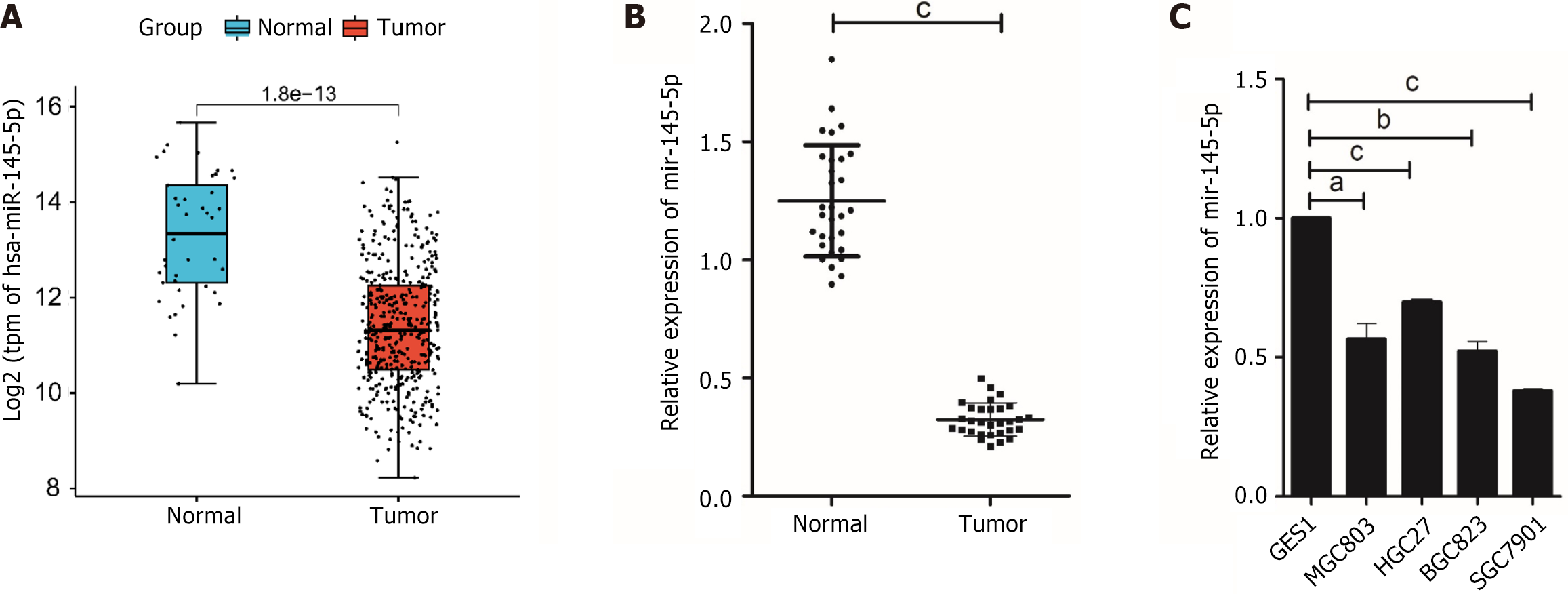
This study used the “edgeR” package to identify DEmiRNAs in TCGA-CESC data. The findings revealed that miR-145-5p was significantly downregulated in tumor tissues (Figure 1A). RT-PCR was then used to evaluate miR-145-5p expression in 30 paired samples of fresh GC tissues and their corresponding non-tumor tissues (P < 0.001). MiR-145-5p was expressed at much lower levels in GC tissues than their non-tumor counterparts (Figure 1B). Moreover, when compared with the immortalized normal gastric mucosal cell line, miR-145-5p expression was markedly lower in MGC803 (P < 0.05), HGC27 (P < 0.001), BGC823 (P < 0.01), and SGC7901 (P < 0.001) cell lines (Figure 1C). Since miR-145-5p expression was lowest in SGC7901 cells and highest in HGC27 cells, these cell lines were selected for the gain-of-function and loss-of-function assays, respectively.
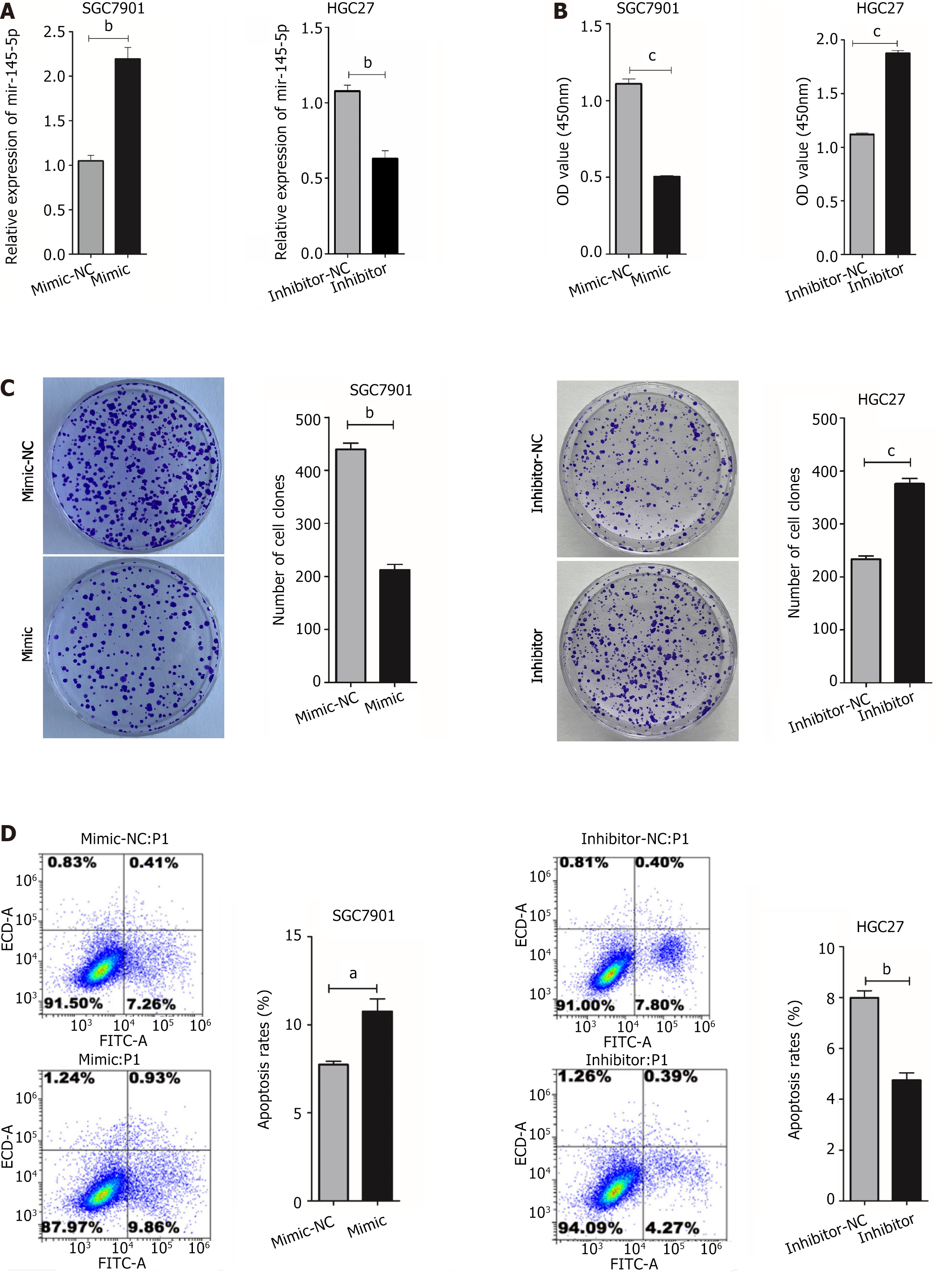
MiR-145-5p downregulation in GC cells suggested that it may play an inhibitory role in this disease. Thus, its mimics and inhibitors were introduced into GC cells (Figure 2A). As expected, miR-145-5p expression was higher in the mimic group (P < 0.01) and lower in the inhibitor group (P < 0.01) (Figure 2A). CCK-8 and colony formation experiments were used to determine if miR-145-5p could regulate GC proliferation. Cell viability was higher in the miR-145-5p inhibition group than in the inhibitor-NC group (P < 0.001) (Figure 2B), and the number of clones formed was also higher (P < 0.001) (Figure 2C). The miR145-5p-mimic group had less cell proliferation than the mimic group (P < 0.001) (Figure 2B), and a fewer number of clones formed (P < 0.01) (Figure 2C).
Flow cytometry was used to measure HGC27 and SGC7901 cell apoptosis after transfection. The number of apoptotic cells was higher in the miR-145-5p mimic group than in the mimic-NC group (P < 0.05). Furthermore, the number of apoptotic cells was lower in the miR-145-5p inhibitor group than in the inhibitor-NC group (P < 0.01) (Figure 2D).
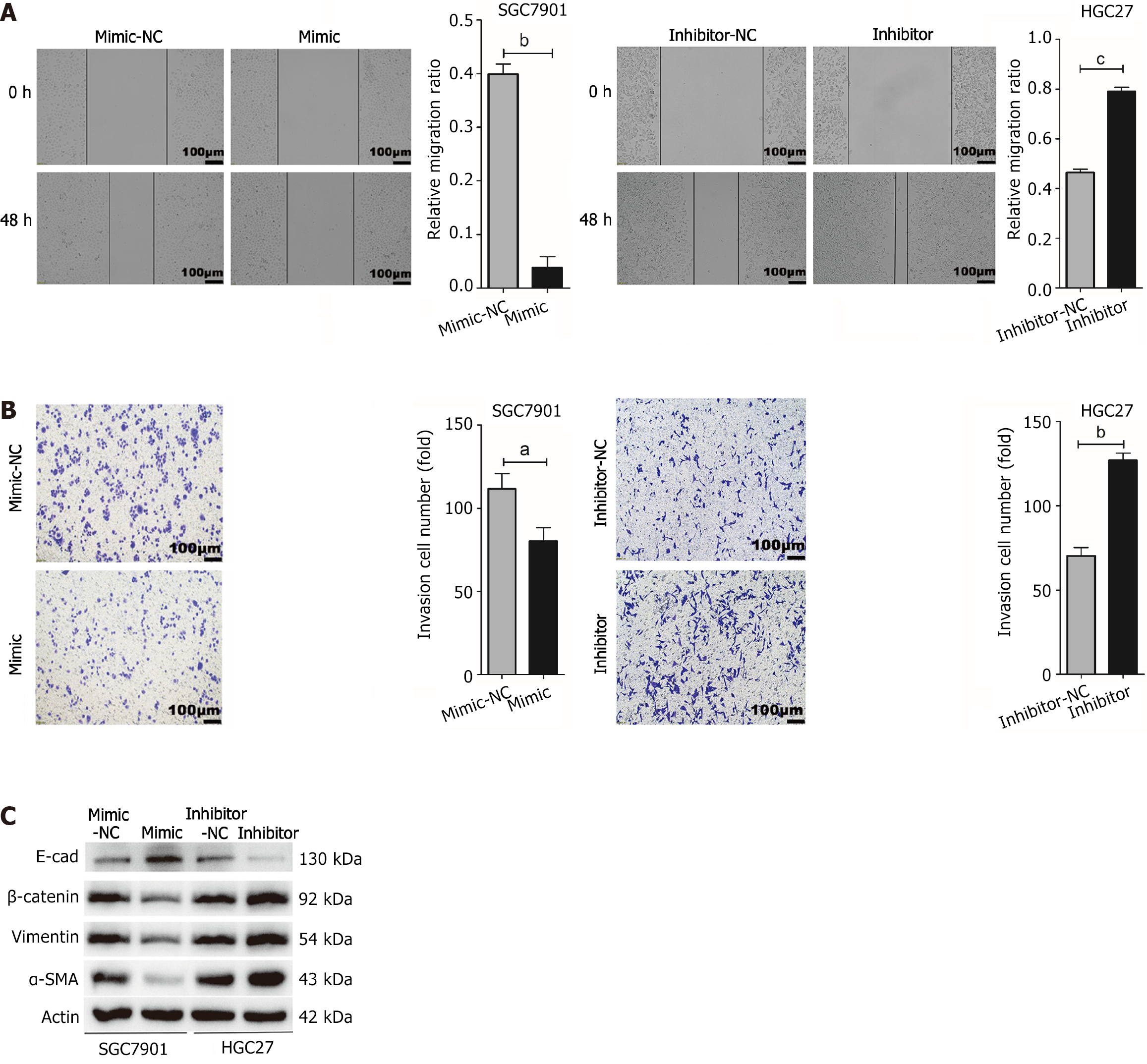
The wound healing assay results indicated that the up-regulation of miR-145-5p suppressed SGC7901 cell migration (P < 0.01). Conversely, the down-regulation of miR-145-5p expression significantly enhanced HGC27 cell migration (P < 0.001) (Figure 3A). The transwell assays showed similar results, with a significantly lower number of invasive cells in the miR-145-5p mimic group than in the mimic-NC group (P < 0.05). Moreover, the number of invasive cells in the miR-145-5p inhibitor group was higher than in the inhibitor-NC group (P < 0.01) (Figure 3B).
To explore whether miR-145-5p could reverse the epithelial-mesenchymal transition (EMT) phenotype, the expression of EMT-related marker proteins (E-cadherin, β-catenin, Vimentin, and α-SMA) was assessed in HGC27 and SGC7901 cells. The upregulation of miR-145-5p significantly increased the expression of E-cadherin and decreased the expression of β-catenin, Vimentin, and α-SMA. Meanwhile, reducing the expression of miR-145-5p had the opposite effect (Figure 3C).
These findings suggest that miR-145-5p acts as a tumor suppressor and can reverse the EMT phenotype, which in turn inhibits GC cell proliferation, invasion, and metastasis.
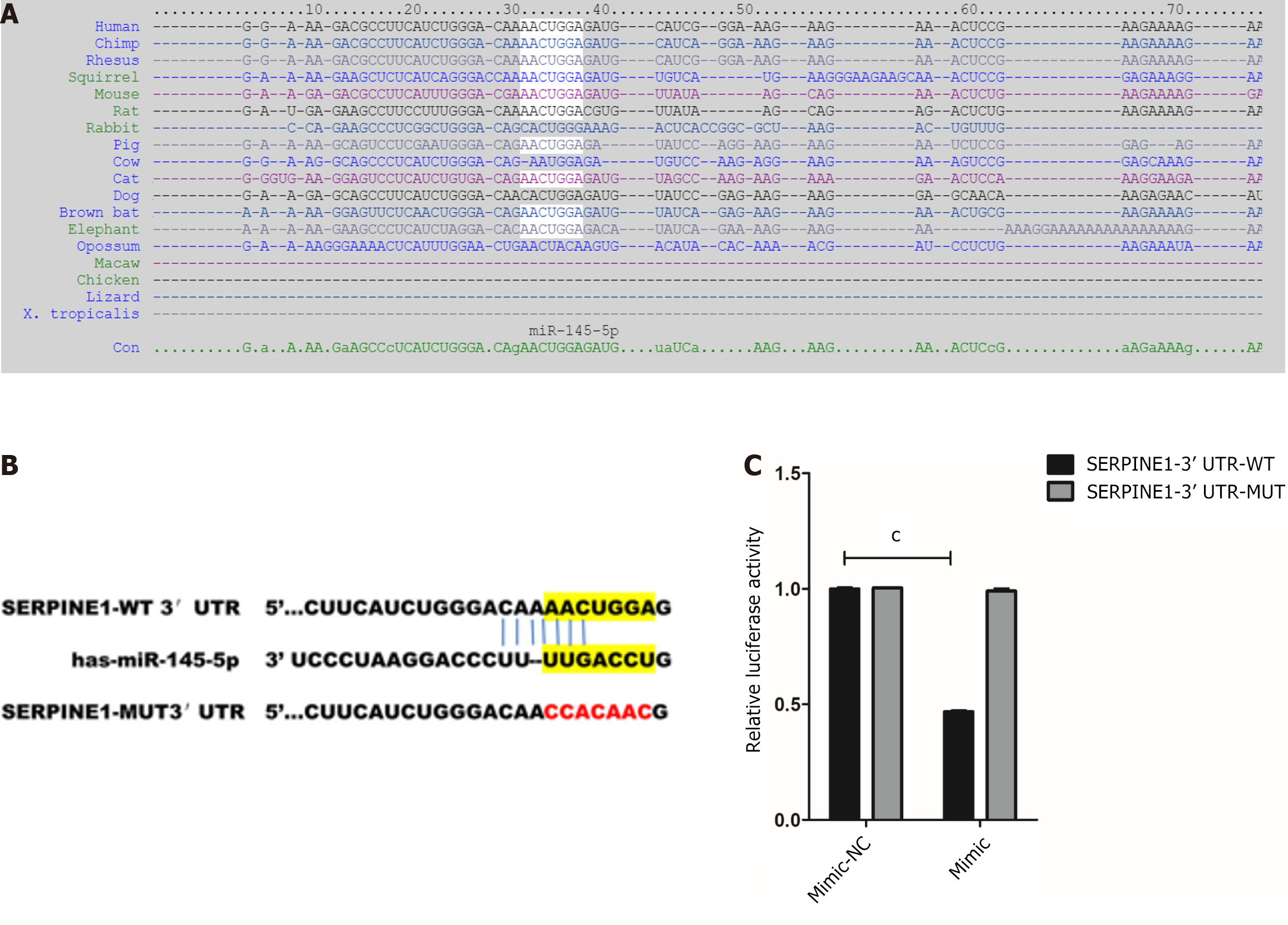
The bioinformatics tool, TargetScan, was used to predict the binding sites of miR-145-5p and SERPINE1 3'- UTR and better understand the molecular mechanism of miR-145-5p action (Figure 4A). Furthermore, to determine whether miR-145-5p directly regulates SERPINE1, SERPINE1 3'-UTR fragments (both wild-type, WT-LZTS1 3'-UTR, and mutated, Mut-LZTS1 3'-UTR) were cloned into a dual luciferase reporter vector (pmirGLO plasmid) and a luciferase reporter assay was conducted (Figure 4B). Co-transfection of 293T cells with miR-145-5p mimic and a vector carrying WT-SERPINE1 3'-UTR led to a significant reduction in luciferase activity (P < 0.001) (Figure 4C). These results suggest that miR-145-5p negatively regulates SERPINE1 expression by directly targeting its 3'-UTR, confirming the predictive findings of TargetScan.
TargetScan and miRDB bioinformatics tools were used to predict the miR-145-5p target gene and better characterize how this miRNA regulates the biological behavior of GC. A Venn diagram was created to visualize the results (Figure 5A). SERPINE1 was identified as a potential target gene of miR-145-5p. This gene is associated with malignant biological behaviors including tumor cell proliferation, invasion, metastasis, and apoptosis, as well as poor clinical prognosis[26-28]. Assessment of TCGA data confirmed that SERPINE1 expression was significantly higher in tumor tissues than in normal tissues (Figure 5B).
Kaplan-Meier plot analysis revealed that patients with high SERPINE1 expression had a shorter survival rate than those with the low SERPINE1 expression (Figure 5C). Both SERPINE1 mRNA and protein expression were also significantly higher in GC tissue (P < 0.0001) as well as the HGC27 (P < 0.01) and SGC7901 (P < 0.01) cell lines (Figure 5D). Furthermore, SERPINE1 expression was significantly lower in cells in the mimic group (P < 0.01) and increased in cells in the inhibitor group (P < 0.01) (Figure 5E). These findings suggest that there is a negative correlation between miR-145-5p and SERPINE1 in GC.
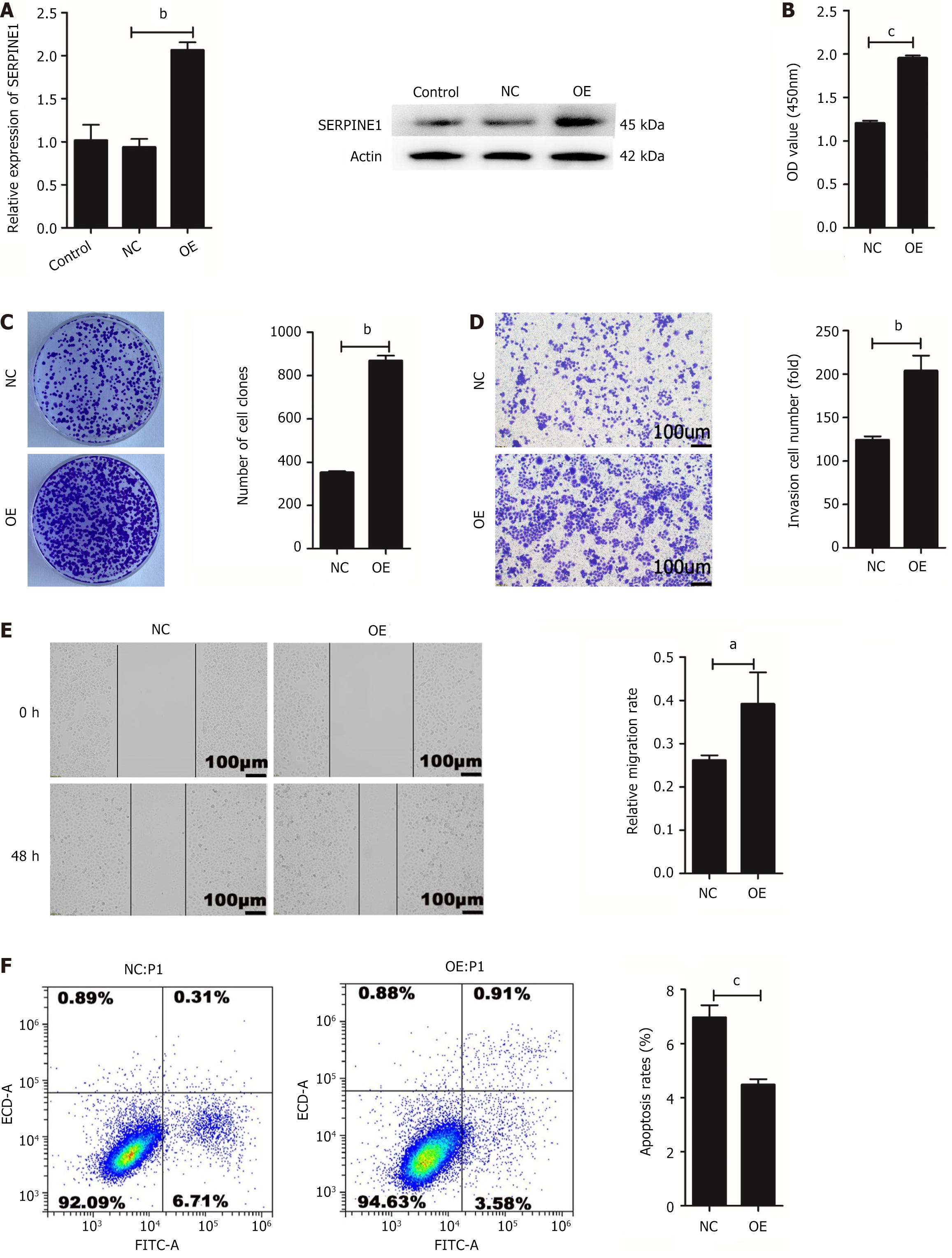
The SGC7901 cell line was used to stably OE and OE-NC-SERPINE1 (OE-NC) for subsequent experiments. The transfection efficiency of SERPINE1 was assessed using qRT-PCR (P < 0.01) and Western blotting (Figure 6A). The CCK-8 (P < 0.001) (Figure 6B) and colony formation assay (P < 0.01) data (Figure 6C) corroborated that the overexpression of SERPINE1 enhances SGC7901 cell proliferation. Meanwhile, transwell invasion (P < 0.01) (Figure 6D) and wound healing assays (P < 0.05) (Figure 6E) performed on the stably transfected cells indicated that SERPINE1 overexpression stimulates GC cell invasion and metastasis. Flow cytometric analysis revealed that SERPINE1 overexpression inhibits SGC7901 cell apoptosis (P < 0.001) (Figure 6F). These findings suggest that SERPINE1 functions as an oncogene in GC cells.
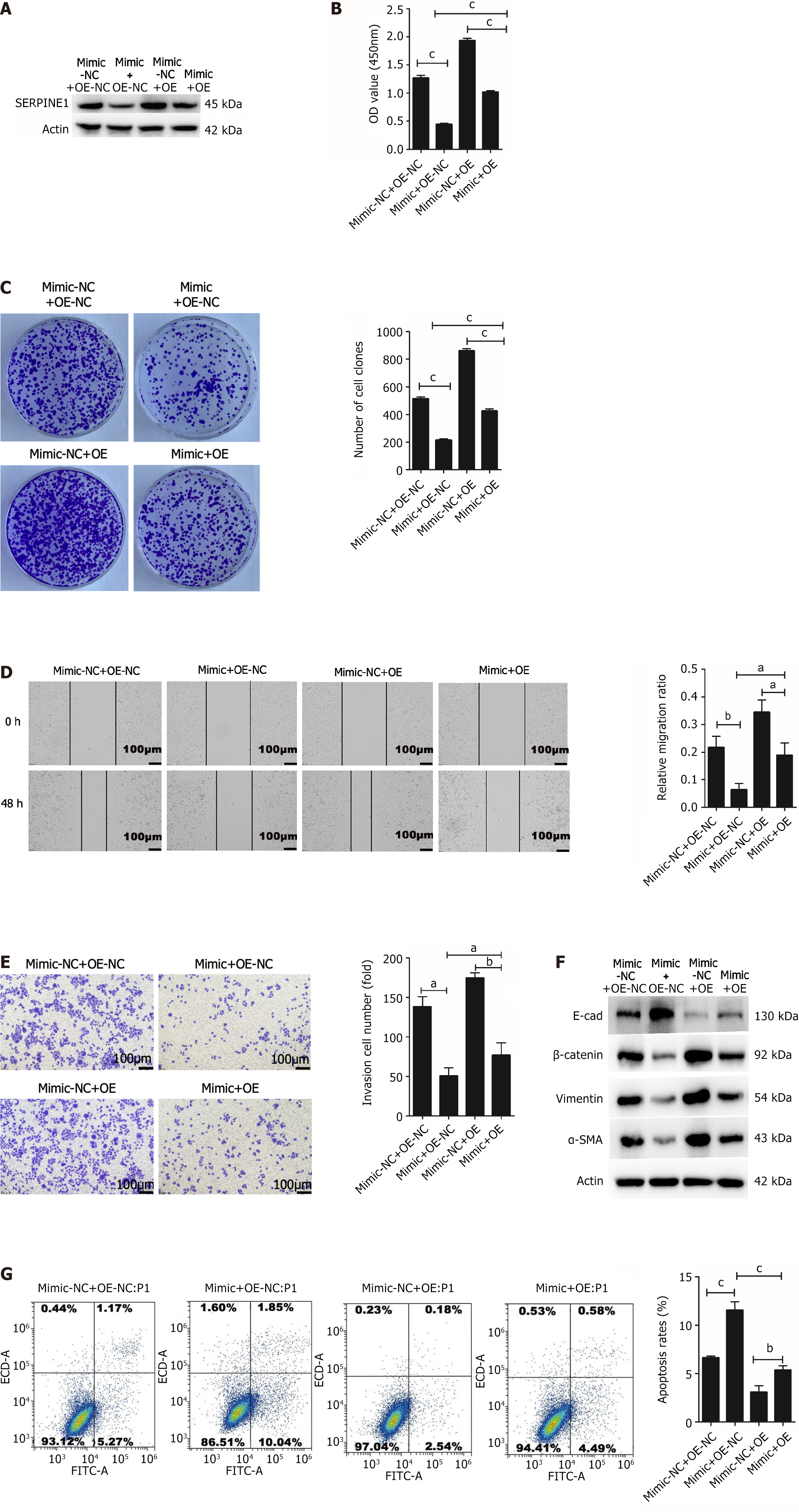
Rescue experiments were conducted to further elucidate the effects of SERPINE1 on miR-145-5p-mediated cell proliferation, apoptosis, invasion, metastasis, and EMT. SGC7901 cells were cotransfected with OE-SERPINE1/OE-NC and miR-145-5p mimic/miR-145-5p mimic NC. Western blotting revealed that the miR-145-5p mimic and OE-SERPINE1+ miR-145-5p mimic groups had lower SERPINE1 protein expression than the miR-145-5p mimic NC and OE-NC+ miR-145-5p mimic groups, respectively (Figure 7A). Overexpression of SERPINE1 in SGC7901 cells partially counteracted the inhibitory effect of miR-145-5p on cell proliferation (P < 0.001) (Figure 7B and C), migration (Figure 7D), invasion (Figure 7E), and EMT (Figure 7F). SERPINE1 also partially reversed miR145-5p-induced apoptosis (Figure 7G). These findings suggest that miR145-5p negatively regulates SERPINE1 in GC, thereby facilitating cell apoptosis, inhibiting cell proliferation, invasion, and metastasis, and reversing EMT.
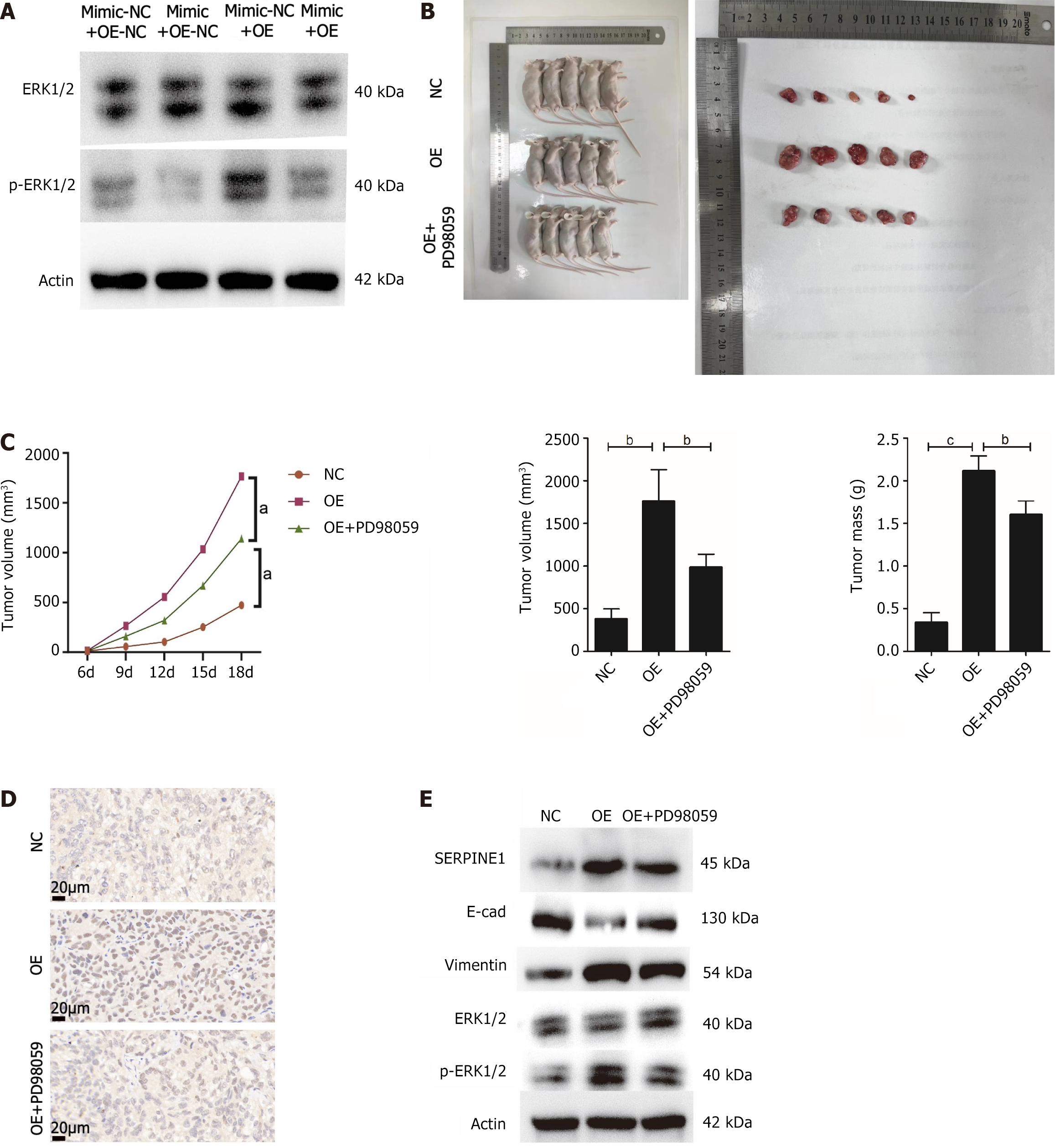
The salvage experiment assessed ERK1/2 and p-ERK1/2 protein expression in the four groups. While miR145-5p and SERPINE1 did not affect ERK1/2 protein expression, miR-145-5p overexpression suppressed p-ERK1/2 expression. Meanwhile, elevated SERPINE1 levels partially counteracted the effect of miR-145-5p (Figure 8A).
To further verify this mechanism, a GC tumor model was established in nude mice. All nude mice were evenly distributed into three groups (NC, OE, and OE + PD98059). Tumor tissues in the mice that overexpressed SERPINE1 (OE group) were heavier and larger in volume than those in the NC group. In contrast, nude mice that received PD98059, an ERK1/2 signal inhibitor, via intraperitoneal administration, had reduced tumor weight (P < 0.01) and volume (P < 0.001) (Figures 8B and C). Immunohistochemical analysis of the tumor tissue revealed that Ki67 was highly expressed in the group with SERPINE1 overexpression. However, its expression was diminished in the tissues from the OE+PD98059 group (Figure 8D). Western blot results revealed that while SERPINE1 overexpression inhibited E-cadherin expression, boosted vimentin expression, induced EMT, and increased ERK1/2 phosphorylation, it had no impact on total ERK1/2 expression. In contrast, PD98059-induced blockage of the ERK1/2 pathway increased E-cadherin and reduced vimentin expression (Figure 8E). In summary, these results indicate that miR-145-5p negatively regulates SERPINE1, inhibiting ERK1/2 signaling, and ultimately suppressing GC proliferation and EMT.
GC is a commonly occurring cancer[29,30] that remains a serious threat to human health, primarily due to its late diagnosis, rapid progression, and high rates of metastasis and postoperative recurrence. The in vivo and in vitro findings of this study indicate that miR-145-5p inhibits GC formation. The results also show that miR-145-5p targets SERPINE1, suggesting that this miRNA inhibits GC development by suppressing the miR-145-5p/SERPINE1/ERK1/2 axis.
Several studies have confirmed that miRNAs, which can function as either proto-oncogenes or tumor suppressor genes, play a pivotal role in the biological behavior of tumors and can thus aid in cancer diagnosis, treatment, and prognosis[31]. MiRNAs also participate in key signaling pathways, including mTOR/P-gp, Wnt/β-catenin, JAK/STAT, KRAS, EGF, and ERK[32,33]. MiR-145-5p is shown to act as a tumor suppressor in various cancers, including breast, cervical, bladder, renal, and gastrointestinal[34]. In non-small cell lung cancer, low expression of miR-145-5p is associated with pemetrexed resistance and EMT[35]. MiR-145-5p suppresses the early stage of colorectal cancer (CRC) but promotes metastasis at a later stage, indicating that it may play a dual role in this disease[36]. In esophageal squamous cell carcinoma, miR-145-5p attenuates proliferation, migration, and invasion[23], and in prostate cancer, miR-145-5p inhibits tumor growth and neuroendocrine differentiation[37]. These findings suggest that the role of miR-145-5p differs by cancer type. The current study demonstrated that miR-145-5p is downregulated in GC. The upregulation of this miRNA was shown to inhibit GC progression by preventing tumor cell migration, proliferation, invasion, and EMT and by promoting apoptosis. These findings are consistent with earlier studies showing that miR-145-5p targets ARF6, ANGPT2, N-cadherin, and ZEB2 and suppresses tumor cell malignancy in GC[38,39]. However, while SERPINE1 was identified as a target for miR-145-5p in oral squamous cell carcinoma (OSCC)[27] the current study was the first to show that miR-145-5p also targets this gene in GC.
SERPINE1, or plasminogen activator inhibitor-1 (PAI-1), is an essential inhibitor of tissue plasminogen activator and urokinase-type[40]. In CRC, overexpression of SERPINE1 is associated with tumor cell proliferation, invasion, and aggressiveness[41]. SERPINE1 expression also correlates with the poor prognosis of head and neck squamous cell carcinoma, esophageal cancer, gastric adenocarcinoma, pancreatic ductal adenocarcinoma, and bladder cancer patients[33,42,43]. The current study confirmed that SERPINE1 fosters GC cell proliferation, invasion, and migration while suppressing apoptosis, thereby reducing patient survival times. However, SERPINE1 expression varies across different tumor tissue types. The protein is dramatically overexpressed in colon, esophageal, GC, breast, and thyroid cancer tissues as well as in clear cell renal cells and head and neck squamous cell carcinomas. Meanwhile, SERPINE1 is notably downregulated in renal papillary cell carcinoma, hepatocellular carcinoma, renal chromophobe cell carcinoma, and endometrial cancer tissue[44]. Several previous studies indicate that SERPINE1 is associated with angiogenesis and tumor progression in GC[45-47]. MAFG-AS1 enhances bladder cancer tumorigenesis by regulating the miR-143-3p/SERPINE1 axis[48], and secretory SERPINE1 expression is increased by a purinergic P2Y12 inhibitor, inducing MMP1 expression and increasing colon cancer metastasis[49]. A study on OSCC found that miR-617-targeted SERPINE1 inhibited OSCC cell proliferation, viability, and apoptosis[50]. Notch1 regulates the aggressive phenotypes of differentiated thyroid cancer, which could also be mediated by SERPINE1 inhibition[51]. Furthermore, SERPINE1 was shown to be upregulated in GC, showing a high diagnostic value, and associated with poorer overall survival and recurrence-free survival[52,53]. SERPINE1 is also a member of the SERM signature and was identified as having great value in predicting GC treatment sensitivity, informing the development of targeted CSC and EMT-related therapies[54]. These studies suggest that SERPINE1 may have functional specificity in different cell types due to the temporal and spatial expression of different upstream regulatory factors. The complex regulatory role of SERPINE1 in vivo and its close association with tumors suggest that identifying the regulatory mechanism of this protein will aid our understanding of GC pathogenesis. The current study confirmed that SERPINE1 expression was higher in several GC cell lines and tissues than in adjacent normal tissues. Further functional experiments demonstrated that SERPINE1 promotes GC cell proliferation, invasion, and metastasis while suppressing apoptosis. SERPINE1 overexpression could partially reverse the anti-tumor effect of miR145-5p mimics and promote EMT. In summary, SERPINE1 is targeted and regulated by miR-145-5p, promotes GC progression, and is associated with poor GC prognosis. These findings inform the development of novel therapeutic approaches or diagnostic tools for this disease.
ERK1/2 is an important subfamily of mitogen-activated protein kinases that control various cellular activities and physiological processes, including promoting cell survival and pro-apoptotic functions under certain conditions[55]. PAI-1 promotes the migration and invasion of ESCC cells and macrophages by activating the Akt and ERK1/2 signaling pathways[56]. The current study indicated that SERPINE1 promoted the phosphorylation of ERK1/2 and expression of EMT-related mesenchymal markers (α-SMA, β-catenin, and vimentin), and inhibited the expression of epidermal marker E-cadherin, thus inducing GC cell proliferation, invasion, and migration. Furthermore, GC tumor growth in nude mice was inhibited after the addition of the PD98059 inhibitor, suggesting that SERPINE1 could promote the growth of GC. SERPINE1 expression was also detected in the tumors, confirming that it promotes tumor growth and EMT by stimulating ERK1/2 phosphorylation, inducing GC invasion and migration. Overexpression of miR-145-5p in SGC7901 cells reduced ERK1/2 phosphorylation, while SERPINE1 partially reversed its effect.
The current study has some important limitations. The relationship between miR-145-5p and different histological types and TNM stages of GC, the impact of Helicobacter pylori infection, and chemotherapeutic resistance were not assessed. The role of these factors will be assessed in follow-up studies to provide more theoretical support for the use of miR-145-5p in a clinical setting.
In conclusion, this is the first study to show that miR-145-5p inhibits GC cell proliferation, invasion, metastasis, and EMT, and promotes apoptosis by regulating the SERPINE1/ERK1/2 axis. The findings suggest that targeting this axis may serve as a potential GC treatment approach.
Gastric cancer (GC), a common malignancy of the digestive system, has an increasing incidence rate and poses a substantial threat to human health.
MicroRNAs (miRNA) play important roles in gene regulation and modulate many physical and pathological processes. The research motivation of this study was to identify the role of miRNA-145-5p (miR-145-5p) in the development of GC and its underlying mechanisms.
This study sought to compare miR-145-5p expression in GC and adjacent normal tissues, clarify the role of miR-145-5p in GC cell proliferation, invasion, metastasis, epithelial-mesenchymal transition (EMT), and apoptosis, and identify the direct target of miR-145-5p in GC cells.
Real-time polymerase chain reaction was performed to detect miRNA expression. Wound-healing and Transwell assays were performed to evaluate cancer cell migration and invasion, respectively. Cell proliferation was examined using cell counting kit-8 and colony formation assays. Cell apoptosis was assessed using flow cytometry. Western blotting analysis was used to identify EMT-associated proteins. A dual-luciferase reporter system was used to validate the target of miR-145-5p. Tumor formation in nude mice was assessed to further explore the mechanism by which miR-145-5p inhibits the progression of GC.
MiR-145-5p was decreased in GC tissues. Serpin family E member 1 (SERPINE1) was characterized as a direct target of miR-363-3p. MiR-145-5p was shown to target SERPINE1 to regulate extracellular signal-regulated kinase-1/2 (ERK1/2) signaling.
MiR-145-5p inhibits GC progression via the SERPINE1-ERK1/2 axis.
The miR-145-5p/SERPINE1/ERK1/2 axis may serve as a GC treatment target.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Exbrayat JM, France S-Editor: Qu XL L-Editor: A P-Editor: Zhang XD
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53206] [Cited by in F6Publishing: 51411] [Article Influence: 8568.5] [Reference Citation Analysis (122)] |
| 2. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50630] [Cited by in F6Publishing: 45688] [Article Influence: 15229.3] [Reference Citation Analysis (47)] |
| 3. | Yeoh KG, Tan P. Mapping the genomic diaspora of gastric cancer. Nat Rev Cancer. 2022;22:71-84. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 67] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 4. | Japanese Gastric Cancer Association. Japanese classification of gastric carcinoma: 3rd English edition. Gastric Cancer. 2011;14:101-112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2390] [Cited by in F6Publishing: 2656] [Article Influence: 204.3] [Reference Citation Analysis (0)] |
| 5. | Thrift AP, Wenker TN, El-Serag HB. Global burden of gastric cancer: epidemiological trends, risk factors, screening and prevention. Nat Rev Clin Oncol. 2023;20:338-349. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 77] [Reference Citation Analysis (0)] |
| 6. | IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Schistosomes, Liver Flukes and Helicobacter pylori. 1994; 61. Available from: https://www.ncbi.nlm.nih.gov/books/NBK487794/. [Cited in This Article: ] |
| 7. | Correa P, Haenszel W, Cuello C, Tannenbaum S, Archer M. A model for gastric cancer epidemiology. Lancet. 1975;2:58-60. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 725] [Cited by in F6Publishing: 682] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 8. | Correa P, Piazuelo MB. Helicobacter pylori Infection and Gastric Adenocarcinoma. US Gastroenterol Hepatol Rev. 2011;7:59-64. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 250] [Cited by in F6Publishing: 267] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 9. | Liu W, Cao Y, Guan Y, Zheng C. BST2 promotes cell proliferation, migration and induces NF-κB activation in gastric cancer. Biotechnol Lett. 2018;40:1015-1027. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 10. | Seeneevassen L, Bessède E, Mégraud F, Lehours P, Dubus P, Varon C. Gastric Cancer: Advances in Carcinogenesis Research and New Therapeutic Strategies. Int J Mol Sci. 2021;22. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 54] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 11. | Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25833] [Cited by in F6Publishing: 26874] [Article Influence: 1343.7] [Reference Citation Analysis (0)] |
| 12. | Wei X, Feng Y, Fu Y, Liu F, Chen Q, Zhang W, Zhao Y, Huang X, Chen Y, Li Q, Zhang Q. miR-100-5p is upregulated in multiple myeloma and involves in the pathogenesis of multiple myeloma through targeting MTMR3. Hematology. 2023;28:2196857. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 13. | Gu Y, Feng X, Jin Y, Liu Y, Zeng L, Zhou D, Feng Y. Upregulation of miRNA-10a-5p promotes tumor progression in cervical cancer by suppressing UBE2I signaling. J Obstet Gynaecol. 2023;43:2171283. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 4] [Reference Citation Analysis (0)] |
| 14. | Chen BJ, Jiang J, Li T, Jiang HJ, Liang XH, Tang YL. miR-183-5p overexpression orchestrates collective invasion in salivary adenoid cystic carcinoma through the FAT1/YAP1 signaling pathway. Biochem Biophys Res Commun. 2023;655:127-137. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 15. | Li M, Lin C, Cai Z. Breast cancer stem cell-derived extracellular vesicles transfer ARRDC1-AS1 to promote breast carcinogenesis via a miR-4731-5p/AKT1 axis-dependent mechanism. Transl Oncol. 2023;31:101639. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 16. | Park CR, Lee M, Lee SY, Kang D, Park SJ, Lee DC, Koo H, Park YG, Yu SL, Jeong IB, Kwon SJ, Kang J, Lee EB, Son JW. Regulating POLR3G by MicroRNA-26a-5p as a promising therapeutic target of lung cancer stemness and chemosensitivity. Noncoding RNA Res. 2023;8:273-281. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 17. | Seok HJ, Choi JY, Yi JM, Bae IH. Targeting miR-5088-5p attenuates radioresistance by suppressing Slug. Noncoding RNA Res. 2023;8:164-173. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 18. | Pan Y, Li K, Tao X, Zhao Y, Chen Q, Li N, Liu J, Go VLW, Guo J, Gao G, Xiao GG. MicroRNA-34a Alleviates Gemcitabine Resistance in Pancreatic Cancer by Repression of Cancer Stem Cell Renewal. Pancreas. 2021;50:1260-1266. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 2] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 19. | Zhang H, Jiang M, Liu Q, Han Z, Zhao Y, Ji S. miR-145-5p inhibits the proliferation and migration of bladder cancer cells by targeting TAGLN2. Oncol Lett. 2018;16:6355-6360. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 20. | Ozen M, Karatas OF, Gulluoglu S, Bayrak OF, Sevli S, Guzel E, Ekici ID, Caskurlu T, Solak M, Creighton CJ, Ittmann M. Overexpression of miR-145-5p inhibits proliferation of prostate cancer cells and reduces SOX2 expression. Cancer Invest. 2015;33:251-258. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 62] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 21. | Tang W, Zhang X, Tan W, Gao J, Pan L, Ye X, Chen L, Zheng W. miR-145-5p Suppresses Breast Cancer Progression by Inhibiting SOX2. J Surg Res. 2019;236:278-287. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 22. | Cao H, Pan G, Tang S, Zhong N, Liu H, Zhou H, Peng Q, Zou Y. miR-145-5p Regulates the Proliferation, Migration and Invasion in Cervical Carcinoma by Targeting KLF5. Onco Targets Ther. 2020;13:2369-2376. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 23. | Lin J, Wu S, Zhu K, Zhang J, Shi X, Shen J, Xu J. The role of miR-145-5p in esophageal squamous cell carcinoma tumor-associated macrophages and selection of immunochemotherapy. J Thorac Dis. 2022;14:2493-2510. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 24. | Chen Q, Zhou L, Ye X, Tao M, Wu J. miR-145-5p suppresses proliferation, metastasis and EMT of colorectal cancer by targeting CDCA3. Pathol Res Pract. 2020;216:152872. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 25. | Zhou T, Chen S, Mao X. miR-145-5p affects the differentiation of gastric cancer by targeting KLF5 directly. J Cell Physiol. 2019;234:7634-7644. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 26. | Hu B, Chen Z, Wang X, Chen F, Song Z, Cao C. MicroRNA-148a-3p Directly Targets SERPINE1 to Suppress EMT-Mediated Colon Adenocarcinoma Progression. Cancer Manag Res. 2021;13:6349-6362. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 2] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 27. | Yu J, Lou Y, Hou M, Ma X, Wang L. Circ_0058063 contributes to oral squamous cell carcinoma development by sponging miR-145-5p and upregulating SERPINE1. J Oral Pathol Med. 2022;51:630-637. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 28. | Guo L, An T, Wan Z, Huang Z, Chong T. SERPINE1 and its co-expressed genes are associated with the progression of clear cell renal cell carcinoma. BMC Urol. 2023;23:43. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 2] [Reference Citation Analysis (0)] |
| 29. | Ajani JA, D'Amico TA, Bentrem DJ, Chao J, Cooke D, Corvera C, Das P, Enzinger PC, Enzler T, Fanta P, Farjah F, Gerdes H, Gibson MK, Hochwald S, Hofstetter WL, Ilson DH, Keswani RN, Kim S, Kleinberg LR, Klempner SJ, Lacy J, Ly QP, Matkowskyj KA, McNamara M, Mulcahy MF, Outlaw D, Park H, Perry KA, Pimiento J, Poultsides GA, Reznik S, Roses RE, Strong VE, Su S, Wang HL, Wiesner G, Willett CG, Yakoub D, Yoon H, McMillian N, Pluchino LA. Gastric Cancer, Version 2.2022, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2022;20:167-192. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 533] [Article Influence: 266.5] [Reference Citation Analysis (0)] |
| 30. | Miller KD, Nogueira L, Devasia T, Mariotto AB, Yabroff KR, Jemal A, Kramer J, Siegel RL. Cancer treatment and survivorship statistics, 2022. CA Cancer J Clin. 2022;72:409-436. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 621] [Cited by in F6Publishing: 800] [Article Influence: 400.0] [Reference Citation Analysis (0)] |
| 31. | Lupini L, Pepe F, Ferracin M, Braconi C, Callegari E, Pagotto S, Spizzo R, Zagatti B, Lanuti P, Fornari F, Ghasemi R, Mariani-Costantini R, Bolondi L, Gramantieri L, Calin GA, Sabbioni S, Visone R, Veronese A, Negrini M. Over-expression of the miR-483-3p overcomes the miR-145/TP53 pro-apoptotic loop in hepatocellular carcinoma. Oncotarget. 2016;7:31361-31371. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 32. | Hegazy M, Elkady MA, Yehia AM, Elsakka EGE, Abulsoud AI, Abdelmaksoud NM, Elshafei A, Abdelghany TM, Elkhawaga SY, Ismail A, Mokhtar MM, El-Mahdy HA, Doghish AS. The role of miRNAs in laryngeal cancer pathogenesis and therapeutic resistance - A focus on signaling pathways interplay. Pathol Res Pract. 2023;246:154510. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 34] [Reference Citation Analysis (0)] |
| 33. | Zhao L, Wu X, Zhang Z, Fang L, Yang B, Li Y. ELF1 suppresses autophagy to reduce cisplatin resistance via the miR-152-3p/NCAM1/ERK axis in lung cancer cells. Cancer Sci. 2023;114:2650-2663. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 34. | Kadkhoda S, Ghafouri-Fard S. Function of miRNA-145-5p in the pathogenesis of human disorders. Pathol Res Pract. 2022;231:153780. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 4] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 35. | Chang WW, Wang BY, Chen SH, Chien PJ, Sheu GT, Lin CH. miR-145-5p Targets Sp1 in Non-Small Cell Lung Cancer Cells and Links to BMI1 Induced Pemetrexed Resistance and Epithelial-Mesenchymal Transition. Int J Mol Sci. 2022;23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 5] [Reference Citation Analysis (0)] |
| 36. | Cheng X, Shen T, Liu P, Fang S, Yang Z, Li Y, Dong J. miR-145-5p is a suppressor of colorectal cancer at early stage, while promotes colorectal cancer metastasis at late stage through regulating AKT signaling evoked EMT-mediated anoikis. BMC Cancer. 2022;22:1151. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 37. | Ji S, Shi Y, Yang L, Zhang F, Li Y, Xu F. miR-145-5p Inhibits Neuroendocrine Differentiation and Tumor Growth by Regulating the SOX11/MYCN Axis in Prostate cancer. Front Genet. 2022;13:790621. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 38. | Zhong X, Wen X, Chen L, Gu N, Yu X, Sui K. Long non-coding RNA KCNQ1OT1 promotes the progression of gastric cancer via the miR-145-5p/ARF6 axis. J Gene Med. 2021;23:e3330. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 39. | Zhou K, Song B, Wei M, Fang J, Xu Y. MiR-145-5p suppresses the proliferation, migration and invasion of gastric cancer epithelial cells via the ANGPT2/NOD_LIKE_RECEPTOR axis. Cancer Cell Int. 2020;20:416. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 40. | Alotaibi FT, Peng B, Klausen C, Lee AF, Abdelkareem AO, Orr NL, Noga H, Bedaiwy MA, Yong PJ. Plasminogen activator inhibitor-1 (PAI-1) expression in endometriosis. PLoS One. 2019;14:e0219064. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 41. | Mazzoccoli G, Pazienza V, Panza A, Valvano MR, Benegiamo G, Vinciguerra M, Andriulli A, Piepoli A. ARNTL2 and SERPINE1: potential biomarkers for tumor aggressiveness in colorectal cancer. J Cancer Res Clin Oncol. 2012;138:501-511. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 86] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 42. | Chan OTM, Furuya H, Pagano I, Shimizu Y, Hokutan K, Dyrskjøt L, Jensen JB, Malmstrom PU, Segersten U, Janku F, Rosser CJ. Association of MMP-2, RB and PAI-1 with decreased recurrence-free survival and overall survival in bladder cancer patients. Oncotarget. 2017;8:99707-99721. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 43. | Ding Y, Niu W, Zheng X, Zhou C, Wang G, Feng Y, Yu B. Plasminogen activator, urokinase enhances the migration, invasion, and proliferation of colorectal cancer cells by activating the Src/ERK pathway. J Gastrointest Oncol. 2022;13:3100-3111. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 44. | Wang Y, Wang J, Gao J, Ding M, Li H. The expression of SERPINE1 in colon cancer and its regulatory network and prognostic value. BMC Gastroenterol. 2023;23:33. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 45. | Zhu Z, Xu J, Li L, Ye W, Chen B, Zeng J, Huang Z. Comprehensive analysis reveals CTHRC1, SERPINE1, VCAN and UPK1B as the novel prognostic markers in gastric cancer. Transl Cancer Res. 2020;9:4093-4110. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 46. | He W, Zhang D, Li D, Zhu D, Geng Y, Wang Q, He J, Wu J. Knockdown of Long Non-coding RNA LINC00200 Inhibits Gastric Cancer Progression by Regulating miR-143-3p/SERPINE1 Axis. Dig Dis Sci. 2021;66:3404-3414. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 47. | Teng F, Zhang JX, Chen Y, Shen XD, Su C, Guo YJ, Wang PH, Shi CC, Lei M, Cao YO, Liu SQ. LncRNA NKX2-1-AS1 promotes tumor progression and angiogenesis via upregulation of SERPINE1 expression and activation of the VEGFR-2 signaling pathway in gastric cancer. Mol Oncol. 2021;15:1234-1255. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 48] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 48. | Sun X, Cai Y, Hu X, Mo M, Zhao C, He W, Li Y. Long noncoding RNA MAFG-AS1 facilitates bladder cancer tumorigenesis via regulation of miR-143-3p/SERPINE1 axis. Transl Cancer Res. 2020;9:7214-7226. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 49. | Kim WT, Mun JY, Baek SW, Kim MH, Yang GE, Jeong MS, Choi SY, Han JY, Leem SH. Secretory SERPINE1 Expression Is Increased by Antiplatelet Therapy, Inducing MMP1 Expression and Increasing Colon Cancer Metastasis. Int J Mol Sci. 2022;23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 8] [Reference Citation Analysis (0)] |
| 50. | Zhao C, Liu Z. MicroRNA 617 Targeting SERPINE1 Inhibited the Progression of Oral Squamous Cell Carcinoma. Mol Cell Biol. 2021;41:e0056520. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 51. | Yu XM, Jaskula-Sztul R, Georgen MR, Aburjania Z, Somnay YR, Leverson G, Sippel RS, Lloyd RV, Johnson BP, Chen H. Notch1 Signaling Regulates the Aggressiveness of Differentiated Thyroid Cancer and Inhibits SERPINE1 Expression. Clin Cancer Res. 2016;22:3582-3592. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 52. | Li XC, Wang S, Zhu JR, Wang YP, Zhou YN. Nomograms combined with SERPINE1-related module genes predict overall and recurrence-free survival after curative resection of gastric cancer: a study based on TCGA and GEO data. Transl Cancer Res. 2020;9:4393-4412. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 53. | Dai W, Xiao Y, Tang W, Li J, Hong L, Zhang J, Pei M, Lin J, Liu S, Wu X, Xiang L, Wang J. Identification of an EMT-Related Gene Signature for Predicting Overall Survival in Gastric Cancer. Front Genet. 2021;12:661306. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 17] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 54. | Jia X, Chen B, Li Z, Huang S, Chen S, Zhou R, Feng W, Zhu H, Zhu X. Identification of a Four-Gene-Based SERM Signature for Prognostic and Drug Sensitivity Prediction in Gastric Cancer. Front Oncol. 2021;11:799223. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 55. | Lu Z, Xu S. ERK1/2 MAP kinases in cell survival and apoptosis. IUBMB Life. 2006;58:621-631. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 432] [Cited by in F6Publishing: 498] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 56. | Sakamoto H, Koma YI, Higashino N, Kodama T, Tanigawa K, Shimizu M, Fujikawa M, Nishio M, Shigeoka M, Kakeji Y, Yokozaki H. PAI-1 derived from cancer-associated fibroblasts in esophageal squamous cell carcinoma promotes the invasion of cancer cells and the migration of macrophages. Lab Invest. 2021;101:353-368. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |