Published online May 15, 2024. doi: 10.4251/wjgo.v16.i5.2018
Peer-review started: October 21, 2023
First decision: January 25, 2024
Revised: January 31, 2024
Accepted: March 8, 2024
Article in press: March 8, 2024
Published online: May 15, 2024
Gastric cancer (GC) is a common malignancy of the digestive system. According to global 2018 cancer data, GC has the fifth-highest incidence and the third-highest fatality rate among malignant tumors. More than 60% of GC are linked to infection with Helicobacter pylori (H. pylori), a gram-negative, active, microaerophilic, and helical bacterium. This parasite induces GC by producing toxic factors, such as cytotoxin-related gene A, vacuolar cytotoxin A, and outer membrane proteins. Ferroptosis, or iron-dependent programmed cell death, has been linked to GC, although there has been little research on the link between H. pylori infection-related GC and ferroptosis.
To identify coregulated differentially expressed genes among ferroptosis-related genes (FRGs) in GC patients and develop a ferroptosis-related prognostic model with discrimination ability.
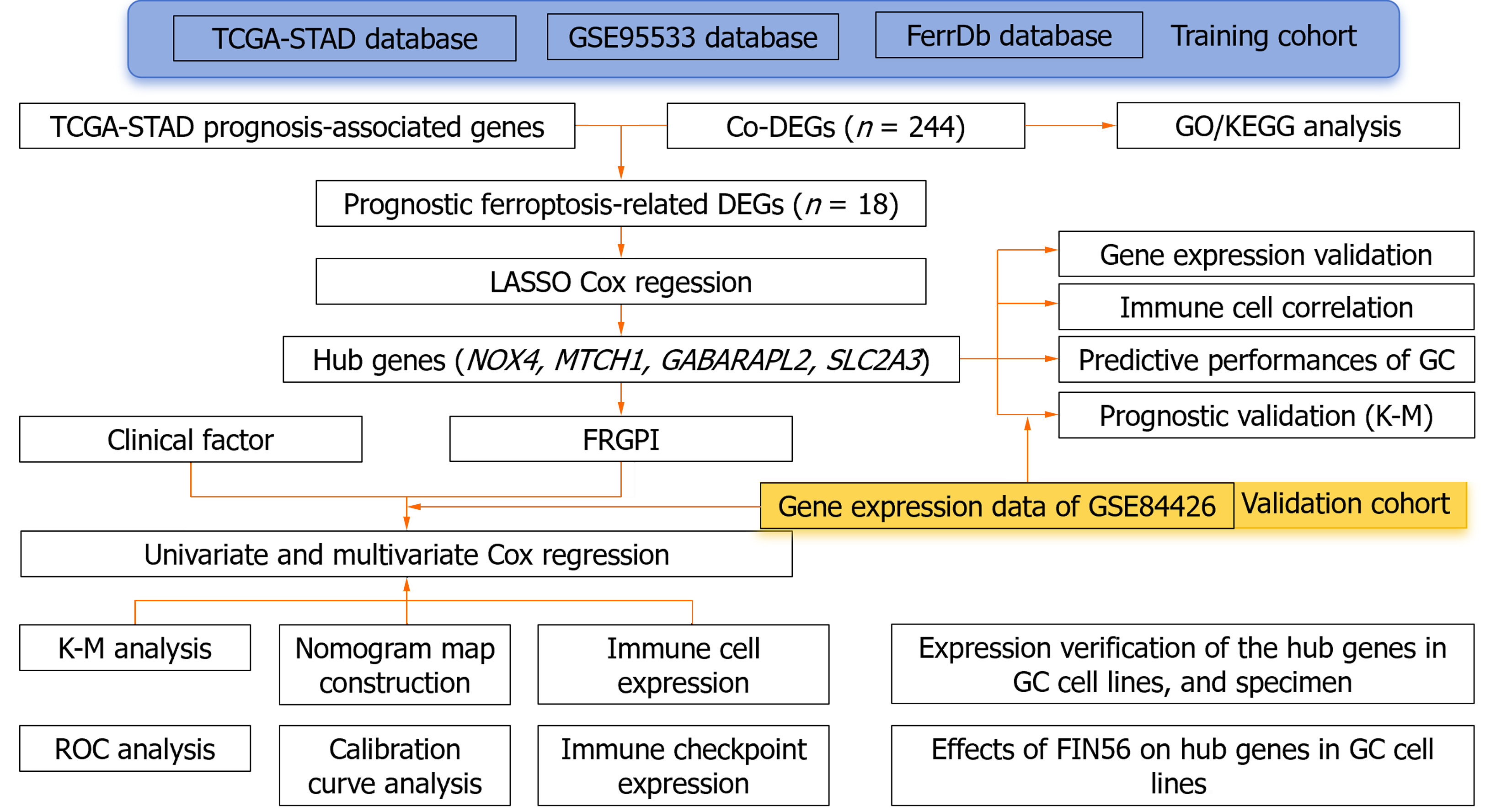
Gene expression profiles of GC patients and those with H. pylori-associated GC were obtained from The Cancer Genome Atlas and Gene Expression Omnibus (GEO) databases. The FRGs were acquired from the FerrDb database. A ferroptosis-related gene prognostic index (FRGPI) was created using least absolute shrinkage and selection operator–Cox regression. The predictive ability of the FRGPI was validated in the GEO cohort. Finally, we verified the expression of the hub genes and the activity of the ferroptosis inducer FIN56 in GC cell lines and tissues.
Four hub genes were identified (NOX4, MTCH1, GABARAPL2, and SLC2A3) and shown to accurately predict GC and H. pylori-associated GC. The FRGPI based on the hub genes could independently predict GC patient survival; GC patients in the high-risk group had considerably worse overall survival than did those in the low-risk group. The FRGPI was a significant predictor of GC prognosis and was strongly correlated with disease progression. Moreover, the gene expression levels of common immune checkpoint proteins dramatically increased in the high-risk subgroup of the FRGPI cohort. The hub genes were also confirmed to be highly overexpressed in GC cell lines and tissues and were found to be primarily localized at the cell membrane. The ferroptosis inducer FIN56 inhibited GC cell proliferation in a dose-dependent manner.
In this study, we developed a predictive model based on four FRGs that can accurately predict the prognosis of GC patients and the efficacy of immunotherapy in this population.
Core Tip: This study aimed to develop a prognostic model based on coregulated differentially expressed genes among ferroptosis-related genes (FRGs) in gastric cancer (GC). Gene expression profiles from GC patients and those with Helicobacter pylori-associated GC were analyzed, along with FRGs obtained from the FerrDb database. The resulting ferroptosis-related gene prognostic index (FRGPI), based on four hub genes (NOX4, MTCH1, GABARAPL2, and SLC2A3), accurately predicted GC patient survival. High-risk individuals had significantly worse overall survival, and the FRGPI was associated with disease progression and increased expression of immune checkpoint proteins. These findings provide insights into GC prognosis and immunotherapy efficacy.
- Citation: Wang L, Gong WH. Predictive model using four ferroptosis-related genes accurately predicts gastric cancer prognosis. World J Gastrointest Oncol 2024; 16(5): 2018-2037
- URL: https://www.wjgnet.com/1948-5204/full/v16/i5/2018.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i5.2018
Gastric cancer (GC) is a common malignancy of the digestive system. According to global 2018 cancer data, GC has the fifth highest incidence and the third highest fatality rate among malignant tumors[1]. More than 60% of GCs are linked to infection with Helicobacter pylori (H. pylori), a gram-negative, active, microaerophilic, and helical bacterium[2]. This parasite induces GC by producing toxic factors, such as cytotoxin-related gene A, vacuolar cytotoxin A, and outer membrane proteins[3]. The World Health Organization classifies H. pylori as a Group I carcinogen[4]. Ferroptosis, or iron-dependent programmed cell death[5], has been linked to GC, although there has been little research on the link between H. pylori infection-related GC and ferroptosis. Therefore, this study investigated the coregulated differentially expressed genes (co-DEGs) among ferroptosis-related genes (FRGs) in H. pylori infection-related GCs and investigated the relationship between the expression of these co-DEGs and clinical prognosis.
We obtained clinical information and molecular data on 375 GC patients from the cancer genome atlas (TCGA) database (241 men and 134 women) to create a training cohort. RNA sequencing (RNA-seq) data and corresponding clinical data for external validation cohorts (GSE99553, n = 42; GSE84426, n = 76) were downloaded from the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo). Additionally, 352 unique FRGs, including driver genes, suppressor genes, and markers, were obtained from the FerrDb database (http://www.zhounan.org/ferrdb/current/)[6]. Figure 1 shows the flow diagram of the data collection and analysis: (1) RNA-seq data in fragments per kilobase of transcript per million fragments mapped (FPKM) format were log2 transformed to generate the downloaded expression profile data; (2) clinical correlation and prognostic analyses: RNA-seq data in the HTSeq-FPKM database were log2 transformed into the transcripts per million format; and (3) clinical information such as tumor stage, metastasis status, and survival status was obtained from the database, and the relevant package in R (version 3.6.3) (http://www. r-project. org/) was used for analysis.
The GC tissues in the TCGA-Stomach Adenocarcinoma (STAD) and GSE99553 datasets were divided into H. pylori-positive and H. pylori-negative groups according to the presence or absence of H. pylori infection, respectively. The limma R package was used to identify DEGs, and genes whose expression was significantly upregulated or downregulated were selected for heatmapping. The survminer package screens for molecules with prognostic value in TCGA-STAD. All the datasets were selected with the criteria set to P < 0.05 and |log2FC| > 1. Venn tools (http://bioinformatics.psb.ugent.be/webtools/Venn/) were used to obtain H. pylori related to cancer of the stomach and FRGS Co-DEGs, and a Venn diagram was drawn.
The clusterProfiler R package was used to conduct GO/KEGG analysis of the Co-DEGs, in which the GO enrichment analysis included biological process (BP), cell composition, and molecular function (MF). KEGG analysis was used to define the pathways related to the Co-DEGs, and the parameters were set to P.adj < 0.05. Immunoinformatic correlation analysis was performed using the GSVA package.
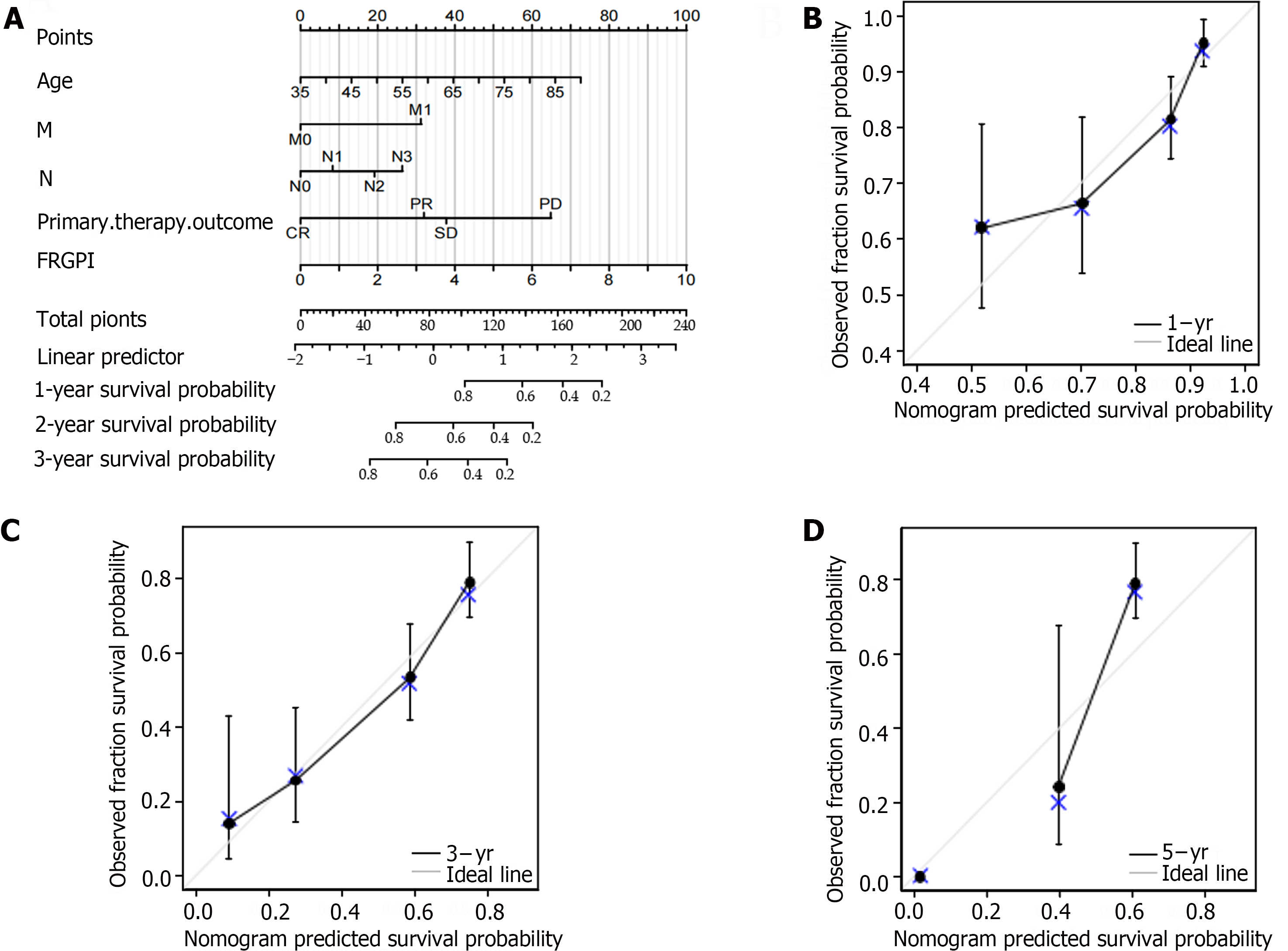
The prognosis-related co-DEGs were chosen as hub genes. The pROC package was used to investigate the diagnostic value of the hub genes for GC and H. pylori-associated GC. Correlations between hub genes and clinicopathological factors were compared in the TCGA and GSE84426 datasets, and univariate and multivariate Cox proportional hazards regression analyses were performed. The ferroptosis-related gene prognostic index (FRGPI) was identified by the minor absolute shrinkage and selection operator (LASSO) and a hazard model via the “glmnet” R package. The predictive value of the FRGPI was determined using Kaplan–Meier survival curves and receiver operating characteristic (ROC) curves generated via the “survival” and “survival ROC” R packages. A nomogram was created using the FRGPI and clinical data, and the predictive efficacy of the FRGPI at 1, 3, and 5 years was evaluated using a prognostic calibration curve. The following formula was used to calculate the FRGPI:
FRGPI = (Expression level of Gene 1 × β coefficient) + (Expression level of Gene 2 × β coefficient) +… + (Expression level of Gene n × β coefficient).
Patients were divided into low-risk and high-risk groups based on the median FRGPI.
The HGC-27 and MGC-803 cell lines were donated by Dr. Wu Zizhen of Peking Medicine and stored in the Clinical Laboratory of the Second Hospital of Zhejiang University. These cells were cultivated in RPMI-1640 medium (Cytiva, China) containing 10% fetal bovine serum (Cytiva, China) at 37 °C and 5% CO2.
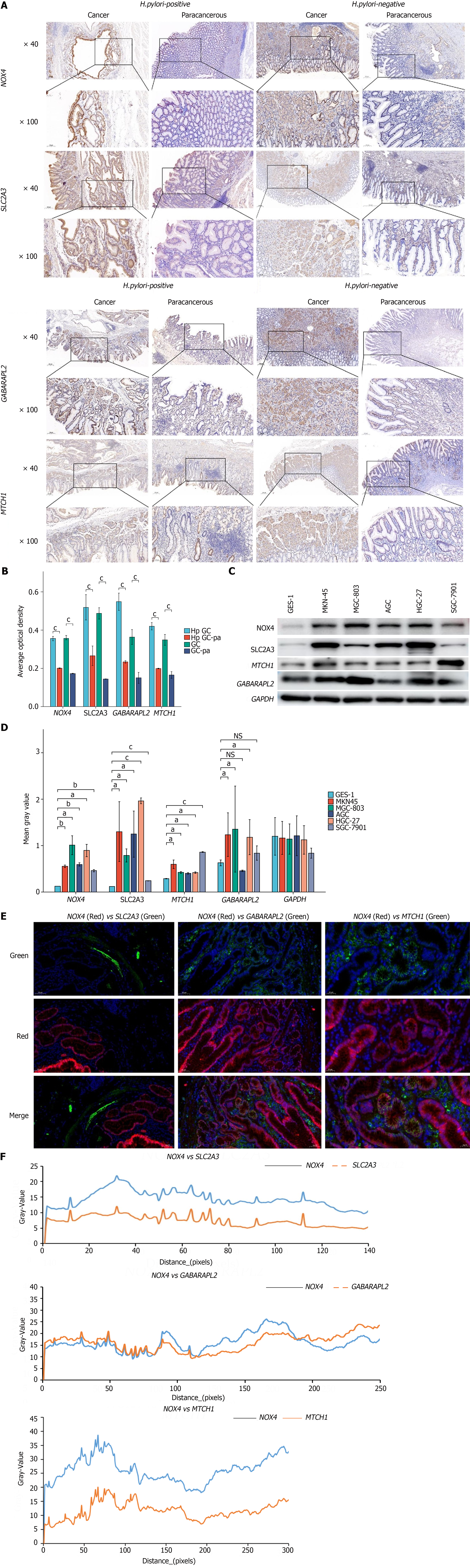
The tumor and neighboring normal tissues were embedded in paraffin after treatment with 10% formalin. Appropriate tissue sections were deparaffinized. The 5-μm-thick paraffin sections were dewaxed with xylene, dehydrated in 100%, 95%, 90%, 80%, and 70% absolute ethanol solutions for 5 min each, and boiled in distilled water for 15 min. After blocking with 10% serum-containing blocking solution at room temperature for 1 h, the sections were incubated with primary antibodies overnight at 4 °C, followed by incubation with secondary antibodies at room temperature for 30 min. The signals from horseradish peroxidase (HRP)-labeled antibodies (Servicebio, China) were developed by diaminobenzidine. After counterstaining with hematoxylin, the sections were dehydrated and sealed. Immunofluorescence analysis was subsequently conducted. ImageJ was used to evaluate the immunohistochemical and immunofluorescence results. The average optical density (OD) and gay value were calculated, and statistical analysis was conducted.
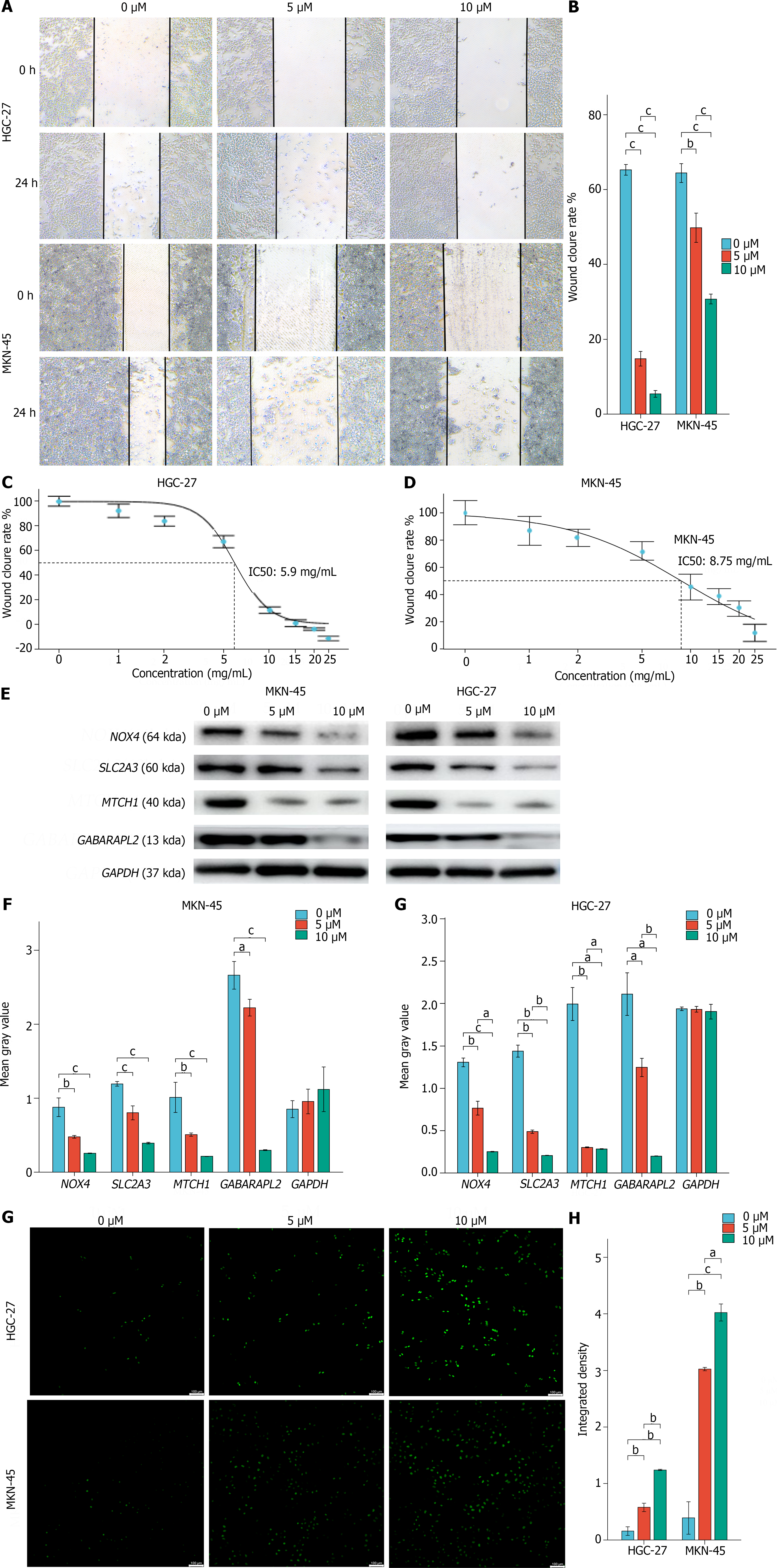
HGC-27 and MGC-803 cells were seeded in 96-well plates at 5000 cells/well and cultured at 37 °C and 5% CO2 for 24 h. The ferroptosis inducer FIN56 (Dojindo, Japan) was dissolved in dimethyl sulfoxide to generate working concentrations of 0, 1, 2, 5, 10, 15, and 20 μM; each concentration was evaluated in six wells. Cell viability was measured using a Cell Counting-8 Kit (CCK-8, Beyotime, China). A microplate reader was used to determine the OD at an absorbance of 450 nm. After calculating the half-maximal inhibitory concentration of each cell line, the inhibition curve was plotted using the ggplot2 R tool.
HGC-27 and MGC-803 cells were grown in 6-well plates for 24 h before being treated with FIN56 (2, 5, and 10 µM) for 24 h. After two washes in phosphate buffered saline (PBS), the cells were incubated for 30 min at 37 °C in new medium supplemented with 10 μM 2',7'-dichlorofluorescein diacetate (DCF; Abcam, ab113851, United States). A fluorescence microscope was used to examine the fluorescence intensity of the reactive oxygen species (ROS) in the cells in the 6-well plate. The images were examined using ImageJ software; the integrated density was calculated, and statistical analysis was carried out.
GC cells were rinsed twice with sterile PBS before being lysed for 5 min on ice in radio immunoprecipitation assay lysis buffer containing 1% phenylmethanesulfonyl fluoride (Beyotime, China). The cell lysates were centrifuged at 12000 pm and 4 °C for 15 min, heated for 5 min in boiling water, and then placed on ice. The bicinchoninic acid technique was used to measure the protein concentration (Beyotime, China). Proteins were separated by electrophoresis at 100 V for 1.5 h and then transferred to a polyvinylidene fluoride membrane, which was washed with phosphate buffered solution, blocked at room temperature for 1 h and incubated overnight at 4 °C with primary antibodies (NOX4: 1:2000; GABARAPL2: 1:1000; MTCH1: 1:1000; and SLC2A3: 1:8000; all from Abcam). After the samples were incubated for 1 h at room temperature with an anti-HRP-conjugated IgG secondary antibody (Service, China), they were detected using chemiluminescence detection equipment (GE, United States). Using ImageJ software, the westernblot images were evaluated, the mean gray value of each band was determined, and the outcomes were statistically examined.
R version 4.0.3 with the R Studio software package and SPSS 23.0 software were used for statistical analysis. overall survival (OS) was analyzed by the K–M method. Univariate and multivariate Cox regression analyses were used to determine the effect of each variable on OS, and LASSO was used to avoid overfitting the multivariate Cox regression model. The generated multivariate Cox regression model was utilized to compute the risk score and develop the prognostic nomogram model. The C-index was calculated, and calibration plots were generated to evaluate the predictive value of the nomogram at 1, 3, and 5 years. Chi-square and Fisher's exact tests were used to assess the associations between clinical features and risk scores, while ROC analysis was utilized to determine sensitivity and specificity. P < 0.05 indicated statistical significance.
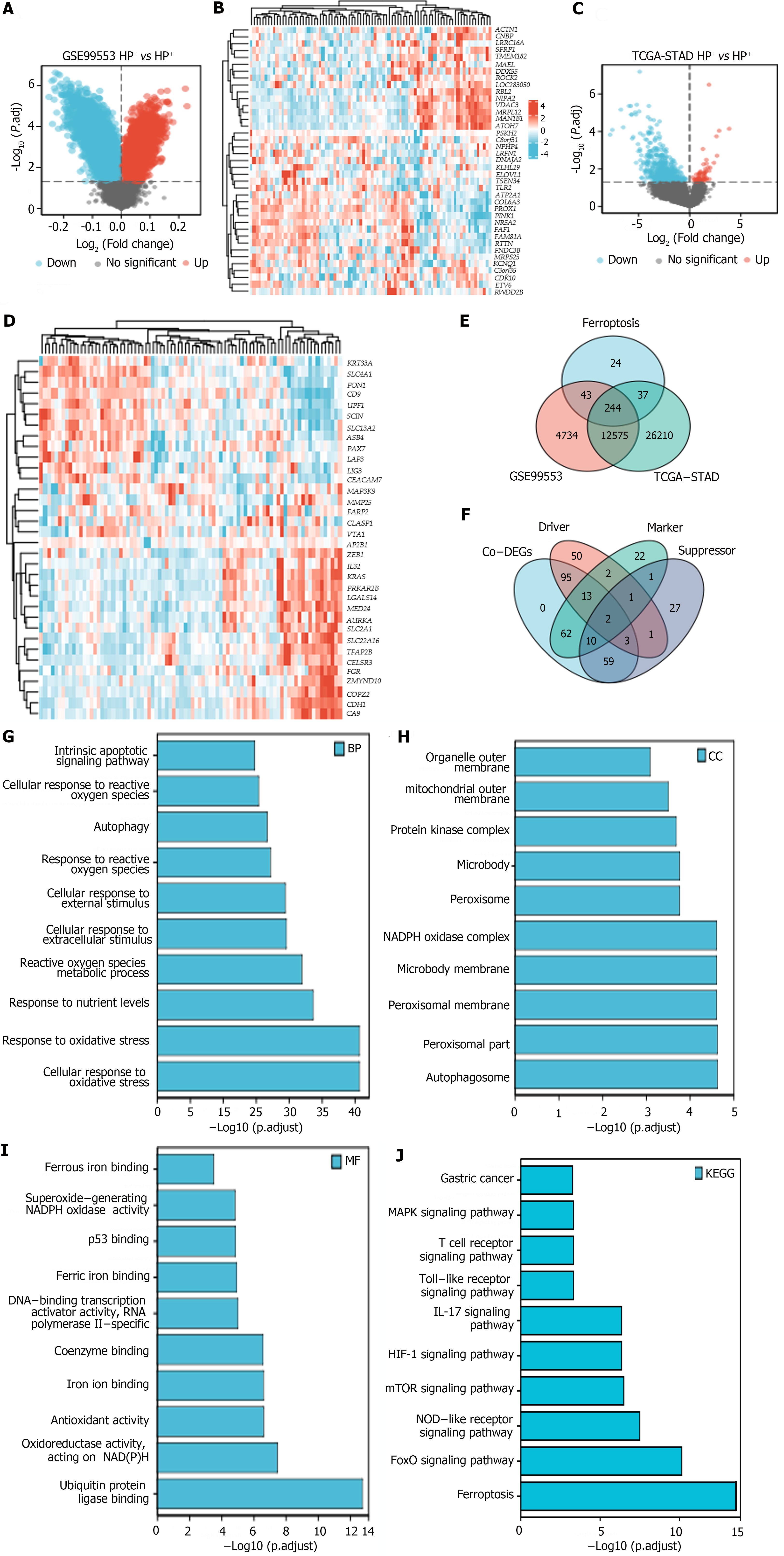
Volcano plots and heatmaps were created to demonstrate the differences in FRG expression between H. pylori+ and H. pylori- GC samples from the TCGA-STAD and GSE99553 cohorts. From these FRGs, we identified 244 ferroptosis-related co-DEGs (Figure 2A-D), including 113 ferroptosis-related driver genes, 74 suppressor genes, and 87 marker genes (Figure 2E and F). GO and KEGG analyses revealed that “cellular response to oxidative stress” and “response to oxidative stress” were the main enriched BP terms. The enriched cellular component terms were related mainly to structures such as autophagosomes, peroxisomes, the peroxisomal membrane, the microbody membrane, and the nicotinamide adenine dinucleotide phosphate (NADPH) oxidase complex. The effective MF terms included ubiquitin protein ligase binding, antioxidant activity, iron ion binding, and ferrous binding. KEGG analysis revealed that the most relevant signaling pathways associated with the Co-DEGs were involved in ferroptosis and the Forkhead box O (FoxO), nucleotide-binding oligomerization domain (NOD)-like receptor, mTOR, hypoxia-inducible factor (HIF)-1, interleukin-17 (IL-17), Toll-like receptor (TLR), and mitogen-activated protein kinase (MAPK) signaling pathways, among others (Figure 2G-J).
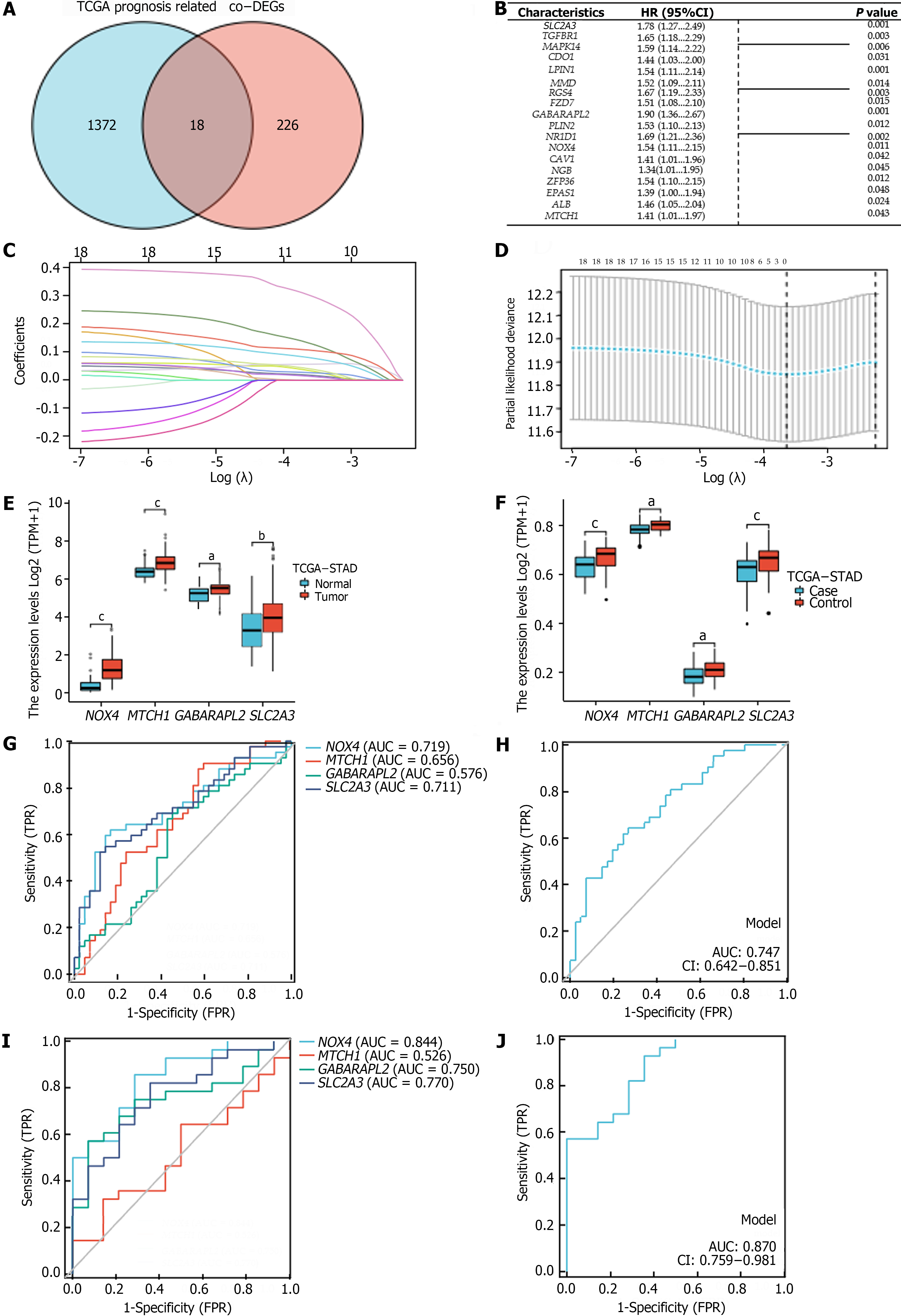
We identified genes that were commonly associated with prognosis among the 1390 oncogenes in the TCGA-STAD cohort and among the 244 co-DEGs; this analysis yielded 18 oncogenes related to OS in GC patients (P < 0.05) (Figure 3A and B). Using LASSO–Cox regression with the coefficient value and hazard ratio (HR), we identified NOX4, MTCH1, GABARAPL2, and SLC2A3 as hub genes (Figure 3C and D) that had higher expression in tumor tissue than in normal tissue (Figure 3E and F). Furthermore, NOX4, MTCH1, GABARAPL2, and SLC2A3 demonstrated some accuracy in predicting GC; NOX4 and SLC2A3 had the highest areas under the curve (AUC) values. The AUCs for predicting GC and H. pylori infection-related GC according to the hub genes were 0.747 and 0.870, respectively (Figure 3G-J). The following formula was used to determine the FRGPIs of the hub genes.
FRGPI = (expression level of NOX4 × 0.432) + (expression level of MTCH1 × 0.344) + (expression level of GABARAPL2 × 0.642) + (expression level of SLC2A3 × 0.577).
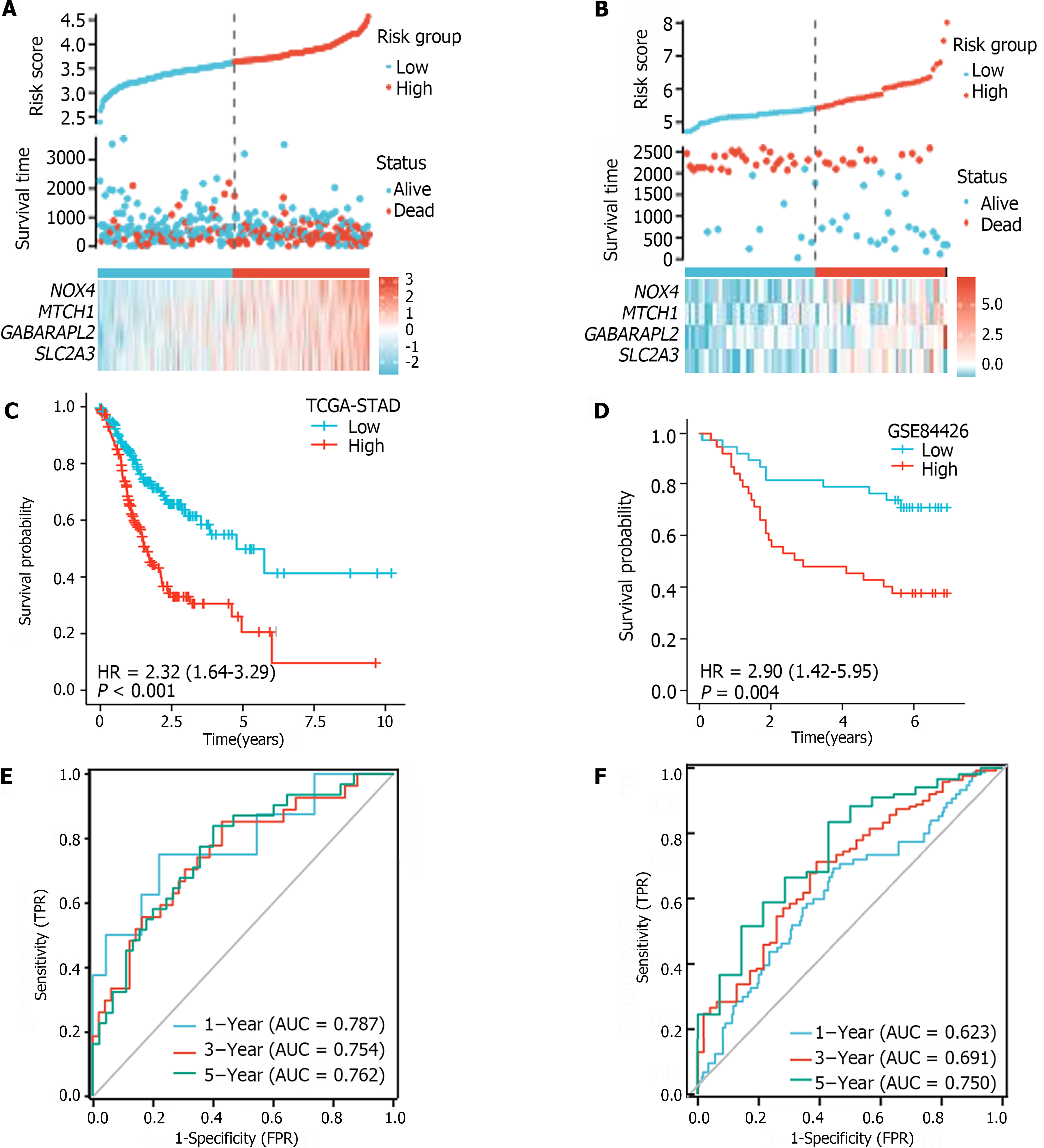
The median FRGPI was used as the cutoff for stratifying patients in the TCGA-STAD and GSE84426 cohorts into high- and low-risk groups. The distributions of the FRGPI and survival status according to the hub genes in the low-risk and high-risk groups are shown in Figure 4A and B. K–M curve analysis revealed that OS was much lower in the high-risk group than in the low-risk group (Figure 4C and D; P < 0.05). The AUCs for 1-year, 3-year, and 5-year survival in the TCGA-STAD dataset were 0.623, 0.691, and 0.750, respectively, while those for the GSE84426 dataset were 0.787, 0.754, and 0.762, respectively; these data demonstrate that the built FRGPI has excellent potential for predicting survival (Figure 4E and F).
For the TCGA-STAD and GSE84426 cohorts, univariate and multivariate Cox proportional hazards regression analyses were performed to verify FRGPI independence. The FRGPI and OS were strongly related (Table 1). In both the TCGA-STAD [HR (95%CI): 2.973 (1.966-4.496), P < 0.01] and GSE84426 [HR (95%CI): 2.001 (1.200-3.501), P < 0.01] cohorts, the FRGPI was an independent predictor of GC prognosis.
| Characteristics | Univariate analysis | Multivariate analysis | ||
| Hazard ratio (95%CI) | P value | Hazard ratio (95%CI) | P value | |
| TCGA cohort | ||||
| Age (≤ 65 vs > 65) | 1.620 (1.154-2.276) | 0.005 | 1.409 (0.943-2.105) | 0.095 |
| Gender (male vs female) | 1.267 (0.891-1.804) | 0.188 | ||
| Pathologic stage (I&II vs III&IV) | 1.947 (1.358-2.793) | < 0.01 | 1.106 (0.614-1.991) | 0.737 |
| T stage (T1&T2 vs T3&T4) | 1.719 (1.131-2.612) | 0.011 | 1.386 (0.763-2.515) | 0.283 |
| N stage (N0 vs N1&N2&N3) | 1.925 (1.264-2.931) | 0.002 | 1.512 (0.776-2.944) | 0.224 |
| M stage (M0 vs M1) | 2.254 (1.295-3.924) | 0.004 | 1.406 (0.679-2.911) | 0.359 |
| Histologic grade (G1 vs G2&G3) | 1.957 (0.484-7.910) | 0.346 | ||
| H. pylori infection | 0.650 (0.279-1.513) | 0.317 | ||
| Primary therapy outcome (CR vs PD&SD&PR) | 4.228 (2.905-6.152) | < 0.01 | 4.453 (2.973-6.672) | < 0.01 |
| FRGPI (high risk vs low risk) | 1.542 (1.105-2.152) | 0.011 | 2.303 (1.531-3.467) | < 0.01 |
| GSE84426 | ||||
| Age (≤ 65 vs > 65) | 1.023 (0.993-1.053) | 0.132 | ||
| Gender (male vs female) | 0.857 (0.401-1.830) | 0.689 | ||
| T stage (T1&T2 vs T3&T4) | 1.392 (0.668-2.900) | 0.677 | ||
| N stage (N0 vs N1&N2&N3) | 1.010 (0.356-2.862) | 0.986 | ||
| FRGPI (high risk vs low risk) | 2.001 (1.000-3.001) | < 0.01 | 2.001 (1.20-3.501) | < 0.01 |
According to the multivariate Cox regression analysis, the primary treatment outcome of tumor progression and the risk score for the FRGPI were considerably greater in the TCGA cohort (P < 0.05; Figure 5A). We created a prognostic nomogram using the above data to improve the prediction of individual patient survival risk, and the results demonstrated high consistency [C-index=0.734 (0.712-0.755)]. Calibration curves showed that the predicted and actual 1-, 3-, and 5-year survival rates of people in the TCGA cohort were similar (Figure 5B-D).
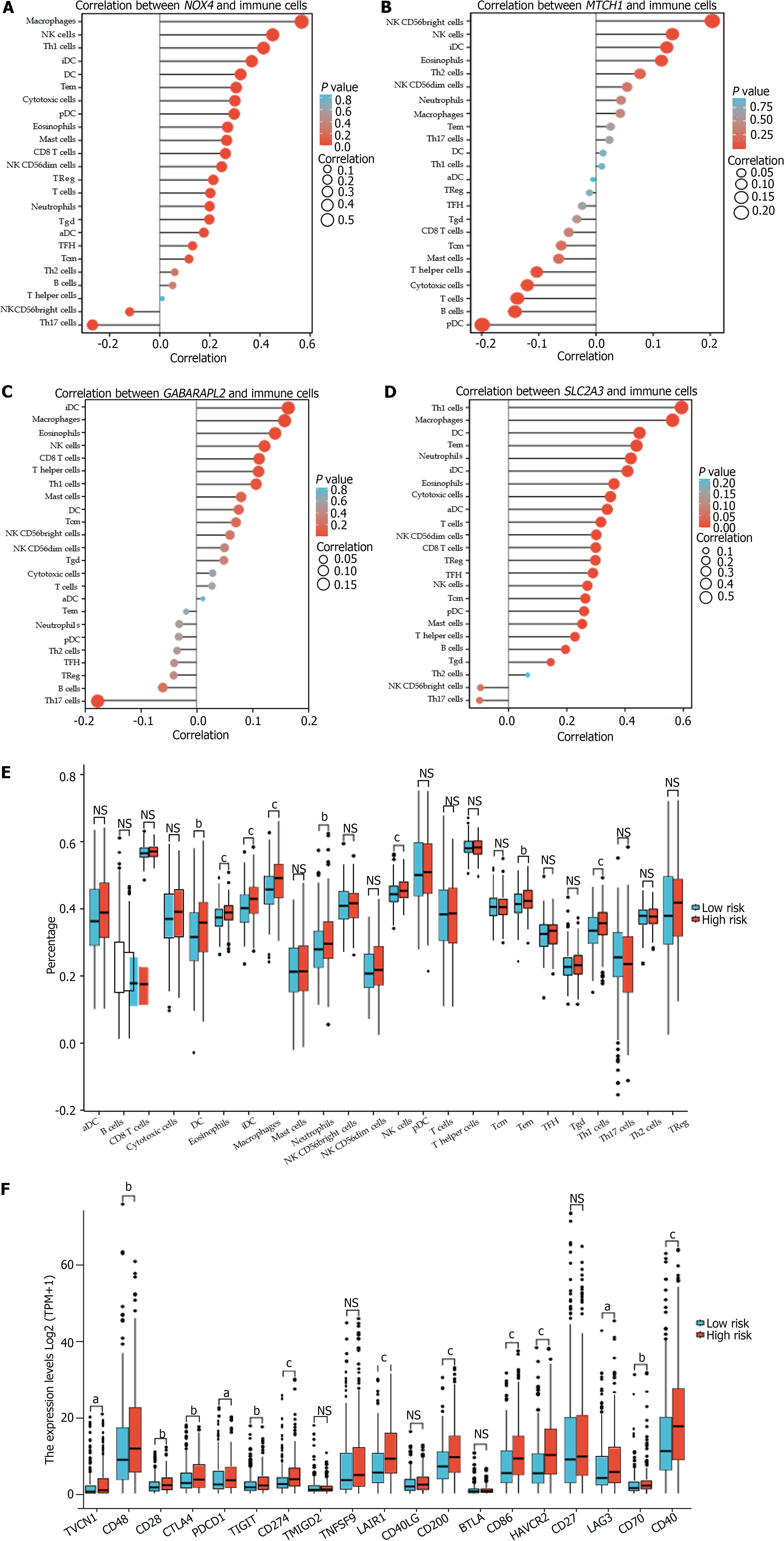
The identified hub genes had various degrees of association with infiltrating immune cells and were positively associated with natural killer (NK) cells, eosinophils, and interdigitating dendritic cells (iDCs) (Figure 6A-D). DCs, eosinophils, iDCs, macrophages, neutrophils, NK cells, effector memory T (Tem) cells, and Type 1 T helper (Th1) cells were considerably upregulated in the high-risk group. Intense infiltration of NK cells, Tem cells, and Th1 cells was observed in the low-risk group (P < 0.05) (Figure 6E).
Additionally, the FRGPI high-risk subgroup exhibited a significant increase in the expression of the most common immune checkpoint proteins (ICPs), including VTCN1, CD48, CD28, CTLA4, PDCD1, TIGHT, and CD274 (PD-L1); LAIR1, CD200, CD86, HAVCR2, LAG3, CD70, and CD40 (Figure 6F).
Immunohistochemistry analysis indicated that the proteins encoded by the hub genes (NOX4, MTCH1, GABARAPL2, and SLC2A3) were strongly expressed in GC and H. pylori-associated GC (Figure 7A and B). Additionally, Western blot analysis revealed that the expression levels of the hub genes were significantly greater in HGC-27 and MGC-803 cells than in normal gastric GES-1 cells (Figure 7C and D). Furthermore, immunofluorescence staining supported this observation by demonstrating hub gene expression in both the nucleus and cytoplasm (Figure 7E and F).
Cytotoxicity assays and scratch experiments indicated that FIN56 inhibited HGC-27 and MGC-803 cells in a dose-dependent manner (Figure 8A and B). The inhibitory concentration values for FIN56 in HGC-27 and MGC-803 cells were 5.9 μM and 8.75 μM, respectively (Figure 8C and D). Moreover, FIN56 treatment decreased the mRNA expression of the hub genes in both HGC-27 and MGC-803 cells; Western blot analyses revealed that hub gene expression decreased in a dose-dependent manner (Figure 8E and F). ROS levels in HGC-27 and MGC-803 cells increased as the FIN56 concentration increased (Figure 8G and H).
Infection with H. pylori is a significant risk factor for distal GC development[1-3]. Ferroptosis is a programmed cell death mechanism associated with fatal lipid peroxidation[7]. Iron metabolism abnormalities are related to both ferroptosis and elevated GC risk[8]. Many mechanisms may be linked to ferroptosis, H. pylori infection, and GC. Nevertheless, few studies have evaluated the relationship between ferroptosis and the prognosis of H. pylori-related GC patients. Therefore, this study aimed to identify Co-DEGs, construct an FRGPI, and validate the predictive significance of the FRGPI in H. pylori-associated GC. The discovered molecular processes and signaling pathways may help explain the relationship between Co-DEGs and H. pylori-associated GC.
This research identified a total of 244 co-DEGs. GO and KEGG analyses revealed that the co-DEGs were involved primarily in ferroptosis and in the FoxO, NOD-like receptor, mTOR, HIF-1, IL-17, TLR, and MAPK signaling pathways, among others; all of these pathways are strongly associated with GC occurrence and development. The FoxO subfamily of the forkhead transcription factor family is involved in cell fate determination and is thought to function as a tumor suppressor in various malignancies[9]. FoxO is implicated in both mitochondria-dependent and mitochondria-independent apoptotic processes by activating death receptor ligands such as the Fas ligand, TNF apoptotic ligand, and Bcl-2 family members[10]. The PI3K/AKT pathway is the most critical pathway through which FoxO1 interacts with many cancers[11]. In addition, numerous other vital pathways, including the Ras–MEK–ERK, IKK, and AMPK pathways, are involved in the mechanism by which FoxOs influence carcinogenesis[10,11]. The inflammasome, which is composed of NOD-like receptors, is also a critical component of innate immunity-induced host defense and inflammation in cancer[12]. Increased inflammasome activity has been linked to various malignancies, including breast, gastric, brain, and malignant prostate cancer, and is protective against colitis-associated cancers[13]. The tumor stage and engagement of different caspase isoforms with the inflammasome pathway impact inflammasome function in cancer. Autophagy inhibition via mTOR-dependent mechanisms such as the AMPK/mTOR and PI3K/Akt/mTOR pathways contributes to the malignant progression of GC cells[14]. Autophagy, which is essential for homeostasis, has dual roles in GC, acting as a tumor suppressor and promoter[15]. Long-term H. pylori infection inhibits autophagy, potentially promoting GC. HIF-1 signaling is vital for metabolism, inflammation, vascular homeostasis, and cancer. HIF-1 activation occurs in several malignancies, including GC[16]. Tumor-associated neutrophils release IL-17a, stimulating GC cell migration and invasion via JAK2/STAT3 activation[17]. TLRs, pattern recognition receptors crucial for H. pylori lipopolysaccharide recognition, are pivotal in GC development, occurrence, and treatment[18,19]. TLR activators hold potential as therapeutic and adjuvant agents in combination with other immunotherapies, suggesting that TLRs are immunotherapy targets for GC[20]. The MAPK regulatory network, which includes many components that initiate a phosphorylation cascade upon activation, elicits precise cellular responses[21]. This network significantly influences GC progression[22].
This study utilized public databases to identify crucial ferroptosis-related DEGs in H. pylori-associated GC. In GC patients, FRGs were differentially expressed between normal and tumor tissues, indicating the substantial involvement of ferroptosis in this disease. Univariate Cox analysis revealed 18 genes linked to OS, highlighting the feasibility and utility of constructing a prognostic signature using the FRGPI. LASSO-Cox analysis identified a novel prognostic signature comprising four FRGs: NOX4, MTCH1, GABARAPL2, and SLC2A3. The four hub genes accurately predicted GC using ROC curves. Functional enrichment and analyses of the tumor microenvironment and immunotherapy response revealed that the hub genes might accurately predict GC patient prognosis and clinical state. To better understand the involvement of these genes in GC, we describe their primary molecular activity.
NOX4 is an NADPH oxidase that promotes the cellular ROS response[23]. When the intracellular iron ion concentration exceeds normal levels, NOX4 is activated, producing a large amount of oxygen free radicals, which combine with iron ions to form highly reactive hydrogen and oxygen free radicals. These radicals then destroy proteins, DNA, and lipid molecular structures in cells. Hydroxyl radical activity causes the release of intracellular iron ions, intensifying the ROS response[23]. Previous studies have shown that it interferes with cancer cell proliferation, apoptosis, and cell cycle progression in conditions such as brain glioma, non-small cell lung cancer, and melanoma[24-26]. Therefore, NOX4 stimulates tumor development through various channels and processes and is thus considered a potential therapeutic target[27]. NOX4 overexpression accelerates cancer development and is correlated with poor prognosis in patients with colorectal cancer[28]. Additionally, the NOX4 gene was deleted in liver cancer patients, demonstrating that it may exert tumor-suppressor effects by facilitating TGF-β1-induced apoptosis[29]. Although the mechanism through which NOX4 promotes GC development and invasion is unknown, studies have revealed that NOX4 may stimulate GC cell proliferation and invasion through epithelial-mesenchymal transition[30,31]. Moreover, Li et al[32] showed that the new biotin derivative XN4 could induce ferroptosis in GC cells by upregulating NOX4 expression[32].
MTCH1 encodes a protein with two proapoptotic domains that localize to the inner mitochondrial membrane, causing apoptosis independently of Bax and Bak[33]. MTCH1 regulates cellular iron levels, maintaining iron within normal limits. When cellular iron levels exceed normal levels, MTCH1 is activated, causing mitochondrial outer membrane permeabilization (MOMP) and the release of mitochondrial molecules into the cytoplasm[34]. MOMP releases iron ions from mitochondria into the cytoplasm, increasing cellular iron. The increased iron can interact with hydroxide ions to produce highly reactive hydroxide radicals. These free radicals can cause cell death by damaging proteins, DNA, and lipids[35]. Although bioinformatics analysis and functional validation suggest that increased MTCH1 expression is correlated with metastasis, cancer stage, and poor survival in liver hepatocellular carcinoma patients[36], no study has validated MTCH1 expression in GC.
SLC2A3, also known as GLUT3, is a transmembrane transporter with high affinity for glucose. It can regulate intracellular glucose metabolism and promote ferroptosis, but little to no expression has been detected in normal tissues[37]. SLC2A3 is activated when the intracellular glucose concentration increases, promoting NOX4 activation, increasing oxygen free radical production, and inducing ferroptosis[38]. Tumor cells enhance glucose consumption through aerobic glycolysis for proliferation. Hence, SLC2A3 is strongly overexpressed in various tumor types[39,40]. SLC2A1 overexpression may increase colorectal cancer cell proliferation and invasion by boosting nucleotide synthesis[41]. Glycolysis control in GC cells promotes macrophage infiltration and tumor development[42,43]. Therefore, SLC2A3 may be a predictive biomarker in these cancers.
GABARAPL2, also known as GATE-16 (16 kDa enhancer of Golgi-associated ATPase), is a member of the autophagy-associated protein 8 subfamily and is involved in the degradation of autophagosome membranes. It promotes intracellular ferroptosis[44]. GABARAPL2 forms a complex with LC3 to operate at various autophagosome phases; the LC3 subfamily promotes phagocyte membrane fusion, while the GABARAP subfamily functions downstream[45]. GABARAPL2 can promote MOMP, causing mitochondrial molecules to escape into the cytoplasm. Under MOMP, iron ions in mitochondria are released into the cytoplasm, increasing cellular iron and inducing ferroptosis[46]. High GABARAPL2 mRNA expression is associated with poorer prognosis in distinct malignancies, such as worse OS in patients with esophageal cancer or GC[47,48] but better OS in patients with kidney cancer[49]. This discrepancy primarily results from the autophagy-dependent and autophagy-independent functions of GABARAPL2.
We utilized multivariate analysis to create a prognostic model and then assessed the accuracy of the individual patient predictions. The prognostic nomogram and calibration curve findings were consistent with the actual results based on the 1-year, 3-year, and 5-year survival curves. Similar curves indicate that the model can accurately predict OS. However, the novel prognostic nomogram has the potential to increase the prediction accuracy and assist in predicting individual patient survival risk, with significant therapeutic implications[50]. According to the FRGPI, which was developed using hub gene characteristics, GC patients can be classified into low-risk and high-risk groups. The low-risk group had a better prognosis and longer OS than did the high-risk group. To further investigate the underlying mechanisms of the hub genes, we determined that they were significantly related to infiltrating immune cells. Moreover, the FRGPI was associated with the tumor microenvironment and immunotherapy response. There were substantial differences in immune cell infiltration between the FRGPI low-risk and high-risk patients. In the high-risk group, DCs, eosinophils, iDCs, macrophages, and neutrophils exhibited considerable infiltration, while in the low-risk group, NK cells, Tem cells, and Th1 cells exhibited substantial infiltration. NK cells inhibit tumor development and promote CD4+ Th1 polarization. Greater increases in the NK cell percentage correlate with prolonged survival in GC patients, suggesting that NK cells may be an independent prognostic biomarker[51]. Central memory T cells activated by tumor antigens create antitumor Tem cells, which destroy cancer cells and bolster immunity[52]. An imbalance in Th1 and Th2 cells is considered significant in GC development[53]. A shift from Th1 to Th2 cytokines can create an immunosuppressive state, compromising antitumor immunity[54]. Additionally, we observed significant differences in patient characteristics and ICPs between the high-risk and low-risk groups. The expression of genes that encode most ICPs was significantly upre
To determine whether the identified hub genes are required for ferroptosis in GC, we utilized FIN56 to trigger ferroptosis in two GC cell lines, HGC-27 and MGC-803. CCK-8 assays revealed that FIN56 significantly inhibited the proliferation of HGC-27 and MGC-803 cells, and low concentrations of FIN56 were fatal to GC cell lines. Lipid peroxides, abundant ROS in living organisms, primarily cause ferroptosis[59]. Thus, we detected significantly elevated ROS levels in FIN56-treated GC cells by flow cytometry and fluorescence microscopy, suggesting that FIN56-induced cell death is due to ferroptosis. Furthermore, both the mRNA and protein levels of the hub genes decreased in a dose-dependent manner in FIN56-treated GC cells, demonstrating that FIN56 regulates hub gene expression during ferroptosis. These data suggest that FIN56 is a potential drug for inhibiting GC cell invasion.
In conclusion, this study identified co-DEGs between H. pylori-associated GC and ferroptosis that promote GC formation and progression through various signaling pathways. The FRGPI, which included NOX4, MTCH1, GABARAPL2, and SLC2A3, demonstrated high predictive accuracy.
Gastric cancer (GC) is a common malignancy of the digestive system. Ferroptosis or iron-dependent programmed cell death, has been linked to GC, although there has been little research on the link between Helicobacter pylori (H. pylori) infection-related GC and ferroptosis.
This study investigated the coregulated differentially expressed genes (co-DEGs) among ferroptosis-related genes (FRGs) in H. pylori infection-related GCs and investigated the relationship between the expression of these co-DEGs and clinical prognosis.
This study developed a prognostic model for GC based on co-DEGs among FRGs. Gene expression profiles from GC patients and H. pylori-associated GC patients were analyzed.
Gene expression profiles of GC patients and those with H. pylori-associated GC were obtained from the cancer genome atlas and gene expression omnibus (GEO) databases. The FRGs were acquired from the FerrDb database. A ferroptosis-related gene prognostic index (FRGPI) was created using least absolute shrinkage and selection operator–Cox regression. The predictive ability of the FRGPI was validated in the GEO cohort. Finally, we verified the expression of the hub genes and the activity of the ferroptosis inducer FIN56 in GC cell lines and tissues.
The four hub genes (NOX4, MTCH1, GABARAPL2, and SLC2A3) accurately predicted GC and H. pylori-associated GC. The FRGPI based on the hub genes independently predicted patient survival. High-risk GC patients had notably worse overall survival. The FRGPI was found to be a significant predictor of GC prognosis and correlated with disease progression. The expression of immune checkpoint proteins increased in the high-risk group. Hub genes were highly overexpressed in GC cell lines and tissues, primarily at the cell membrane. The ferroptosis inducer FIN56 inhibited GC cell proliferation in a dose-dependent manner.
The predictive model provides an accurate prognosis for GC patients and helps evaluate immunotherapy efficacy.
The mechanism of the hub gene in the occurrence and development of GC was studied, and its effect on iron death in GC cells was analyzed.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Yap RV, Philippines S-Editor: Liu H L-Editor: A P-Editor: Cai YX
| 1. | Alipour M. Molecular Mechanism of Helicobacter pylori-Induced Gastric Cancer. J Gastrointest Cancer. 2021;52:23-30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 85] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 2. | Matsunaga S, Nishiumi S, Tagawa R, Yoshida M. Alterations in metabolic pathways in gastric epithelial cells infected with Helicobacter pylori. Microb Pathog. 2018;124:122-129. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 3. | Bakhti SZ, Latifi-Navid S, Zahri S. Unique constellations of five polymorphic sites of Helicobacter pylori vacA and cagA status associated with risk of gastric cancer. Infect Genet Evol. 2020;79:104167. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 4. | FitzGerald R, Smith SM. An Overview of Helicobacter pylori Infection. Methods Mol Biol. 2021;2283:1-14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 5. | Wang L, Wang H. The putative role of ferroptosis in gastric cancer: a review. Eur J Cancer Prev. 2023;32:575-583. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 6. | Zhou N, Bao J. FerrDb: a manually curated resource for regulators and markers of ferroptosis and ferroptosis-disease associations. Database (Oxford). 2020;2020. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 212] [Cited by in F6Publishing: 424] [Article Influence: 141.3] [Reference Citation Analysis (0)] |
| 7. | Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, Fulda S, Gascón S, Hatzios SK, Kagan VE, Noel K, Jiang X, Linkermann A, Murphy ME, Overholtzer M, Oyagi A, Pagnussat GC, Park J, Ran Q, Rosenfeld CS, Salnikow K, Tang D, Torti FM, Torti SV, Toyokuni S, Woerpel KA, Zhang DD. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell. 2017;171:273-285. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4107] [Cited by in F6Publishing: 3763] [Article Influence: 537.6] [Reference Citation Analysis (0)] |
| 8. | Wei J, Gao X, Qin Y, Liu T, Kang Y. An Iron Metabolism-Related SLC22A17 for the Prognostic Value of Gastric Cancer. Onco Targets Ther. 2020;13:12763-12775. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 9. | Jiramongkol Y, Lam EW. FOXO transcription factor family in cancer and metastasis. Cancer Metastasis Rev. 2020;39:681-709. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 121] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 10. | Farhan M, Wang H, Gaur U, Little PJ, Xu J, Zheng W. FOXO Signaling Pathways as Therapeutic Targets in Cancer. Int J Biol Sci. 2017;13:815-827. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 208] [Cited by in F6Publishing: 286] [Article Influence: 40.9] [Reference Citation Analysis (0)] |
| 11. | Ramezani A, Nikravesh H, Faghihloo E. The roles of FOX proteins in virus-associated cancers. J Cell Physiol. 2019;234:3347-3361. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 12. | Cao X, Xu J. Insights into inflammasome and its research advances in cancer. Tumori. 2019;105:456-464. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 13. | Perera AP, Sajnani K, Dickinson J, Eri R, Körner H. NLRP3 inflammasome in colitis and colitis-associated colorectal cancer. Mamm Genome. 2018;29:817-830. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 14. | Chen JF, Wu P, Xia R, Yang J, Huo XY, Gu DY, Tang CJ, De W, Yang F. STAT3-induced lncRNA HAGLROS overexpression contributes to the malignant progression of gastric cancer cells via mTOR signal-mediated inhibition of autophagy. Mol Cancer. 2018;17:6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 130] [Cited by in F6Publishing: 172] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 15. | Schmitt-Ney M. The FOXO's Advantages of Being a Family: Considerations on Function and Evolution. Cells. 2020;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 38] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 16. | Li H, Jia Y, Wang Y. Targeting HIF-1α signaling pathway for gastric cancer treatment. Pharmazie. 2019;74:3-7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 12] [Reference Citation Analysis (0)] |
| 17. | Li S, Cong X, Gao H, Lan X, Li Z, Wang W, Song S, Wang Y, Li C, Zhang H, Zhao Y, Xue Y. Tumor-associated neutrophils induce EMT by IL-17a to promote migration and invasion in gastric cancer cells. J Exp Clin Cancer Res. 2019;38:6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 128] [Article Influence: 25.6] [Reference Citation Analysis (1)] |
| 18. | Liu YD, Yu L, Ying L, Balic J, Gao H, Deng NT, West A, Yan F, Ji CB, Gough D, Tan P, Jenkins BJ, Li JK. Toll-like receptor 2 regulates metabolic reprogramming in gastric cancer via superoxide dismutase 2. Int J Cancer. 2019;144:3056-3069. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 19. | Meliț LE, Mărginean CO, Mărginean CD, Mărginean MO. The Relationship between Toll-like Receptors and Helicobacter pylori-Related Gastropathies: Still a Controversial Topic. J Immunol Res. 2019;2019:8197048. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 20. | Cui L, Wang X, Zhang D. TLRs as a Promise Target Along With Immune Checkpoint Against Gastric Cancer. Front Cell Dev Biol. 2020;8:611444. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 22] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 21. | Magnelli L, Schiavone N, Staderini F, Biagioni A, Papucci L. MAP Kinases Pathways in Gastric Cancer. Int J Mol Sci. 2020;21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 22. | Wei X, Liu X, Liu H, He X, Zhuang H, Tang Y, Wang B. BRCA1-associated protein induced proliferation and migration of gastric cancer cells through MAPK pathway. Surg Oncol. 2020;35:191-199. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 23. | Huang Y, Li Y, Lou A, Wang GZ, Hu Y, Zhang Y, Huang W, Wang J, Zhu X, Chen T, Lin J, Meng Y, Li X. Alamandine attenuates hepatic fibrosis by regulating autophagy induced by NOX4-dependent ROS. Clin Sci (Lond). 2020;134:853-869. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 24. | Lin L, Li X, Zhu S, Long Q, Hu Y, Zhang L, Liu Z, Li B. Ferroptosis-related NFE2L2 and NOX4 Genes are Potential Risk Prognostic Biomarkers and Correlated with Immunogenic Features in Glioma. Cell Biochem Biophys. 2023;81:7-17. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 25. | Li J, Lan T, Zhang C, Zeng C, Hou J, Yang Z, Zhang M, Liu J, Liu B. Reciprocal activation between IL-6/STAT3 and NOX4/Akt signalings promotes proliferation and survival of non-small cell lung cancer cells. Oncotarget. 2015;6:1031-1048. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 60] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 26. | Tang CT, Lin XL, Wu S, Liang Q, Yang L, Gao YJ, Ge ZZ. NOX4-driven ROS formation regulates proliferation and apoptosis of gastric cancer cells through the GLI1 pathway. Cell Signal. 2018;46:52-63. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 27. | Ju HQ, Ying H, Tian T, Ling J, Fu J, Lu Y, Wu M, Yang L, Achreja A, Chen G, Zhuang Z, Wang H, Nagrath D, Yao J, Hung MC, DePinho RA, Huang P, Xu RH, Chiao PJ. Mutant Kras- and p16-regulated NOX4 activation overcomes metabolic checkpoints in development of pancreatic ductal adenocarcinoma. Nat Commun. 2017;8:14437. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 68] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 28. | Lin XL, Yang L, Fu SW, Lin WF, Gao YJ, Chen HY, Ge ZZ. Overexpression of NOX4 predicts poor prognosis and promotes tumor progression in human colorectal cancer. Oncotarget. 2017;8:33586-33600. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 49] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 29. | Herranz-Itúrbide M, López-Luque J, Gonzalez-Sanchez E, Caballero-Díaz D, Crosas-Molist E, Martín-Mur B, Gut M, Esteve-Codina A, Jaquet V, Jiang JX, Török NJ, Fabregat I. NADPH oxidase 4 (Nox4) deletion accelerates liver regeneration in mice. Redox Biol. 2021;40:101841. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 30. | Du S, Miao J, Lu X, Shi L, Sun J, Xu E, Wang X, Zhao M, Chen H, Wang F, Kang X, Ding J, Guan W, Xia X. NADPH oxidase 4 is correlated with gastric cancer progression and predicts a poor prognosis. Am J Transl Res. 2019;11:3518-3530. [PubMed] [Cited in This Article: ] |
| 31. | Gao X, Sun J, Huang C, Hu X, Jiang N, Lu C. RNAi-mediated silencing of NOX4 inhibited the invasion of gastric cancer cells through JAK2/STAT3 signaling. Am J Transl Res. 2017;9:4440-4449. [PubMed] [Cited in This Article: ] |
| 32. | Li R, Yin B, Zeng D, Liu Z. A novobiocin derivative, XN4, triggers ferroptosis in gastric cancer cells via the activation of NOX4. Pharm Biol. 2022;60:1449-1457. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 1] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 33. | Lamarca V, Sanz-Clemente A, Pérez-Pé R, Martínez-Lorenzo MJ, Halaihel N, Muniesa P, Carrodeguas JA. Two isoforms of PSAP/MTCH1 share two proapoptotic domains and multiple internal signals for import into the mitochondrial outer membrane. Am J Physiol Cell Physiol. 2007;293:C1347-C1361. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 20] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 34. | Bonora M, Giorgi C, Pinton P. Molecular mechanisms and consequences of mitochondrial permeability transition. Nat Rev Mol Cell Biol. 2022;23:266-285. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 174] [Article Influence: 87.0] [Reference Citation Analysis (0)] |
| 35. | Zheng J, Conrad M. The Metabolic Underpinnings of Ferroptosis. Cell Metab. 2020;32:920-937. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 314] [Cited by in F6Publishing: 535] [Article Influence: 133.8] [Reference Citation Analysis (0)] |
| 36. | Chen G, Mo S, Yuan D. Upregulation Mitochondrial Carrier 1 (MTCH1) Is Associated with Cell Proliferation, Invasion, and Migration of Liver Hepatocellular Carcinoma. Biomed Res Int. 2021;2021:9911784. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 37. | Koutsioumpa M, Hatziapostolou M, Polytarchou C, Tolosa EJ, Almada LL, Mahurkar-Joshi S, Williams J, Tirado-Rodriguez AB, Huerta-Yepez S, Karavias D, Kourea H, Poultsides GA, Struhl K, Dawson DW, Donahue TR, Fernández-Zapico ME, Iliopoulos D. Lysine methyltransferase 2D regulates pancreatic carcinogenesis through metabolic reprogramming. Gut. 2019;68:1271-1286. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 38. | Nascè A, Gariani K, Jornayvaz FR, Szanto I. NADPH Oxidases Connecting Fatty Liver Disease, Insulin Resistance and Type 2 Diabetes: Current Knowledge and Therapeutic Outlook. Antioxidants (Basel). 2022;11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 19] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 39. | Kelemen O, Pla I, Sanchez A, Rezeli M, Szasz AM, Malm J, Laszlo V, Kwon HJ, Dome B, Marko-Varga G. Proteomic analysis enables distinction of early- vs advanced-stage lung adenocarcinomas. Clin Transl Med. 2020;10:e106. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 40. | Gao H, Hao Y, Zhou X, Li H, Liu F, Zhu H, Song X, Niu Z, Ni Q, Chen MS, Lu J. Prognostic value of glucose transporter 3 expression in hepatocellular carcinoma. Oncol Lett. 2020;19:691-699. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 41. | Dai W, Xu Y, Mo S, Li Q, Yu J, Wang R, Ma Y, Ni Y, Xiang W, Han L, Zhang L, Cai S, Qin J, Chen WL, Jia W, Cai G. GLUT3 induced by AMPK/CREB1 axis is key for withstanding energy stress and augments the efficacy of current colorectal cancer therapies. Signal Transduct Target Ther. 2020;5:177. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 47] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 42. | Yao X, He Z, Qin C, Deng X, Bai L, Li G, Shi J. SLC2A3 promotes macrophage infiltration by glycolysis reprogramming in gastric cancer. Cancer Cell Int. 2020;20:503. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 43] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 43. | Chen D, Wang H, Chen J, Li Z, Li S, Hu Z, Huang S, Zhao Y, He X. MicroRNA-129-5p Regulates Glycolysis and Cell Proliferation by Targeting the Glucose Transporter SLC2A3 in Gastric Cancer Cells. Front Pharmacol. 2018;9:502. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 54] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 44. | Nguyen TN, Padman BS, Usher J, Oorschot V, Ramm G, Lazarou M. Atg8 family LC3/GABARAP proteins are crucial for autophagosome-lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J Cell Biol. 2016;215:857-874. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 350] [Cited by in F6Publishing: 427] [Article Influence: 53.4] [Reference Citation Analysis (0)] |
| 45. | Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29:1792-1802. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 508] [Cited by in F6Publishing: 575] [Article Influence: 41.1] [Reference Citation Analysis (0)] |
| 46. | Du J, Zhou Y, Li Y, Xia J, Chen Y, Chen S, Wang X, Sun W, Wang T, Ren X, An Y, Lu K, Hu W, Huang S, Li J, Tong X, Wang Y. Identification of Frataxin as a regulator of ferroptosis. Redox Biol. 2020;32:101483. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 121] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 47. | Chen C, Chen S, Cao H, Wang J, Wen T, Hu X, Li H. Prognostic significance of autophagy-related genes within esophageal carcinoma. BMC Cancer. 2020;20:797. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 48. | Qiu J, Sun M, Wang Y, Chen B. Identification and validation of an individualized autophagy-clinical prognostic index in gastric cancer patients. Cancer Cell Int. 2020;20:178. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 31] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 49. | Chan JCY, Gorski SM. Unlocking the gate to GABARAPL2. Biol Futur. 2022;73:157-169. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 50. | Li Y, He X, Fan L, Zhang X, Xu Y, Xu X. Identification of a novel immune prognostic model in gastric cancer. Clin Transl Oncol. 2021;23:846-855. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 51. | Xie MZ, Tang YP, Hu BL, Li KZ, Li JL, Liang XQ. Percentage of Natural Killer (NK) Cells in Peripheral Blood Is Associated with Prognosis in Patients with Gastric Cancer: A Retrospective Study from a Single Center. Med Sci Monit. 2021;27:e927464. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 52. | Zhang R, Li F, Li H, Yu J, Ren X. The clinical significance of memory T cells and its subsets in gastric cancer. Clin Transl Oncol. 2014;16:257-265. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 53. | Rezalotfi A, Ahmadian E, Aazami H, Solgi G, Ebrahimi M. Gastric Cancer Stem Cells Effect on Th17/Treg Balance; A Bench to Beside Perspective. Front Oncol. 2019;9:226. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 54. | Shin SJ, Kim SY, Choi YY, Son T, Cheong JH, Hyung WJ, Noh SH, Park CG, Kim HI. Mismatch Repair Status of Gastric Cancer and Its Association with the Local and Systemic Immune Response. Oncologist. 2019;24:e835-e844. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 55. | Xu H, Ye D, Ren M, Zhang H, Bi F. Ferroptosis in the tumor microenvironment: perspectives for immunotherapy. Trends Mol Med. 2021;27:856-867. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 125] [Article Influence: 41.7] [Reference Citation Analysis (0)] |
| 56. | Wang W, Green M, Choi JE, Gijón M, Kennedy PD, Johnson JK, Liao P, Lang X, Kryczek I, Sell A, Xia H, Zhou J, Li G, Li J, Li W, Wei S, Vatan L, Zhang H, Szeliga W, Gu W, Liu R, Lawrence TS, Lamb C, Tanno Y, Cieslik M, Stone E, Georgiou G, Chan TA, Chinnaiyan A, Zou W. CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569:270-274. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 769] [Cited by in F6Publishing: 1427] [Article Influence: 285.4] [Reference Citation Analysis (0)] |
| 57. | Sun Y, Berleth N, Wu W, Schlütermann D, Deitersen J, Stuhldreier F, Berning L, Friedrich A, Akgün S, Mendiburo MJ, Wesselborg S, Conrad M, Berndt C, Stork B. Fin56-induced ferroptosis is supported by autophagy-mediated GPX4 degradation and functions synergistically with mTOR inhibition to kill bladder cancer cells. Cell Death Dis. 2021;12:1028. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 98] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 58. | Forcina GC, Dixon SJ. GPX4 at the Crossroads of Lipid Homeostasis and Ferroptosis. Proteomics. 2019;19:e1800311. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 199] [Cited by in F6Publishing: 445] [Article Influence: 89.0] [Reference Citation Analysis (0)] |
| 59. | Latunde-Dada GO. Ferroptosis: Role of lipid peroxidation, iron and ferritinophagy. Biochim Biophys Acta Gen Subj. 2017;1861:1893-1900. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 315] [Cited by in F6Publishing: 340] [Article Influence: 48.6] [Reference Citation Analysis (0)] |