MATERIALS AND METHODS
Materials
Human hepatocarcinoma cell (HHCC) was grown in RPMI1640 medium supplemented with 100 mL/L newborn calf serum. PV I tenuate vaccinias line (Third Department of vaccinia, Chinese Drug and Bioproduct Identified Agent) was amplified in Hep2 cells, and the virus titer was calculated by plaque assay.
Methods
Plasmid construction By using subcloning methods, the IRNA and mIRNA sequence were cloned into the pcDNA3 vector, yielding pcRz-IRNA and pcRz-mIRNA which introduced the ribozyme sequence over the both sites of IRNA and mIRNA to generate the correct site of IRNA and mIRNA. In summary, by using PCR methods the sequences of target RNA were generated from pGRz-IRNA or pGRz-mIRNA which were constructed by our laboratory. Then the PCR product was cloning into the BamH I-Apa I sites of the pcDNA3 vector.
Construction of HHCCs expressing IRNA or mIRNA Plasmids pcRz-IRNA and pcRz-mIRNA were transfected into HHCC cells respectively by using Lipofectamine 2000 reagent (GIbco) and screened for neomycin resistance with 300 μg of G418 (Invitrogen) per milliliter for 4 weeks. The antibiotic-resistant cell clones were harvested and further screened by dilution titer. The control cells were also prepared by using similar method using plasmid pcDNA3.
Detection of IRNA/mIRNA in the cells IRNA or mIRNA expression in the cells was measured by isolating total RNA from these cells and the IRNA or mIRNA were detected by reverse transcriptase (RT)-mediated PCR (RT-PCR) by using IRNA or mIRNA specific oligonucleotide primers.1 to 2 μg of total RNA isolated from the IRNA or mIRNA expressing cells, and 2 μg total RNA from HHCC control cells were reversely transcribed by murine leukemia virus RT using random hexamer primers in a 20 μl reaction mixture according to the TaKaRa RNA PCR kit protol. 20pmol of each primer (corresponding to 5’ nt 1 to 20 and 3’ nt 1 to 20 of the IRNA or mIRNA sequence) was used to amplify the 60-nt fragement in a 100 μl PCR reaction. The cycling parameters were as follows: denaturation, 95 °C for 1 min; annealing, 65 °C for 1 min; extrension, 72 °C for 1 min, total extrension, 72 °C for 10 mins; a total of 50 cycles.Twenty microliters of each reaction product were loaded onto 20 g/L gel and visualized by ethidium brominde staining.
Detection of cell protein levels by laser confocal microscopy Monolayers of HHCCs were plated on cover glass and fixed with pure ethanol for 10 mins. Monoclonal antibody of β-actin was properly diluted and covered on the glass with HHCC cells for 1h at 37 °C, and then the glass was washed with PBS 3 times (10 mins each). Then FITC labeled second antibody was covered on the glass at 37 °C for 1 h and the glass was washed with PBS 3 times again. At last the cells were examined by using the laser confocal microscopy.
Plaque assay Plaque assay were performed as described below. HHCCs (106 cells) were infected with PV I tenuate line, and after 72 h, cytopathic effect was observed and cell extracts were prepared. Two hundred microliters of cell extracts was used to further infect Hep2 cells (5 × 106 cells in 60 millimeter diameter plates). After 3 days of inoculation at 37 °C, the plaques were developed by staining with 10 g/L crystal violet.
Mobility shift electrophoresis assay Fifty micrograms of rabbit reticulocyte lysate (RRL) was preincubated at 30 °C for 10 mins with 4 μg of poly (dI-dC) (Pharmacia) in a 15 μl reaction mixture containing 5 mmol/L HEPES (N-2-hydroxyethylpiperazina-N’-2-ethanesulfonic acide) (pH7.5), 25 mmol/L KCl2, 2 mmol/L MgCl2, 38g/L glycerol, and 2 mmol/L dithiothreitol. For competition experiments, 25 to 50-fold excess of unlabeled competitor RNAs were added to the reaction mixture and incubated for 10 min at 30 °C. Finally, 5 to 10 fmol of labeled RNA probes were added to respective reaction mixture and incubation was continued for another 30 min at 30 °C. The nonspecific RNA used in competition assay was the sequence of the polylinker region of the pGEM 3Z vector (Promega). Three microliters of gel loading dye was added to the reaction mixture to a final concentration of 100 g/L glyceol and 2 g/L each of bromophenol blue and xylene cyanol. RNA-protein complexes were then analyzed on a 40 g/L polyacrylamide gel (39:1 ration of acrylanide: bis) in a 0.5 × TBE.
RESULTS
Construction of hepatoma cells expressing IRNA or mIRNA constitutionally
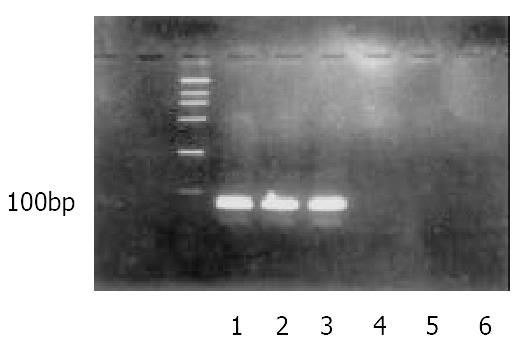
To determine the long-term effect of expression of IRNA in HHCC cells, the cells constitutionally expressing IRNA were generated by using a pcDNA-based vector as described in Material and Methods. In order to obtain the correct and stable both sides of the expressed IRNA, the ribozyme sequences were introduced into the both sides of the IRNA and mIRNA. The IRNA or mIRNA was examined by RT-PCR using appropriate primers. As can be seen in Figure 1, IRNA and mIRNA was expressed stably in the cloning cells.
Figure 1 Detection IRNA or mIRNA in the total RNA of HHCCs which constitutionally expressing IRNA or mIRNA.
1: DNA Marker DL2000; 2 and 3: RT-PCR of cell lines expressing IRNA; 4: RT-PCR of cell lines expressing mIRNA; 5 and 6: PCR of cell lines expressing IRNA or mIRNA.
Effect of long-term IRNA expression on cellular protein expression
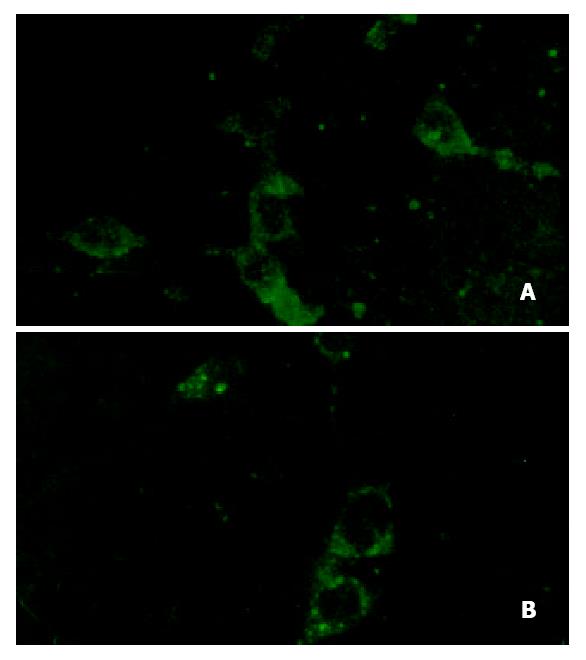
To determine whether constitutionally expressing of IRNA interfered with cellular protein translation, levels of cellular protein β-actin was determined by laser confocal microscopy. As can be seen in Figure 2A and Figure 2B, no differences were detected in the overall expression of the cellular protein between HHCC (control) and IRNA-expressing cells (11.74 ± 6.52 vs 10.06 ± 5.75, P > 0.05).
Figure 2 The translation level of β-actin in HHCC and IRNA-expressing lines.
(Laser confocal, 400 × 1.00). A: IRNA-express-ing cell lines, B: Control cell lines.
Inhibition of long-term expressing IRNA on HCV IRES mediated translation
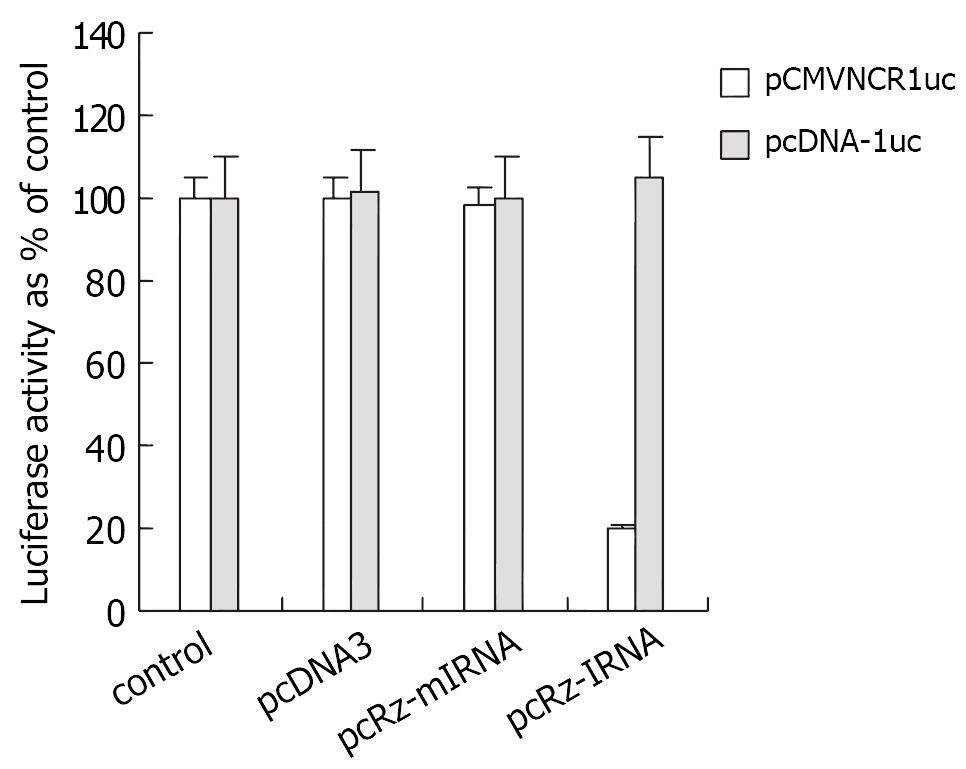
To test the inhibitor activity of long-term expressing IRNA on HCV IRES mediated translation, the control cells (HHCC) and cells expressing IRNA or mIRNA were transiently cotransfected with pCMVNCRluc and pSV40/β-Gal. Additionally, in order to exclude the nonspecific effect of nonspecific small RNA on the protein translation, the cells expressing pcDNA3 were cotransfected with pCMVNCRluc and pSV40/β-Gal as well. Cell extracts were used to measure both luciferase and β-Gal activities. The results were plotted as percent of control after normalizing for β-Gal activity and protein concentration. Maximum inhibition of luciferase expression was 80% in the cells expressing IRNA compared to the control, but no differences were observed in the cells expressing mIRNA and pcDNA3.
To determine the effect of long-term expressing IRNA on cap-dependent translation, the cells were transfected with pcDNA-luc. As seen in Figure 3, no significant inhibition of cap-dependent translation from the pcDNA-luc construct was observed in cells expressing IRNA compared to the control cells.
Figure 3 The effect of long-term expressing IRNA on cap-independent and cap-dependent protein translation.
Hepatoma cells constitutionally expressing IRNA are refractory to infection by a PV tenuate virus
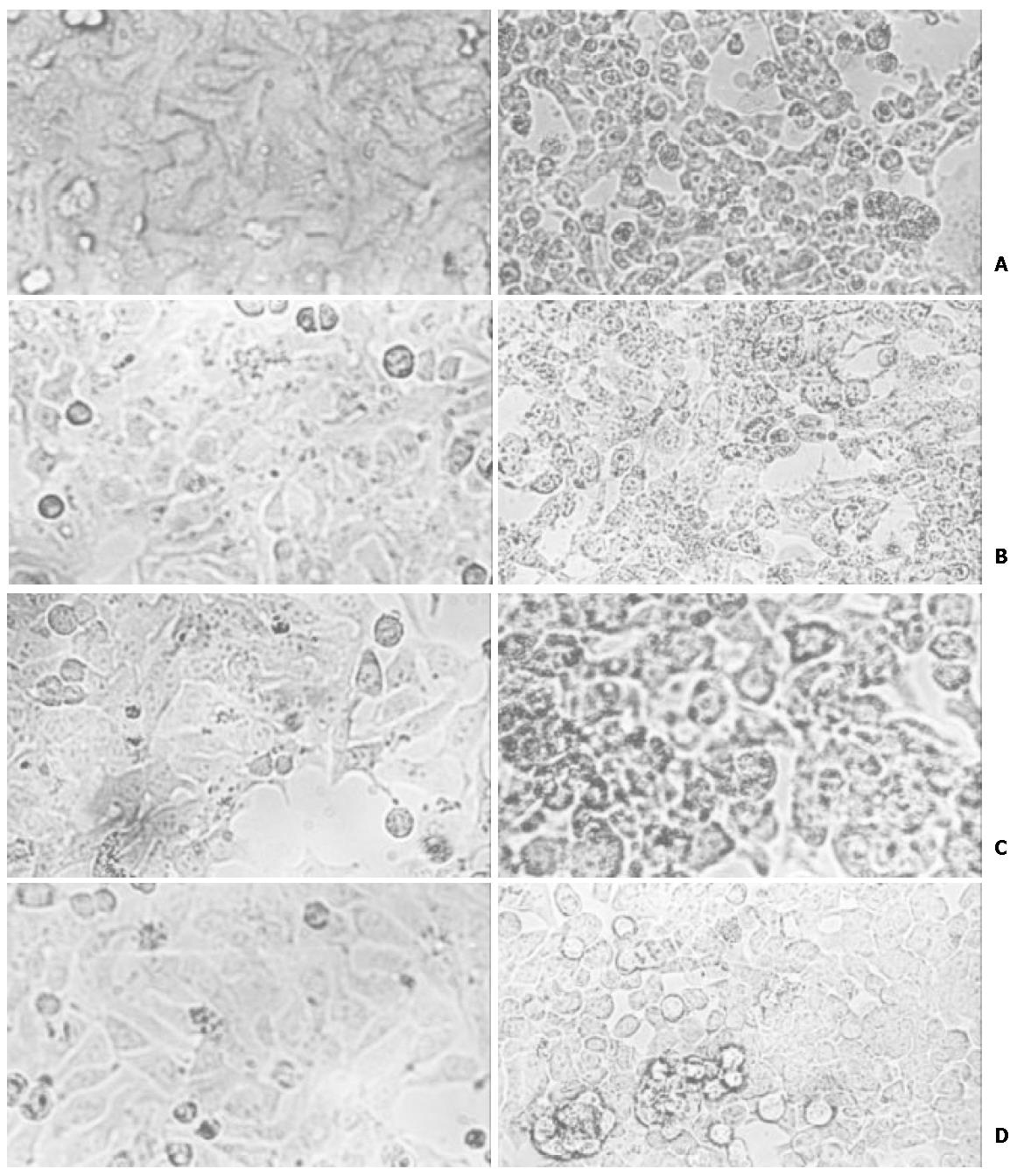
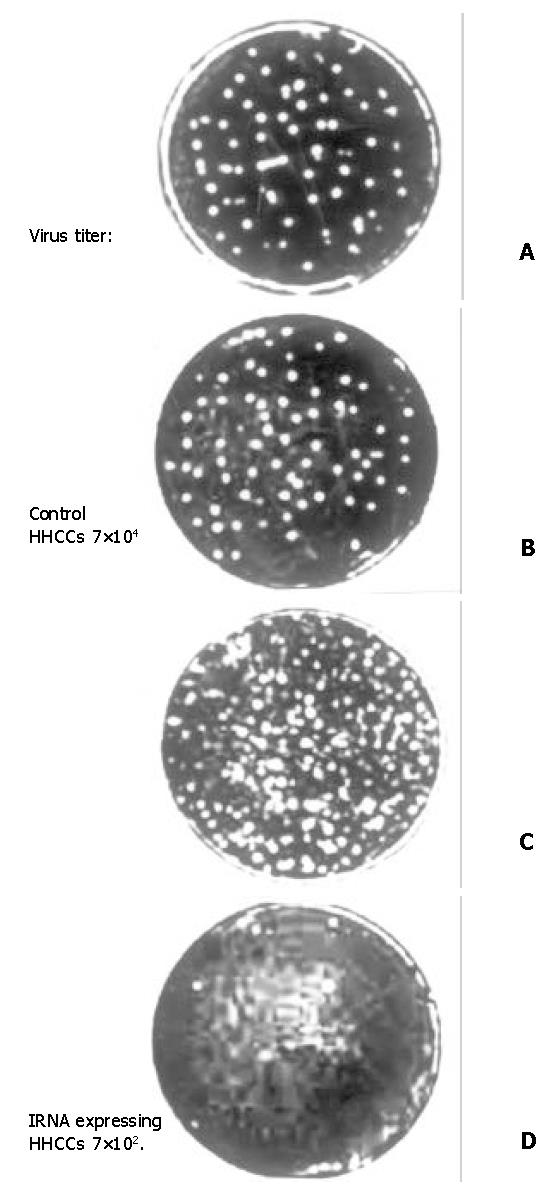
To determine the inhibit effect of IRNA on IRES mediated protein translation during virus infection, the pcRz-IRNA and pcRz-mIRNA cells were infected with a vaccinia PV. After infection with PV, the cells were stained and the cytopathic effects were observed and we found that the pcRz-mIRNA cells and control cells were appeared marked cytopathic effect compared to the cells prior to the infection, but the cells expressing IRNA were almost totally protected from the cytopathic effect of PV (Figure 4). Following the infection, cell extracts were prepared from infected cells which were then used to further infect Hep2 monolayer cells. Plaques characteristic of PV were apparent in Hep2 cells infected with cell extract from control hepatoma cells and pcRz-mIRNA cells (Figure 5A, Figure 5B). Evidently viral replication was drastically affected in the cels expressing IRNA (Figure 5D).
Figure 4 The cytopathic effect of different HHCC cells infected with PV.
A: control HHCC cells; B: mIRNA cells; C: pcDNA3 cells; D: IRNA cells.
Figure 5 Plaque characteristic of different HHCC infected of PV.
A: control HHCC; B: mIRNA cells; C: pcDNA3 cells; D: IRNA cells.
To rule out the possibility that the nonspecific small RNA expressing cells affected the viral replication, the pcDNA3 cells were infected under the same condition. As can be seen in the Figure 4C and Figure 5C, the PV replication was not affected in the pcDNA3 cells.
IRNA forms a gel-retarded complex that is inhibited by UTR
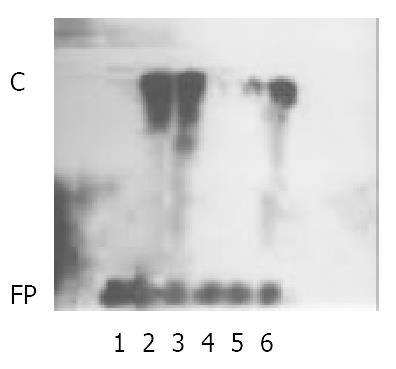
In order to study the mechanism of IRNA inhibitor activity on IRES mediated protein translation, 32P-body-labeled IRNA probe was prepared and mixed with RRL extract, and the resulting RNA-protein complexes were analyzed by nondenaturing polyacrylamide gel electrophoresis. As shown in Figure 6, lane 2, a single complexes (denoted C).
Figure 6 IRNA forms a gel-retarded complex which is inhibited by 5’ UTR of HCV RNA.
32P-labeled IRNA was incubated with RRL extract in the absence (lane 2) or presence of an unla-beled 25- or 50-fold molar excess of IRNA (lane3 and lane 4, respectively) or 5’ UTR (lane 5) or of a 50-fold molar excess of nonspecific RNA (lane 6). Protein-RNA complexes were analyzed on a nondenaturing gel. In lane 1, RRL extract was not added. FP: free probe; C: complexes.
DISCUSSION
IRES-dependent protein translation mechanism was first discovered in picornavirus, including PV, rhinovirus and hepatitis A virus, as well as certain flavivirus, such as hepatitis C virus[9-12]. Although there is very little sequence homology between these different IRES elements, structural similarity do appear to exist. In fact for keeping the activity of IRES, it is more important to maintain the secondary structure than to maintain the integrality of certain genome sequences[1-3,7,8]. IRES is the key structure for some virus RNA replication, so it has been the target for antiviralo infection[13-20]. We have constructed the self-cleavage plasmid of IRNA, and affirmed that IRNA can inhibit IRES-dependent protein translation both in vivo and in vitro[21]. In order to further confirm the long-term expressing IRNA effect on cellular protein and virus protein translation, we cloned the HHCCs expressing IRNA, and confirmed that long-term expressing IRNA could inhibit IRES mediated protein translation significantly compared to the control cells and mIRNA expressing cells. Das and his collaborates prepared the a number of human hepatoma (Huh-7) cell lines expressing IRNA by using the similar methods. They found that HCV IRES-mediated cap-independent translation was markedly inhibited in cells constitutively expressing IRNA compared to control hepatoma cells[22].
Additionally, We have shown here that the long-term expression of IRNA did not affect the overall translation of cellular protein as shown by laser confocal microscopy. This was surprising since some cellular mRNAs such as the immunoglobulin heavy-chain-binding protein, mouse androgen receptor, and Drosophilia antennapedia mRNAs has been shown to use IRES-mediated translation. It is possible that cellular mRNAs having IRES elements are also translated in a cap-dependent manner, as almost all mRNAs synthesized in vivo are capped. Das and his collaborates also confirmed that the constitutive expression of IRNA was not detrimental to cell growth[22].
Recent studies on picornavirus-IRES-mediated translation have demonstrated that cellular trans-acting proteins distinct from canonical translation initiation factors play an important role in IRES-mediated translation. These proteins bind to the IRES and presumably help to facilitate the binding of ribosomes to the IRES. Some of the trans-acting proteins have been identified as the La autoantigen, polypyrtimidine tract-binding (PTB) protein, glyceraldehydes 3-phosphate dehydrogenase (GAPDH). The La polypeptide binds to both PV and HCV IRES elements and stimulates IRES-mediated translation. Similarly, the PTB protein interacts with HCV and other picornavirus IRES sequences and stimulated viral 5’UTR mediated translation[23-25]. Because HCV IRES and PV IRES bind the similar cellular protein, and we lack effective HCV culture system in vitro, we studied the long-term expressing IRNA blocking viral replication ability by using PV infection in vitro. Our results demonstrated that long-term expressing IRNA blocks the PV replication effectively. Das and his collaborates contructed PV-HCV chimera in which PV IRES was replaced by the HCV IRES. They demonstrated that Huh-7 cells constitutively expressing IRNA became refractory to infection by the PV-HCV chimera, but the replication of a PV-encephalomyocarditis virus (ECMV) chimera containing the ECMV IRES element was not affected significantly in the IRNA-producing cell line[22].
At low mulitiplicities of infection of the virus, cells expressing IRNA showed significant resistance to virus infection compared to control cells. This could be due to inhibition of primary translation of the input viral RNA in cells expressing IRNA. Multiple rounds of replication and reinfection of control cells, but not of IRNA-expressing cells, resulting in an amplified effect seen in the plaque assay was shown in Figure 5. At higher mulitiplicities of infections, the virus titer was approximately 10-fold lower in IRNA cells compared to control cells. This could be due to one or more of the following reasons: (1) the level of IRNA expression was relatively low in the cell line; (2) the viral 5’ UTR, which competed with IRNA for binding of relevant protein factors, has significantly higher affinity for these proteins than IRNA; (3) the amount of IRNA in the cytoplasma was low compared to the total amount of expressed IRNA; and finally (4) all of the IRNA molecules in the cell line might not have been properly folded to assume the right secondary structure required for its translation-inhibitory activity.
Theoretically, IRNA could inhibit IRES-mediated translation by two possible mechanisms. It could bind to some sequences in the UTR as antisense RNA or it could bind protein factors necessary for the internal entry of ribosome, thus inhibiting IRES-dependent translation. The possibility that the IRNA acted by binding protein factors required for IRES-mediated translation was plausible for the following reasons. First, purified RNA was not complementary to 5’ UTR sequences nor did it hybridize with virus 5’ UTR. Secondly, inhibition of the translation of PV RNA could be overcome by the addition of increasing concentrations of HeLa extract but not of PV RNA or 5’ UTR sequence. In fact, in order to identify the mechanism of IRNA inhibitor activity, several laboratories by using the UV-crossing linking methods and Gel retardation methods studied the mechanism respectively. They found that IRNA specifically binds proteins which interact with RNA secondary structures within the virus 5’UTR presumably involved in internal initiation. Furthermore, they demonstrated that purified IRNA specifically competed with virus RNA structures within the 5’UTR which bind a cellular protein with an approximate molecular mass of 52 KD, 57 KD. Then they determined that p52 protein appears to be identical to human La autoantigen and the binding of the La autoantigen to the HCV IRES element was specifically and efficiently competed by IRNA. The data present here (Figure 6) also suggested that the IRNA inhibit viral IRES mediated protein translation through binding specific cellular factors[22,26-31]. Additionally, Das and his collaborates determined the secondary structure of IRNA and founded that both a stem and a loop formed by intramolecular folding of IRNA were important for inhibition of viral IRES mediated translation. Venkatesan had also predicted the secondary structure of the IRNA and its complementary RNA (cIRNA) using biosoftware MFOLD by energy minimal principle and further determined the structure using enzyme methods and chemical modification methods. They found that IRNA and cIRNA have not only same secondary structure but also similar inhibitor activity on PV IRES mediated protein translation[31]. Our laboratory have also modeled the secondary structure of IRNA and cIRNA and got the similar results to the Venkatesan (data reported in other file).
In summary, we could get the conclusion that (1) IRNA expressing cells could significantly anti polio virus infection; (2) IRNA long-term expression has no significant affect on cellular protein translation; (3) IRNA expressing cells significantly inhibit the HCV IRES mediated protein translation.