Published online Oct 7, 2020. doi: 10.3748/wjg.v26.i37.5617
Peer-review started: July 8, 2020
First decision: August 8, 2020
Revised: September 1, 2020
Accepted: September 18, 2020
Article in press: September 18, 2020
Published online: October 7, 2020
Although artificial intelligence (AI) was initially developed many years ago, it has experienced spectacular advances over the last 10 years for application in the field of medicine, and is now used for diagnostic, therapeutic and prognostic purposes in almost all fields. Its application in the area of hepatology is especially relevant for the study of hepatocellular carcinoma (HCC), as this is a very common tumor, with particular radiological characteristics that allow its diagnosis without the need for a histological study. However, the interpretation and analysis of the resulting images is not always easy, in addition to which the images vary during the course of the disease, and prognosis and treatment response can be conditioned by multiple factors. The vast amount of data available lend themselves to study and analysis by AI in its various branches, such as deep-learning (DL) and machine learning (ML), which play a fundamental role in decision-making as well as overcoming the constraints involved in human evaluation. ML is a form of AI based on automated learning from a set of previously provided data and training in algorithms to organize and recognize patterns. DL is a more extensive form of learning that attempts to simulate the working of the human brain, using a lot more data and more complex algorithms. This review specifies the type of AI used by the various authors. However, well-designed prospective studies are needed in order to avoid as far as possible any bias that may later affect the interpretability of the images and thereby limit the acceptance and application of these models in clinical practice. In addition, professionals now need to understand the true usefulness of these techniques, as well as their associated strengths and limitations.
Core Tip: The biological variability in the behavior of hepatocellular carcinoma (HCC), conditioned by multiple factors, makes it difficult to establish general standards of action applicable equally to all patients. Analysis of the vast amount of data now available and their relation with tumor behavior is fundamental to be able to establish an efficient approach to HCC. It is here that the computational power of artificial intelligence can play a determining role, though it is necessary to understand the strengths and limitations of this technology before it can be applied in clinical practice.
- Citation: Jiménez Pérez M, Grande RG. Application of artificial intelligence in the diagnosis and treatment of hepatocellular carcinoma: A review. World J Gastroenterol 2020; 26(37): 5617-5628
- URL: https://www.wjgnet.com/1007-9327/full/v26/i37/5617.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i37.5617
In the era of Big Data, the need for the efficient management of the great amount of information available has led to the development and application of artificial intelligence (AI) and its various techniques in the general field of medicine. Although the concept of AI arose in the 1950s[1], it was not until a few years ago that it really came to experience its breakthrough.
The term AI refers to those computer programs that try to reproduce human cognitive functions, like learning or problem solving. Initially, machine learning (ML), developed as a branch of AI, analyzed data in order to create algorithms that can detect patterns of behavior from which predictive models can be established. Different ML techniques, such as support vector machines (SVM), artificial neural networks (ANNs) or classification and regression trees, have all been used in multiple studies in the field of medicine[2]. Technological advances over the last ten years have resulted in the appearance of deep learning (DL) as a new model of ML to develop multi-layered neural network algorithms, using such techniques as convolutional neural network (CNN), a multilayer of ANN, that has proven of great use in the analysis of radiological images[3,4].
Although the application of AI in various fields of medicine has shown promising results, we should nevertheless be aware of its limitations. The retrospective manner of many of these studies and the use of not particularly suitable databases, with their inherent bias, can affect the accuracy of AI. Thus, it is necessary to draw up prospective, well-designed, multicenter studies that are free from bias which could affect their interpretability and thereby limit their acceptance and application in clinical practice. And of course, we must not forget such other aspects as cost-effectiveness, regulations by the health authorities and ethical considerations.
AI has been used in the field of hepatology for the diagnosis, treatment and prognostic prediction of various different disorders, though with special relevance in the study of hepatocellular carcinoma (HCC) as this is a very common tumor. Estimates by the American Cancer Society put the number of new cases of liver and intrahepatic bile duct cancer during 2020 to be 42810, with 30160 deaths[5]. HCC has particular radiological features that enable its diagnosis without the need for any histological study. Accordingly, the analysis of imaging tests takes on special relevance as their interpretation is not always easy, in addition to which they vary over the course of the disease, as do the prognosis and response to treatment, which are all affected by multiple factors. This all results in a vast amount of data whose integration and efficient analysis lend themselves to study by AI. Indeed, several studies have recently been undertaken to aid in the decision-making process and overcome the limitations of human evaluation[4,6].
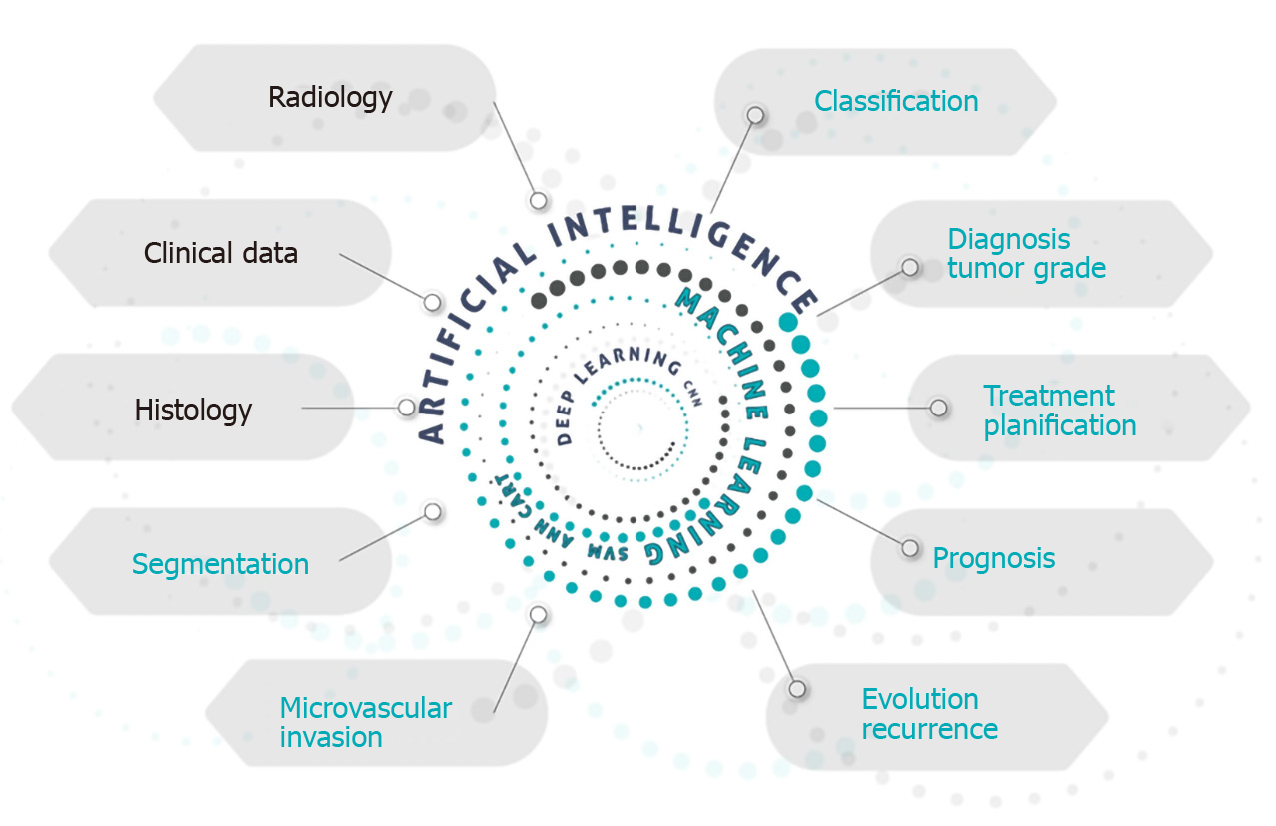
Figure 1 gives a schematic idea of how AI, with its variants, could be used to study a patient with HCC, whether diagnosed or suspected. AI is able to perform a combined analysis of radiological, clinical and histological data, producing information that can aid in the diagnostic accuracy, tumor staging, treatment planning using methods of segmentation and evaluation of the presence of microvascular invasion, in addition to giving a prognostic estimate.
In the field of liver cancer, the use of AI techniques to aid traditional diagnostic techniques is promising. CNN is a multilayer ANN interconnected in such a way that all input data traverse all the various layers during which the information is processed to produce output data. It can be considered an advanced form of DL with its own learning capacity. CNN can increase the diagnostic yield of ultrasound studies, abdominal computerized tomography or abdominal magnetic resonance imaging (MRI), positron emission tomography (PET) and histology.
HCC usually, though not always, develops in a cirrhotic liver. Accordingly, clinical practice guidelines recommend regular abdominal ultrasound in patients with hepatic cirrhosis; indeed, it is considered the method of choice for screening of space-occupying lesions. Ultrasound is therefore the main tool to evaluate liver disease and detect new lesions. However, image interpretation is not always easy and may present interobserver variability.
To assess the underlying disease, Bharti et al[7] proposed an ANN model to differentiate four stages of liver disease using data obtained from ultrasound images: normal liver, chronic liver disease, cirrhosis, and HCC. The classification accuracy of the model was 96.6%[7]. Liu et al[8] designed an algorithm to classify ultrasound images. They selected the liver capsule to determine the presence of cirrhosis, even in early stages when the usual findings reported by the radiologist, such as a nodular liver outline, enlarged porta or splenomegaly, are still not obvious. Using their analysis of the morphology of the liver capsule, they were able to determine the presence or absence of cirrhosis, with an area under the curve of 0.968[8].
The human yield when characterizing a liver lesion from ultrasound images is limited. Schmauch et al[9] designed a DL system able to detect and classify space-occupying lesions in the liver as benign or malignant. After a supervised training using a database of 367 images together with the radiological reports, the resulting algorithm detected and characterized the lesions with a mean receiver operating characteristic of 0.93 and 0.916, respectively. Although the system requires validation, it could increase the diagnostic yield of ultrasound and warn of possibly malignant lesions[9].
AI has also been used in contrast-enhanced ultrasound (C-US), improving its ability to identify characteristics suggestive of cancer. Guo et al[10] demonstrated that DL applied to the behavior of liver lesions seen on C-US in three phases (arterial, portal and late) increased the accuracy, sensitivity and specificity of the study[10].
When a follow-up ultrasound shows a new liver lesion, other imaging studies are undertaken, mainly dynamic contrast-enhanced computed tomography (CT) or MRI, to obtain a more precise evaluation. The radiological behavior of liver lesions in dynamic CT or MRI studies is useful for characterization of the lesion. If a liver lesion > 1 cm fulfils certain radiological criteria, almost pathognomonic for HCC such as the presence of hyperenhancement at arterial phase and washout at portal or late phases in a cirrhotic patient, no further studies are needed for its diagnosis or histological confirmation. However, liver nodules often present an indeterminate behavior on CT and a biopsy of the lesion is required, as recommended in the European Association for the Study of the Liver guidelines[11]. This, though, requires assuming the risks involved in the procedure, or close follow-up, as indicated in the American Association for the Study of Liver Diseases guidelines[12], with a high number of studies and the possibility of not diagnosing a malignant lesion in time. Mokrane et al[13] undertook a retrospective analysis of 178 patients with cirrhosis and liver nodules in whom the Liver Reporting and Data System criteria were unable to distinguish the neoplastic from the non-neoplastic lesions, thereby necessitating a biopsy; 77% proved to be malignant on biopsy. Using DL techniques to classify nodules as HCC or non-HCC achieved an area under the curve (AUC) of 0.70. Another retrospective study, by Yasaka et al[14], analyzed the yield of an ANN, composed of three layers, classifying liver masses using contrast-enhanced CT into five categories: A, classic HCC; B, malignant tumors apart from HCC (cholangiocarcinoma, hepatocholangiocarcinoma or metastasis); C, indeterminate masses, dysplastic nodules or early HCC and benign masses other than cysts or hemangiomas; D, hemangiomas; E, cysts. After supervised training using over 55000 image sets the authors obtained a high accuracy for the classification of liver lesions, especially for the differentiation between categories A-B and C-D.
Quantifying the tumor load may be useful, particularly for the detection of tumor recurrence in follow-up CT studies. As tumor relapses can be small and go unnoticed, Vivanti et al[15] described an automated detection method of recurrence, based on the initial appearance of the tumor, its CT behavior, and the quantification of the tumor load at baseline and during the follow-up. The technique had a high rate of true positives in the identification of tumor recurrence, with an accuracy of 86%.
Liver segmentation is of great importance to assess liver lesions and for planning the ideal treatment. However, manual segmentation is made more difficult by the heterogeneity of the lesions or their diffuse borders. Li et al[16] proposed a CNN that can segment liver tumors based on CT images, with an accuracy of 82.67% ± 1.43%, better than that of traditional techniques, thereby favoring suitable treatment planning.
CNN applied to MRI has also been analyzed. Hamm et at[17] developed and validated a DL system based on a CNN that classifies MRI liver lesions, with an accuracy of 92%, a sensitivity of 92% and a specificity of 98%; and an average computation time of 5.6 ms.
Other studies have associated additional MRI sequences and risk factors plus the patient’s clinical data to apply an automated classification system cataloguing liver lesions as adenoma, cyst, hemangioma, HCC and metastasis, with a sensitivity and specificity of 0.8/0.78, 0.93/0.93, 0.84/0.82, 0.73/0.56 and 0.62/0.77, respectively[18]. Zhang et al[19] described a training model using MRI in 20 patients to classify liver tissues. Their results were promising, improving the yield of the reference models used.
Preis et al[20] evaluated the yield of fluorine 18 fluorodeosyglucose positron emission tomography/computed tomography (18F-FDG PET/CT) using a neural network to analyze liver uptake of 18F together with patient and laboratory data. They achieved a high sensitivity and specificity to detect liver malignancy unidentified visually, showing that this technique could complement the radiologist in the interpretation of PET, though their main aim was to evaluate metastatic liver disease, where 18F-FDG PET/CT has greater applicability.
The histopathological classification of a liver lesion and the differentiation of the tumor strain is crucial for treatment planning and a prognostic estimate of the disease, though this can sometimes prove challenging even for expert pathologists. Kiani et al[21] used AI as a support for the pathologists, focusing on the histological differentiation between HCC and cholangiocarcinoma. They analyzed prospectively the impact of this aid in the diagnostic yield of 11 pathologists, finding that it made no change to their mean accuracy.
Others have described how a deep CNN using previous histopathological images of HCC can make an automated diagnosis of HCC and distinguish healthy tissue from tumor tissue, in addition to identifying certain biological predictors[22]. Table 1 shows the main studies using AI techniques in the diagnosis of HCC.
| Ref. | Title | Aim of the study of the use of AI in imaging techniques | Diagnostic technique studied | AI tool used | University/department |
| Bharti et al[7] 2018 | Preliminary study of chronic liver classification on ultrasound images using an ensemble model | Classification of liver disease in four stages; normal liver, chronic liver disease, cirrhosis and HCC | Ultrasound | CNN | Thapar Institute of Engineering & Technology, Patiala, India |
| Liu et al[8] 2017 | Accurate prediction of responses to transarterial chemoembolization for patients with hepatocellular carcinoma by using artificial intelligence in contrast-enhanced ultrasound | Early identification of the presence of cirrhosis | Ultrasound | ML | Sun Yat-sen University, Guangzhou, China |
| Schmauch et al[9] 2019 | Diagnosis of focal liver lesions from ultrasound using deep learning | Classify liver lesions as benign or malignant | Ultrasound | DL | Owkin Inc, Research and Development Laboratory, Paris, France |
| Guo et al[10] 2018 | A two-stage multi-view learning framework based computer-aided diagnosis of liver tumors with contrast enhanced ultrasound images | Characterize liver lesions and identify data of malignancy | C-US | ML | University School of Medicine, Shanghai, China |
| Mokrane et al[13] 2020 | Radiomics machine-learning signature for diagnosis of hepatocellular carcinoma in cirrhotic patients with indeterminate liver nodules | Identify malignancy in hepatic space-occupying lesions catalogued as indeterminate | CT | Radiomics | Department of Radiology, New York Presbyterian Hospital, Columbia University Vagelos College of Physicians and Surgeons, New York City, NY, United States |
| Yasaka et al[14] 2018 | Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast-enhanced CT: A preliminary study | Classification of liver lesions in five categories | CT | CNN | Department of Radiology, The University of Tokyo Hospital, Tokyo, Japan |
| Vivanti et al[15] 2017 | Automatic detection of new tumors and tumor burden evaluation in longitudinal liver CT scan studies | Detection of tumor recurrence analyzing volume/tumor load | CT | CNN | The Rachel and Selim Benin School of Computer Science and Engineering, The Hebrew University of Jerusalem, Jerusalem, Israel |
| Li et al[16] 2015 | Automatic segmentation of liver tumor in CT Images with deep convolutional neural networks | Liver tumor segmentation | CT | CNN | Research Lab for Medical Imaging and Digital Surgery, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China |
| Hamm et al[17] 2019 | Deep learning for liver tumor diagnosis part I: development of a convolutional neural network classifier for multi-phasic MRI | Classification of liver lesions | MRI | DL | Department of Radiology and Biomedical Imaging, Yale School of Medicine, United States |
| Jansen et al[18] 2019 | Automatic classification of focal liver lesions based on MRI and risk factors | Classification of liver lesions in: Adenomas, cysts, hemangiomas, HCC and metastasis | MRI | ML | Image Sciences Institute, University Medical Center Utrecht & Utrecht University, Utrecht, the Netherlands |
| Zhang et al[19] 2018 | Liver tissue classification using an auto-context-based deep neural network with a multi-phase training framework | Classification of liver tissue | MRI | CNN | Department of Biomedical Engineering, Yale University, New Haven, CT, United States |
| Preis et al[20] 2011 | Neural network evaluation of pet scans of the liver: A potentially useful adjunct in clinical interpretation | Identify metastatic liver disease | PET | CNN | Department of Radiology, Massachusetts General Hospital and Harvard Medical School, Boston |
| Kiani et al[21] 2020 | Impact of a deep learning assistant on the histopathologic classification of liver cancer | Differentiate HCC from cholangiocarcinoma | Histology | DL | Department of Computer Science, Stanford University, Stanford, CA, United States |
| Liao et al[22] 2020 | Deep learning-based classification and mutation prediction from histopathological images of hepatocellular carcinoma | Automated identification of liver tumor tissue, differentiating it from healthy tissue | Histology | DL | Department of Liver Surgery & Liver Transplantation, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University and Collaborative Innovation Center of Biotherapy, Chengdu, China |
The individual biological variability between patients in the behavior of HCC hinders evidence-based clinical assessment that is applicable to all patients. Accordingly, powerful, standardized risk stratification systems are needed in order to optimize treatment strategies and assess their effects. It is here that AI can play an important role in the therapeutic approach to HCC. Most studies on the use of AI in the treatment of HCC are aimed at the analysis of certain particular tumor characteristics, such as radiological, histological or genetic features, or a combination of the clinical data in order to predict the response to a particular treatment. This, in turn, will allow for the suitable selection of patients for particular treatment options.
In usual clinical practice the diagnosis and treatment assessment of HCC are commonly done with such imaging techniques as C-US, CT and MRI, after analyzing certain tumor features like vascularization or behavior after the administration of contrast material[23]. However, these characteristics are liable to subjectivity in their interpretation by the radiologist, in addition to the lack of high resolution dimensional images. But a new technology has recently appeared in the area of radiology and cancer, radiomics[24]. Although it is not yet in extensive use in clinical practice, it has nevertheless awoken great interest. This technology allows for the extraction of a great amount of quantifiable objective data contained in the radiological images and their later association with the underlying biological processes. Analysis of all these data with AI software can provide useful diagnostic and prognostic information with predictive accuracy[24,25].
Early tumor recurrence after surgical resection is associated with a poor prognosis. The preoperative identification of patients at high risk of recurrence is fundamental to avoid unnecessary treatment. Computer models have been developed that analyze certain tumor characteristics and aid in the preoperative prediction of the risk of recurrence or the evaluation of survival after resection.
Vascular microinvasion (VMI) has been established as an independent predictive factor of recurrence, associated with poor results after tumor resection[26]. Although the preoperative availability of information about VMI would be of much benefit, radiological techniques currently in clinical use do not provide for an adequate direct diagnosis.
Several studies have managed to elaborate radiomic signatures that enable prediction of the preoperative status of VMI, based on contrast-enhanced CT[27,28] or MRI[29]. However, these techniques involve radiological exposure, and are laborious to perform and costly. Recently, Dong et al[30] published a study using radiomic algorithms based on grayscale ultrasound images to elaborate radiomic signatures with the potential to aid in the prediction of VMI, with promising results. Ji et al[31] created predictive models for recurrence after surgical resection using radiomic techniques to analyze contrast-enhanced CT images, with a C-index of 0.633-0.699. In conjunction with the inclusion of clinical data, the model can be used to establish a personalized risk stratification facilitating the individual management of HCC.
Survival after surgical resection has also been assessed in several studies using ML techniques[32-34], and more recently with more advanced DL models based on digitalized histological images of the tumor. Saillard et al[35] drew up a predictive model of survival after resection, attaining a C-index for survival prediction of 0.78. A recent prospective study by Schoenberg et al[36] involving 180 patients also led to a predictive model based on the analysis of 26 preoperative routine clinical variables, obtaining a predictive value of 0.78.
Transcatheter arterial chemoembolization (TACE) is the treatment of choice for intermediate stage B HCC, in the Barcelona Clinical Liver Cancer (BCLC) classification[37]. Adequate selection of patients who might benefit from this treatment is vital in order to avoid unnecessary examinations that can sometimes have undesirable secondary effects for the patient and waste costs for the health system. Studies have been developed based on AI techniques to attempt to predict the response to treatment with TACE and aid adequate patient selection. Most of these studies are based on imaging analysis, though some have used genomic signatures. Morshid et al[38] elaborated a fully automated ML algorithm using the combination of quantitative characteristics of CT images plus pretreatment patient clinical data to predict the response to TACE. They achieved a prediction accuracy rate of 74.2% using a combination of the BCLC stage plus quantitative image features vs using just the BCLC stage alone. Peng et al[39] validated a DL model to predict the response to TACE using CT images from a total of 789 patients in three different hospitals. They obtained an accuracy of 84% and an AUC of 0.97 to predict complete response. Liu et al[40] constructed and validated a DL model (DL radiomics-based C-US model) but based on the quantitative analysis of C-US cine recordings. It was highly reproducible and had an AUC of 0.93 (95%CI: 0.80-0.98) to predict the response to TACE.
Other studies have used ML techniques combining MRI with clinical data to predict the response to TACE. Abajian et al[41] studied 36 patients who underwent MRI before TACE. They developed a predictive model of response with an accuracy of 78%, sensitivity of 62.5% and specificity of 82%.
The efficacy of TACE has also been examined by survival analysis of the patients after its application. Mähringer-Kunz et al[42] developed a prediction model of survival after TACE by constructing an ANN, using all the parameters of the main conventional prediction scores (ART[43], ABCR[44] and SNACOR[45]). They predicted a one-year survival with an AUC of 0.77, sensitivity of 78% and specificity of 81%, better results compared to those of the conventional scores mentioned.
Although most studies evaluating the use of AI to assess TACE have used radiomics, some have also evaluated the prediction of response to TACE using genetic analysis. Ziv et al[46] studied genetic mutations using SVM techniques to predict the tumor response after TACE, though it was a retrospective study with a low number of cases.
Radiofrequency ablation (RFA) as a therapy aimed at curing HCC in early stages[37] has also been evaluated. Liang et al[47] drew up a predictive model of HCC recurrence based on SVM. They studied 83 patients with HCC who underwent RFA, obtaining an AUC of 0.69, sensitivity of 67% and specificity of 86%, with which they were able to identify patients at a high risk of recurrence. Table 2 summarizes the studies using AI techniques in the treatment of HCC.
| Ref. | Tittle | n | Study type | Study aim | AI tool used | Outcome | University/department |
| Dong et al[30] 2020 | Preoperative prediction of microvascular invasion in hepatocellular carcinoma: initial application of a radiomic algorithm based on grayscale ultrasound images | 322 | Retrospective | Prediction of VMI using C-US | Radiomics | AUC: 0.73; Sen: 0.919; Spe: 0.359 | Department of Ultrasound, Zhongshan Hospital, Fudan University, Shanghai, China |
| Xu et al[28] 2019 | Radiomic analysis of contrast-enhanced CT predicts microvascular invasion and outcome in hepatocellular carcinoma | 495 | Retrospective | Prediction of VMI using C-CT | Radiomics | AUC: 0.90 | Department of Radiology, The First Affiliated Hospital with Nanjing Medical University, Nanjing, Jiangsu Province, China |
| Ma et al[27] 2019 | Preoperative radiomics nomogram for microvascular invasion prediction in hepatocellular carcinoma using contrast-enhanced CT | 157 | Retrospective | Prediction of VMI using C-CT | Radiomics | AUC: 0.73 | Department of Diagnostic Radiology, National Cancer Center/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, China |
| Zhou et al[29] 2017 | Malignancy characterization of hepatocellular carcinomas based on texture analysis of contrast-enhanced MR images | 46 | Retrospective | Prediction of VMI using C-MRI | AUC: 0.918; Sen: 92%; Spe: 66% | Laboratory for Health Informatics, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China | |
| Ziv et al[46] 2017 | Gene signature associated with upregulation of the Wnt/β-Catenin signaling pathway predicts tumor response to transarterial embolization | 17 | Retrospective | Prediction of response to TACE using signature gene | Prediction accuracy: 70% | Interventional Radiology Service, Department of Radiology, Memorial Sloan Kettering Cancer Center, New York, NY, United States | |
| Morshid et al[38] 2019 | A machine learning model to predict hepatocellular carcinoma response to transcatheter arterial chemoembolization | 105 | Retrospective | Prediction of response to TACE using CT | ML | Acc: 74% | Departments of Imaging Physics, Diagnostic Radiology, Gastrointestinal Oncology and Interventional Radiology, The University of Texas, MD Anderson Cancer Center, Houston |
| Liu et al[40] 2020 | Accurate prediction of responses to transarterial chemoembolization for patients with hepatocellular carcinoma by using artificial intelligence in contrast-enhanced ultrasound | 130 | Retrospective | Prediction of response to TACE using C-US | DL | AUC: 0.93 | Department of Medical Ultrasonics, Institute of Diagnostic and Interventional Ultrasound, The First Affiliated Hospital of Sun Yat-sen University, China |
| Peng et al[39] 2020 | Residual convolutional neural network for predicting response of transarterial chemoembolization in hepatocellular carcinoma from CT imaging | 789 | Retrospective | Prediction of response to TACE using CT | CNN | AUC: 0.97; Acc: 84.3% | Hepatology Unit and Department of Infectious Diseases, Nanfang Hospital, Southern Medical University, China |
| Abajian et al[41] 2018 | Predicting treatment response to intra-arterial therapies for hepatocellular carcinoma with the use of supervised machine learning-an artificial intelligence concept | 36 | Retrospective | Prediction of response to TACE using MRI | ML | Acc: 78%; Sen: 62%; Spe: 82% | Yale School of Medicine, Department of Radiology and Biomedical Imaging, United States |
| Mähringer-Kunz et al[42] 2020 | Predicting survival after transarterial chemoembolization for hepatocellular carcinoma using a neural network: A pilot study | 282 | Retrospective | Prediction of survival after TACE | CNN | Acc: 0.77; Sen: 78%; Spe: 81% | Department of Diagnostic and Interventional Radiology, University Medical Center of the Johannes Gutenberg-University Mainz, Mainz, Germany |
| Saillard et al[35] 2020 | Predicting survival after hepatocellular carcinoma resection using deep-learning on histological slides | 194 | Retrospective | Prediction of survival after surgical resection | DL | C-index: 0.78 | Owkin Lab, Owkin |
| Liang et al[47] 2014 | Recurrence predictive models for patients with hepatocellular carcinoma after radiofrequency ablation using support vector machines with feature selection methods | 83 | Prospective | Prediction of recurrence after RFA | ML | AUC: 67%; Sen: 86%; Spe: 82% | Department of Internal Medicine, National Taiwan University Hospital and National Taiwan University College of Medicine, Taipei, Taiwan |
| Ji et al[31] 2019 | Machine-learning analysis of contrast-enhanced CT radiomics predicts recurrence of hepatocellular carcinoma after resection: A multi-institutional study | 470 | Retrospective | Prediction of recurrence after resection | ML | C-index: 0.633-0.699 | Hepatobiliary Center, The First Affiliated Hospital of Nanjing Medical University, Nanjing, PR China |
The prediction of overall survival of HCC, apart from the application of any therapy, has also been assessed using AI techniques. Current evidence on the relation of abnormalities in DNA methylation and HCC[48-50] was the basis for the study by Dong et al[51]. These authors used ML techniques (SVM) to analyze DNA methylation data from 377 HCC samples, and constructed three risk categories to predict overall survival, with a mean 10-fold cross-validation score of 0.95.
Larger studies are needed comparing the yield of medical professionals with the support of AI vs other professionals without such support in order to demonstrate its benefit as an aid in medicine. In particular, to assess liver masses and study HCC these trials should focus on aspects related to treatment and prognosis, such as the characterization of hepatic lesions catalogued as indeterminate, the presence of vascular invasion and the response to percutaneous therapy. Another important aspect is the use of AI in the analysis of the behavior of HCC in cirrhotic and non-cirrhotic patients, as well as the differentiation of primary and metastatic liver lesions[52] and especially the differential diagnosis with cholangiocarcinoma that can be complicated with currently available techniques and whose management and prognosis are completely different from those of HCC. At the same time, it is also necessary to start training health-care professionals to be prepared for the future incorporation of AI in daily practice in the field of liver cancer.
The incorporation of AI technologies in medicine has represented one of the most relevant advances in recent years. It will doubtless experience a progressively increasing rise, due to its usefulness in the processing and analysis of the enormous amount of data currently available. Nevertheless, we should be aware that certain constraints still exist that can limit its acceptance and applicability in clinical practice. Health care professionals must learn the true usefulness of AI and accept the need for its coexistence with the indispensable need for human evaluation, accepting that AI is here to support human intelligence, never to replace it. Despite the great progress represented by AI, it is nevertheless vital to guarantee that medical protocols remain rigorously transparent.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Spain
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Azer S, Du Z, Gao YT S-Editor: Yan JP L-Editor: A P-Editor: Ma YJ
| 1. | Turing AM. I.—Computing Machinery and Intelligence. Mind. 1950;LIX:433-460. [DOI] [Cited in This Article: ] |
| 2. | Kaul V, Enslin S, Gross SA. History of artificial intelligence in medicine. Gastrointest Endosc. 2020;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 125] [Cited by in F6Publishing: 165] [Article Influence: 41.3] [Reference Citation Analysis (1)] |
| 3. | Yang YJ, Bang CS. Application of artificial intelligence in gastroenterology. World J Gastroenterol. 2019;25:1666-1683. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 166] [Cited by in F6Publishing: 135] [Article Influence: 27.0] [Reference Citation Analysis (4)] |
| 4. | Le Berre C, Sandborn WJ, Aridhi S, Devignes MD, Fournier L, Smaïl-Tabbone M, Danese S, Peyrin-Biroulet L. Application of Artificial Intelligence to Gastroenterology and Hepatology. Gastroenterology. 2020;158:76-94.e2. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 230] [Cited by in F6Publishing: 259] [Article Influence: 64.8] [Reference Citation Analysis (0)] |
| 5. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7-30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12667] [Cited by in F6Publishing: 13879] [Article Influence: 3469.8] [Reference Citation Analysis (4)] |
| 6. | Briceño J. Artificial intelligence and organ transplantation: challenges and expectations. Curr Opin Organ Transplant. 2020;25:393-398. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 7. | Bharti P, Mittal D, Ananthasivan R. Preliminary Study of Chronic Liver Classification on Ultrasound Images Using an Ensemble Model. Ultrason Imaging. 2018;40:357-379. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 8. | Liu X, Song JL, Wang SH, Zhao JW, Chen YQ. Learning to Diagnose Cirrhosis with Liver Capsule Guided Ultrasound Image Classification. Sensors (Basel). 2017;17:149. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 9. | Schmauch B, Herent P, Jehanno P, Dehaene O, Saillard C, Aubé C, Luciani A, Lassau N, Jégou S. Diagnosis of focal liver lesions from ultrasound using deep learning. Diagn Interv Imaging. 2019;100:227-233. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 67] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 10. | Guo LH, Wang D, Qian YY, Zheng X, Zhao CK, Li XL, Bo XW, Yue WW, Zhang Q, Shi J, Xu HX. A two-stage multi-view learning framework based computer-aided diagnosis of liver tumors with contrast enhanced ultrasound images. Clin Hemorheol Microcirc. 2018;69:343-354. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 11. | European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J Hepatol. 2018;69:182-236. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3934] [Cited by in F6Publishing: 4903] [Article Influence: 817.2] [Reference Citation Analysis (0)] |
| 12. | Heimbach JK, Kulik LM, Finn RS, Sirlin CB, Abecassis MM, Roberts LR, Zhu AX, Murad MH, Marrero JA. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2018;67:358-380. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2107] [Cited by in F6Publishing: 2578] [Article Influence: 429.7] [Reference Citation Analysis (2)] |
| 13. | Mokrane FZ, Lu L, Vavasseur A, Otal P, Peron JM, Luk L, Yang H, Ammari S, Saenger Y, Rousseau H, Zhao B, Schwartz LH, Dercle L. Radiomics machine-learning signature for diagnosis of hepatocellular carcinoma in cirrhotic patients with indeterminate liver nodules. Eur Radiol. 2020;30:558-570. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 90] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 14. | Yasaka K, Akai H, Abe O, Kiryu S. Deep Learning with Convolutional Neural Network for Differentiation of Liver Masses at Dynamic Contrast-enhanced CT: A Preliminary Study. Radiology. 2018;286:887-896. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 293] [Cited by in F6Publishing: 318] [Article Influence: 45.4] [Reference Citation Analysis (0)] |
| 15. | Vivanti R, Szeskin A, Lev-Cohain N, Sosna J, Joskowicz L. Automatic detection of new tumors and tumor burden evaluation in longitudinal liver CT scan studies. Int J Comput Assist Radiol Surg. 2017;12:1945-1957. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 45] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 16. | Li W, Jia F, Hu Q. Automatic Segmentation of Liver Tumor in CT Images with Deep Convolutional Neural Networks. J Comput Commun. 2015;3:146-151. [DOI] [Cited in This Article: ] [Cited by in Crossref: 151] [Cited by in F6Publishing: 155] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 17. | Hamm CA, Wang CJ, Savic LJ, Ferrante M, Schobert I, Schlachter T, Lin M, Duncan JS, Weinreb JC, Chapiro J, Letzen B. Deep learning for liver tumor diagnosis part I: development of a convolutional neural network classifier for multi-phasic MRI. Eur Radiol. 2019;29:3338-3347. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 152] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 18. | Jansen MJA, Kuijf HJ, Veldhuis WB, Wessels FJ, Viergever MA, Pluim JPW. Automatic classification of focal liver lesions based on MRI and risk factors. PLoS One. 2019;14:e0217053. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 19. | Zhang F, Yang J, Nezami N, Laage-Gaupp F, Chapiro J, De Lin M, Duncan J. Liver Tissue Classification Using an Auto-context-based Deep Neural Network with a Multi-phase Training Framework. Patch Based Tech Med Imaging (2018). 2018;11075:59-66. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 20. | Preis O, Blake MA, Scott JA. Neural network evaluation of PET scans of the liver: a potentially useful adjunct in clinical interpretation. Radiology. 2011;258:714-721. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 21. | Kiani A, Uyumazturk B, Rajpurkar P, Wang A, Gao R, Jones E, Yu Y, Langlotz CP, Ball RL, Montine TJ, Martin BA, Berry GJ, Ozawa MG, Hazard FK, Brown RA, Chen SB, Wood M, Allard LS, Ylagan L, Ng AY, Shen J. Impact of a deep learning assistant on the histopathologic classification of liver cancer. NPJ Digit Med. 2020;3:23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 136] [Cited by in F6Publishing: 108] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 22. | Liao H, Xiong T, Peng J, Xu L, Liao M, Zhang Z, Wu Z, Yuan K, Zeng Y. Classification and Prognosis Prediction from Histopathological Images of Hepatocellular Carcinoma by a Fully Automated Pipeline Based on Machine Learning. Ann Surg Oncol. 2020;27:2359-2369. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 23. | Villanueva A. Hepatocellular Carcinoma. N Engl J Med. 2019;380:1450-1462. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2066] [Cited by in F6Publishing: 2565] [Article Influence: 513.0] [Reference Citation Analysis (0)] |
| 24. | Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2016;278:563-577. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4541] [Cited by in F6Publishing: 4480] [Article Influence: 560.0] [Reference Citation Analysis (2)] |
| 25. | Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, Sanduleanu S, Larue RTHM, Even AJG, Jochems A, van Wijk Y, Woodruff H, van Soest J, Lustberg T, Roelofs E, van Elmpt W, Dekker A, Mottaghy FM, Wildberger JE, Walsh S. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749-762. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1825] [Cited by in F6Publishing: 2716] [Article Influence: 388.0] [Reference Citation Analysis (0)] |
| 26. | Erstad DJ, Tanabe KK. Prognostic and Therapeutic Implications of Microvascular Invasion in Hepatocellular Carcinoma. Ann Surg Oncol. 2019;26:1474-1493. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 139] [Cited by in F6Publishing: 230] [Article Influence: 46.0] [Reference Citation Analysis (0)] |
| 27. | Ma X, Wei J, Gu D, Zhu Y, Feng B, Liang M, Wang S, Zhao X, Tian J. Preoperative radiomics nomogram for microvascular invasion prediction in hepatocellular carcinoma using contrast-enhanced CT. Eur Radiol. 2019;29:3595-3605. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 96] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 28. | Xu X, Zhang HL, Liu QP, Sun SW, Zhang J, Zhu FP, Yang G, Yan X, Zhang YD, Liu XS. Radiomic analysis of contrast-enhanced CT predicts microvascular invasion and outcome in hepatocellular carcinoma. J Hepatol. 2019;70:1133-1144. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 278] [Cited by in F6Publishing: 388] [Article Influence: 77.6] [Reference Citation Analysis (0)] |
| 29. | Zhou W, Zhang L, Wang K, Chen S, Wang G, Liu Z, Liang C. Malignancy characterization of hepatocellular carcinomas based on texture analysis of contrast-enhanced MR images. J Magn Reson Imaging. 2017;45:1476-1484. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 80] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 30. | Dong Y, Zhou L, Xia W, Zhao XY, Zhang Q, Jian JM, Gao X, Wang WP. Preoperative Prediction of Microvascular Invasion in Hepatocellular Carcinoma: Initial Application of a Radiomic Algorithm Based on Grayscale Ultrasound Images. Front Oncol. 2020;10:353. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 31. | Ji GW, Zhu FP, Xu Q, Wang K, Wu MY, Tang WW, Li XC, Wang XH. Machine-learning analysis of contrast-enhanced CT radiomics predicts recurrence of hepatocellular carcinoma after resection: A multi-institutional study. EBioMedicine. 2019;50:156-165. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 107] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 32. | Ho WH, Lee KT, Chen HY, Ho TW, Chiu HC. Disease-free survival after hepatic resection in hepatocellular carcinoma patients: a prediction approach using artificial neural network. PLoS One. 2012;7:e29179. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 33. | Chiu HC, Ho TW, Lee KT, Chen HY, Ho WH. Mortality predicted accuracy for hepatocellular carcinoma patients with hepatic resection using artificial neural network. ScientificWorldJournal. 2013;2013:201976. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 34. | Shi HY, Lee KT, Lee HH, Ho WH, Sun DP, Wang JJ, Chiu CC. Comparison of artificial neural network and logistic regression models for predicting in-hospital mortality after primary liver cancer surgery. PLoS One. 2012;7:e35781. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 53] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 35. | Saillard C, Schmauch B, Laifa O, Moarii M, Toldo S, Zaslavskiy M, Pronier E, Laurent A, Amaddeo G, Regnault H, Sommacale D, Ziol M, Pawlotsky JM, Mulé S, Luciani A, Wainrib G, Clozel T, Courtiol P, Calderaro J. Predicting survival after hepatocellular carcinoma resection using deep-learning on histological slides. Hepatology. 2020;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 95] [Cited by in F6Publishing: 120] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 36. | Schoenberg MB, Bucher JN, Koch D, Börner N, Hesse S, De Toni EN, Seidensticker M, Angele MK, Klein C, Bazhin AV, Werner J, Guba MO. A novel machine learning algorithm to predict disease free survival after resection of hepatocellular carcinoma. Ann Transl Med. 2020;8:434. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 37. | Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301-1314. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2800] [Cited by in F6Publishing: 3379] [Article Influence: 563.2] [Reference Citation Analysis (3)] |
| 38. | Morshid A, Elsayes KM, Khalaf AM, Elmohr MM, Yu J, Kaseb AO, Hassan M, Mahvash A, Wang Z, Hazle JD, Fuentes D. A machine learning model to predict hepatocellular carcinoma response to transcatheter arterial chemoembolization. Radiol Artif Intell. 2019;1:e180021. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 39. | Peng J, Kang S, Ning Z, Deng H, Shen J, Xu Y, Zhang J, Zhao W, Li X, Gong W, Huang J, Liu L. Residual convolutional neural network for predicting response of transarterial chemoembolization in hepatocellular carcinoma from CT imaging. Eur Radiol. 2020;30:413-424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 93] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 40. | Liu D, Liu F, Xie X, Su L, Liu M, Xie X, Kuang M, Huang G, Wang Y, Zhou H, Wang K, Lin M, Tian J. Accurate prediction of responses to transarterial chemoembolization for patients with hepatocellular carcinoma by using artificial intelligence in contrast-enhanced ultrasound. Eur Radiol. 2020;30:2365-2376. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 74] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 41. | Abajian A, Murali N, Savic LJ, Laage-Gaupp FM, Nezami N, Duncan JS, Schlachter T, Lin M, Geschwind JF, Chapiro J. Predicting Treatment Response to Image-Guided Therapies Using Machine Learning: An Example for Trans-Arterial Treatment of Hepatocellular Carcinoma. J Vis Exp. 2018;140:e58382. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 42. | Mähringer-Kunz A, Wagner F, Hahn F, Weinmann A, Brodehl S, Schotten S, Hinrichs JB, Düber C, Galle PR, Pinto Dos Santos D, Kloeckner R. Predicting survival after transarterial chemoembolization for hepatocellular carcinoma using a neural network: A Pilot Study. Liver Int. 2020;40:694-703. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 26] [Article Influence: 6.5] [Reference Citation Analysis (2)] |
| 43. | Sieghart W, Hucke F, Pinter M, Graziadei I, Vogel W, Müller C, Heinzl H, Trauner M, Peck-Radosavljevic M. The ART of decision making: retreatment with transarterial chemoembolization in patients with hepatocellular carcinoma. Hepatology. 2013;57:2261-2273. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 239] [Cited by in F6Publishing: 253] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 44. | Adhoute X, Penaranda G, Naude S, Raoul JL, Perrier H, Bayle O, Monnet O, Beaurain P, Bazin C, Pol B, Folgoc GL, Castellani P, Bronowicki JP, Bourlière M. Retreatment with TACE: the ABCR SCORE, an aid to the decision-making process. J Hepatol. 2015;62:855-862. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 115] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 45. | Kim BK, Shim JH, Kim SU, Park JY, Kim DY, Ahn SH, Kim KM, Lim YS, Han KH, Lee HC. Risk prediction for patients with hepatocellular carcinoma undergoing chemoembolization: development of a prediction model. Liver Int. 2016;36:92-99. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 56] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 46. | Ziv E, Yarmohammadi H, Boas FE, Petre EN, Brown KT, Solomon SB, Solit D, Reidy D, Erinjeri JP. Gene Signature Associated with Upregulation of the Wnt/β-Catenin Signaling Pathway Predicts Tumor Response to Transarterial Embolization. J Vasc Interv Radiol. 2017;28:349-355.e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 47. | Liang JD, Ping XO, Tseng YJ, Huang GT, Lai F, Yang PM. Recurrence predictive models for patients with hepatocellular carcinoma after radiofrequency ablation using support vector machines with feature selection methods. Comput Methods Programs Biomed. 2014;117:425-434. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 48. | Xu RH, Wei W, Krawczyk M, Wang W, Luo H, Flagg K, Yi S, Shi W, Quan Q, Li K, Zheng L, Zhang H, Caughey BA, Zhao Q, Hou J, Zhang R, Xu Y, Cai H, Li G, Hou R, Zhong Z, Lin D, Fu X, Zhu J, Duan Y, Yu M, Ying B, Zhang W, Wang J, Zhang E, Zhang C, Li O, Guo R, Carter H, Zhu JK, Hao X, Zhang K. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater. 2017;16:1155-1161. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 478] [Cited by in F6Publishing: 508] [Article Influence: 72.6] [Reference Citation Analysis (0)] |
| 49. | Villanueva A, Portela A, Sayols S, Battiston C, Hoshida Y, Méndez-González J, Imbeaud S, Letouzé E, Hernandez-Gea V, Cornella H, Pinyol R, Solé M, Fuster J, Zucman-Rossi J, Mazzaferro V, Esteller M, Llovet JM; HEPTROMIC Consortium. DNA methylation-based prognosis and epidrivers in hepatocellular carcinoma. Hepatology. 2015;61:1945-1956. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 266] [Cited by in F6Publishing: 281] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 50. | Yeh CC, Goyal A, Shen J, Wu HC, Strauss JA, Wang Q, Gurvich I, Safyan RA, Manji GA, Gamble MV, Siegel AB, Santella RM. Global Level of Plasma DNA Methylation is Associated with Overall Survival in Patients with Hepatocellular Carcinoma. Ann Surg Oncol. 2017;24:3788-3795. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 51. | Dong RZ, Yang X, Zhang XY, Gao PT, Ke AW, Sun HC, Zhou J, Fan J, Cai JB, Shi GM. Predicting overall survival of patients with hepatocellular carcinoma using a three-category method based on DNA methylation and machine learning. J Cell Mol Med. 2019;23:3369-3374. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 52. | Azer SA. Deep learning with convolutional neural networks for identification of liver masses and hepatocellular carcinoma: A systematic review. World J Gastrointest Oncol. 2019;11:1218-1230. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 60] [Cited by in F6Publishing: 44] [Article Influence: 8.8] [Reference Citation Analysis (4)] |