Abstract
Background: Pseudomonas aeruginosa is one of the most prevalent opportunistic pathogens in humans that has thrived and proved to be difficult to control in this “post-antibiotic era.” Antibiotic alternatives are necessary for fighting against this resilient bacterium. Even though phages might not be “the wonder drug” that solves everything, they still provide a viable option to combat P. aeruginosa and curb the threat it imposes.
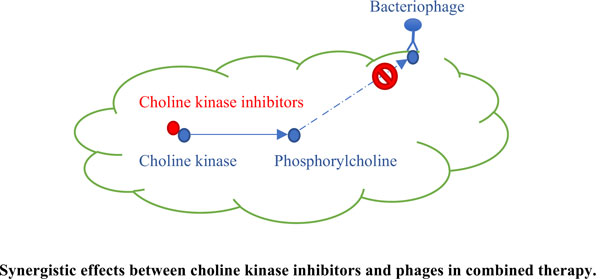
Main Findings: The combination of antibiotics with phages, however, poses a propitious treatment option for P. aeruginosa. Choline kinase (ChoK) is the enzyme that synthesizes phosphorylcholine subsequently incorporated into lipopolysaccharide located at the outer membrane of gram-negative bacteria. Recently, inhibition of ChoKs has been proposed as a promising antibacterial strategy. Successful docking of Hemicholinium-3, a choline kinase inhibitor, to the model structure of P. aeruginosa ChoK also supports the use of this inhibitor or its derivatives to inhibit the growth of this microorganism.
Conclusion: Therefore, the combination of the novel antimicrobial “choline kinase inhibitors (ChoKIs)” with a phage cocktail or synthetic phages as a potential treatment for P. aeruginosa infection has been proposed.
Keywords: Choline kinase inhibitor, phage therapy, Pseudomonas aeruginosa, antibiotic resistance, infection, combined therapy.
[http://dx.doi.org/10.1128/CMR.00019-18] [PMID: 30158299]
[http://dx.doi.org/10.1093/gbe/evy139] [PMID: 29982603]
[http://dx.doi.org/10.1186/s13756-015-0067-2] [PMID: 26075066]
[http://dx.doi.org/10.1001/jama.2009.1754] [PMID: 19952319]
[http://dx.doi.org/10.1128/AAC.48.4.1175-1187.2004] [PMID: 15047518]
[http://dx.doi.org/10.3389/fmicb.2019.00913] [PMID: 31130925]
[http://dx.doi.org/10.1111/2049-632X.12033] [PMID: 23620179]
[http://dx.doi.org/10.1080/09168451.2014.877828] [PMID: 25036502]
[http://dx.doi.org/10.3390/molecules24173152] [PMID: 31470632]
[http://dx.doi.org/10.1016/j.micpath.2020.104048] [PMID: 32035104]
[http://dx.doi.org/10.1016/j.chom.2019.01.018] [PMID: 30763535]
[http://dx.doi.org/10.1038/nrmicro2315] [PMID: 20348932]
[http://dx.doi.org/10.5772/intechopen.87247]
[http://dx.doi.org/10.1002/bies.201000071] [PMID: 20979102]
[http://dx.doi.org/10.1089/sur.2018.184] [PMID: 30256176]
[http://dx.doi.org/10.3390/v10040177] [PMID: 29621149]
[http://dx.doi.org/10.3390/v5030806] [PMID: 23478639]
[http://dx.doi.org/10.1155/2012/863945] [PMID: 23316235]
[http://dx.doi.org/10.1128/AEM.01546-14] [PMID: 25107968]
[http://dx.doi.org/10.1016/S1473-3099(18)30482-1] [PMID: 30292481]
[http://dx.doi.org/10.2174/138920110790725401] [PMID: 20214609]
[http://dx.doi.org/10.3389/fphar.2019.00513] [PMID: 31139086]
[http://dx.doi.org/10.1038/nrmicro3564] [PMID: 26548913]
[http://dx.doi.org/10.1038/nrd2201] [PMID: 17159923]
[http://dx.doi.org/10.3389/fcimb.2018.00376] [PMID: 30406049]
[http://dx.doi.org/10.1042/ETLS20170002] [PMID: 33525818]
[http://dx.doi.org/10.1016/j.cell.2019.09.015] [PMID: 31585083]
[http://dx.doi.org/10.1146/annurev-virology-101416-041616] [PMID: 28749735]
[http://dx.doi.org/10.1073/pnas.93.8.3188] [PMID: 8622911]
[http://dx.doi.org/10.1016/j.virusres.2005.05.014] [PMID: 16055223]
[http://dx.doi.org/10.1016/j.surg.2005.02.012] [PMID: 15933632]
[http://dx.doi.org/10.1186/1471-2180-11-195] [PMID: 21880144]
[http://dx.doi.org/10.1046/j.1472-765X.2003.01400.x] [PMID: 12969496]
[http://dx.doi.org/10.1073/pnas.76.12.6147] [PMID: 160562]
[http://dx.doi.org/10.1073/pnas.0800442106] [PMID: 19255432]
[http://dx.doi.org/10.1101/cshperspect.a023879] [PMID: 27481531]
[http://dx.doi.org/10.1016/j.tim.2018.09.006] [PMID: 30322741]
[http://dx.doi.org/10.1016/j.copbio.2020.11.003] [PMID: 33310655]
[http://dx.doi.org/10.1038/srep28115] [PMID: 27301427]
[http://dx.doi.org/10.1371/journal.pone.0106628] [PMID: 25259735]
[http://dx.doi.org/10.3389/fmicb.2020.00327] [PMID: 32194532]
[http://dx.doi.org/10.1128/mBio.01652-19] [PMID: 31551330]
[http://dx.doi.org/10.1186/s12967-019-2120-z] [PMID: 31727099]
[http://dx.doi.org/10.3390/antibiotics9110827] [PMID: 33227949]
[http://dx.doi.org/10.1128/mBio.00240-17] [PMID: 28377527]
[http://dx.doi.org/10.1128/mSystems.00756-19] [PMID: 32019835]
[http://dx.doi.org/10.1038/srep26717] [PMID: 27225966]
[http://dx.doi.org/10.1371/journal.pone.0000799] [PMID: 17726529]
[http://dx.doi.org/10.1093/infdis/jiw632] [PMID: 28007922]
[http://dx.doi.org/10.1038/s41598-018-37636-x] [PMID: 30728389]
[http://dx.doi.org/10.1371/journal.pone.0168615] [PMID: 28076361]
[http://dx.doi.org/10.3389/fcimb.2019.00022] [PMID: 30834237]
[http://dx.doi.org/10.1093/femsec/fiy107] [PMID: 29878184]
[http://dx.doi.org/10.1007/s00705-018-3811-0] [PMID: 29550930]
[http://dx.doi.org/10.3390/antibiotics6040020] [PMID: 28946671]
[http://dx.doi.org/10.1046/j.1365-2958.2000.01825.x] [PMID: 10760154]
[http://dx.doi.org/10.1016/j.micpath.2009.08.001] [PMID: 19682567]
[http://dx.doi.org/10.1111/j.1574-6968.1996.tb08147.x] [PMID: 9026440]
[http://dx.doi.org/10.1042/bj1100573] [PMID: 4387389]
[http://dx.doi.org/10.1128/JB.01795-07] [PMID: 18245291]
[http://dx.doi.org/10.1371/journal.ppat.1002521] [PMID: 22396641]
[http://dx.doi.org/10.1128/IAI.68.3.1664-1671.2000] [PMID: 10678986]
[http://dx.doi.org/10.1038/srep33189] [PMID: 27616047]
[http://dx.doi.org/10.1128/AAC.00920-13] [PMID: 24041883]
[http://dx.doi.org/10.1021/acs.biochem.7b01257] [PMID: 29389115]
[http://dx.doi.org/10.1038/cddis.2013.453] [PMID: 24287694]
[http://dx.doi.org/10.3389/fmicb.2019.02146] [PMID: 31681254]
[http://dx.doi.org/10.18632/oncotarget.9466] [PMID: 27206796]
[http://dx.doi.org/10.1038/s41598-019-40885-z] [PMID: 30911014]
[http://dx.doi.org/10.1158/1535-7163.MCT-14-0531] [PMID: 25487918]
[http://dx.doi.org/10.1038/s41598-020-72165-6] [PMID: 32963303]
[http://dx.doi.org/10.4172/2155-9872.1000356]
[http://dx.doi.org/10.1186/s12860-019-0214-3] [PMID: 31387520]
[http://dx.doi.org/10.1002/jcc.21797] [PMID: 21541955]
[http://dx.doi.org/10.1093/nar/gkr366] [PMID: 21624888]
[http://dx.doi.org/10.1002/elps.200900140] [PMID: 19517507]
[http://dx.doi.org/10.1093/nar/gky427] [PMID: 29788355]
[http://dx.doi.org/10.1093/bioinformatics/btq662] [PMID: 21134891]
[http://dx.doi.org/10.1093/bioinformatics/btz828] [PMID: 31697312]
[http://dx.doi.org/10.1021/ci049714+] [PMID: 15667143]
[http://dx.doi.org/10.1021/acs.jcim.5b00559] [PMID: 26479676]
[http://dx.doi.org/10.1002/jcc.20084] [PMID: 15264254]
[PMID: 16015482]
[http://dx.doi.org/10.1021/acs.jcim.6b00174] [PMID: 27391578]
[PMID: 2555674]
[http://dx.doi.org/10.1007/s11030-020-10134-x] [PMID: 32880078]
[http://dx.doi.org/10.3390/v10080396] [PMID: 30060520]
[http://dx.doi.org/10.4161/bact.28281] [PMID: 24616838]