Manuscript accepted on : 08-05-2020
Published online on: 21-05-2020
Plagiarism Check: Yes
Reviewed by: Kiranmayi Patnala
Second Review by: Nagendra Sastry Yarla
Final Approval by: Dr. Eugene A. Silow ![]()
![]()
Imran Zafar1*
![]() , Muhammad Tariq Pervez1
, Muhammad Tariq Pervez1 , Mohd Ashraf Rather2
, Mohd Ashraf Rather2
![]() , Masroor Ellahi Babar3
, Masroor Ellahi Babar3![]() , Mehar Ali Raza4
, Mehar Ali Raza4 , Rida Iftikhar,1
, Rida Iftikhar,1  and Maryam Fatima3
and Maryam Fatima3![]()
1Department of Bioinformatics and Computational Biology, Virtual University Pakistan
2Division of Fish Genetics and Biotechnology, Faculty of Fisheries, Rangil- Gandarbal (SKAUST-K)
3Department of Biotechnology, Virtual University of Pakistan
4Department of Bioinformatics & Biotechnology, Government College University Faisalabad
Corresponding Author E-mail :bioinfo.pk@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/2834
ABSTRACT: Genome wide analysis of the genes responsible for polyphenol oxidase (PPO) or peroxidase (POX) enzymes was performed in nine plant species namely (Carica papaya), model plant dicot (Selaginella moellendorffi), monocot (Zea mays), legumes (Pisum sativum, Vigna angularis), cereals (Avena sativa, Setaria italic and Oryza sativa) and fruit (Morus notabilis). We identified 135 PPO and 1645 POX genes in the selected plant species during the genome wide distribution analysis. The result showed that A.sativa and P. sativum have no PPO genes and a few number of POX genes were found in these plants. The synteny analysis of PPO genes in seven plant species excluding A.sativa and P.sativum revealed higher degree of conservation, recommending that these genes occurred before monocot/ eudicot divergence. The gene ontology (GO) analysis showed that PPO gene (Accession no DQ532396.1; 577 amino acids) is localized maximum in chloroplast (43.1%) with score of 2.156, molecular function (47.4%), biological process (53.7%) and cellular component (13.9%). While the POX protein (Accession no DQ855429.1; 335 amino acids) is highly founded in the extracellular part of the cell membrane (62.2%) with score 3.108, molecular function 49.8%, biological process 50.9% and cellular component 61.3%. Among the identified PPO and POX genes were reported to be (91.15%) and (85.21) moderately or highly expressed. The network analysis showed that PPO and POX genes had 21, 21 nodes, 100, 97 edges, 9.52, 9.24 average nodes and 0.876, 0.539 average local cluster coefficient. The POX genes were located in sub-telomeric section of chromosomes in the form of clusters. This study about genome-wide investigation and consideration of gene-expression patterns generated the valuable evidence on PPO and POX gene families that will be helpful for improving crop production in monocot or dicot plants.
KEYWORDS: Comparative Genomics; Polyphenol Oxidase; Peroxidase; Plant Species
Download this article as:| Copy the following to cite this article: Zafar I, Pervez M. T, Rather M. A, Babar M. E, Raza M. A, Iftikhar R, Fatima M. Genome-Wide Identification and Expression Analysis of PPOs and POX Gene Families in the Selected Plant Species. Biosci Biotech Res Asia 2020;17(2). |
| Copy the following to cite this URL: Zafar I, Pervez M. T, Rather M. A, Babar M. E, Raza M. A, Iftikhar R, Fatima M. Genome-Wide Identification and Expression Analysis of PPOs and POX Gene Families in the Selected Plant Species. Biosci Biotech Res Asia 2020;17(2). Available from: https://bit.ly/2V66kti |
Introduction
All plant species have a biological system to constantly fighting with biotic and abiotic stress1. The stress like alteration in environmental conditions, ultraviolet radiation, and microbial pathogen attacks plays a dynamic role for cell retardation to damage their structural and functional measurements, which possesses the functionality of death mechanism in plants. Moreover, every plant species has a self-defence mechanism by nature against various types of stresses. In several circumstances, plants protect themselves against pathogen attack by activating the Polyphenol oxidases (PPOs) pathway. The PPOs are copper-binding enzymes extensively scattered in whole genome of plants. In nature, the PPO gene assembly is a conflicting group with various enzymes which catalyse exclusively the dehydrogenation of catechols to the corresponding o-quinones or show performance as bi-functional enzymes to mobilized decomposition of mono phenols to o-diphenol intermediates (recognized as a tyrosinase / cresolase / monophenolase activity) thus following the decomposition of o-diphenols to the corresponding o-quinones.2-3 These o-quinones along with diverse useful clusters of proteins can yield red, brown and black pigments.4 The brown discoloration produced by PPO activity has significant influences on the appearance and processing qualities of many crop products.5 For instance, in case of wheat grains high PPO activity can result in brown or dark discoloration of flour and extremely declines its quality for noodle making.6 In higher plants, PPOs genes have been found to involve in diverse biological and molecular processes, such as discoloration of cereal spikes and grains.7 PPO genes have been identified in green plants as well as in animals, fish, shrimp and fungi.8 The PPOs play a vital role in protecting from insect and pathogen attacks in plants.9-10 They have also involved in the synthesis of antimicrobial unit in a series of monocotyledons and dicotyledonous plants to show frequent responses to the tyrosinases and appear to involved in pigment formation pathogens.11
The genetic factors of plant peroxidase (POX) genes are heme-containing glycoprotein showing relation with the different classes of pathogen-related (PR) protein.12 These genes take part in defence mechanism by producing ethylene to protect plant from pathogen or insect’s attack.13-14 These POX genes associated with a superfamily to consisting of three distinct classes based on their structural differences such as class I, II and III15. The superfamily also consists of two basic wound inducible proteins such as POX and POX N already reported which are consist of 47 amino acids in length with no signal peptide, and one of POX shown conservation between monocot and dicot species with specific expression to conform function of transgenic rice.16 Therefore, contemporary analyses were focus to study peroxidase gene for reconnoitring and recognition of synteny association between monocot or dicot species.
The whole genome sequencing gives a chance to find the genes correlations with respect to the relative positions in the genome of the species. These processes of understanding unable to detect the ratio of orthology shared in the genome and mechanism of phylogenetic relations. In the field of bioinformatics, advanced data analysis is a major part to do comparative genomics and synteny analysis. Thus, the focus for detection of a gene family depends upon the species of interest and these relationships consist of mainly two important concepts of evolutionary genomics such as homologous genes which are categorised as paralogs or orthologs.17 The homology is the blanket term for orthologs and paralogs. However, the orthologs are corresponding genes in multiple different lineages and diverged due to a speciation event, because paralogs result from duplication.18 Hence, the orthologs are genes that are identified in number of species but recently have originated from single gene and keep identical biological function.19 Some studies have reported that they have been preserved in the middle of evolutionarily correlated species. Thus, the defined percentage of this conservation between linked species of each plant is possible to use for finding their synteny interactions. The progressive proportion of orthology existence for distinct gene families in the middle of two species imitates maximum preservation of their role in those species. In this research, we accomplished the genome wide study of selected monocot and dicot plant species. WE investigated PPO and POX genes by performing following tasks: (I) Comparative analysis of whole genome expansion of PPO and POX genes by way of reverence to their place in the genome (II) Phylogenetic analysis (III) Comparison among monocot and dicot based on orthologous relation (IV) Protein-protein interaction network analysis and gene ontology of PPO and POX genes families in selected plant species.
The results of this study indicated that, the total of 135 PPO and 1645 POX genes are identified in selected plant species such as (Carica papaya), model plant dicot (Selaginella moellendorffi), monocot (Zea mays), legumes (Pisum sativum, Vigna angularis), cereals (Avena sativa, Setaria italic and Oryza sativa) and fruit (Morus notabilis) for the period of genome wide distribution analysis besides the A.sativa and P. sativum had no PPO genes and an insufficient number of POX genes were found in these plants. The synteny analysis of PPO genes in seven plant species excluding A.sativa and P.sativum exposed sophisticated degree of conservation, mentioning that these genes arisen before monocot/ eudicot divergence. The gene ontology (GO) investigation presented that PPO gene (Accession no DQ532396.1; 577 amino acids) contained maximum in chloroplast region (43.1%) with 2.156 score values, molecular function (47.4%), biological process (53.7%) and cellular part (13.9%). While the POX protein (Accession no DQ855429.1; 335 amino acids) is highly founded in the extracellular part of the cell membrane (62.2%) with score 3.108, molecular function 49.8%, biological process 50.9% and 61.3% of cellular part. Among the identified PPO and POX genes were testified to be (91.15%) and (85.21) moderately or highly expressed. The network investigation show that PPO and POX genes had 21, 21 nodes, 100, 97 edges, 9.52, 9.24 average nodes and 0.876, 0.539 average local cluster coefficient. The POX gene was located in sub-telomeric section of chromosomes in the form of clusters. This study about genome-wide investigation and consideration of gene-expression patterns generated the valuable evidence on PPO and POX gene families that will be helpful for improving crop production in monocot or dicot plants.
Material and Methods
Dataset
All genomic and proteomic sequences for comparative analyses of selected plant species (five dicots and four monocots) used in this study was downloaded from the National Centre of Biotechnological Information (NCBI. https://www.ncbi.nlm.nih.gov/), Plant Genome Data Bank (PlantGDB: http://plantgdb.org/) and Phytozome (http://www.phytozome.net/). Detail information of downloaded data presented in Table 1.
Table 1: The Selected plant list including with total of chromosomes, individual Genome size, and complete number of genes as well as EST sequences
| S.no | Yield Name | Scientific Name | Category | Chromosomes | Genome size | Genes | ESTs |
| 1 | Spikemoss | Selaginella moellendorffi | Model dicot | 27 | 212.5 Mb | 34,782 | 97,512 |
| 2 | Peas | Pisum sativum | Legumes
(dicot) |
14 | 4275.93 Mb | 218 | 21,69 |
| 3 | Adzuki bean | Vigna angularis | Legumes
(Dicot) |
11 | 449.74 | 312 | 11,199 |
| 4 | Mulberries | Morus notabilis | Fruit
(dicot) |
7 | 304.11 Mb Mb | 28,572 | 11,265 |
| 5 | Rice | Oryza sativa | Model
(monocot) |
12 | 372.31 Mb | 95,489 | 1,274,742 |
| 6 | Oat | Avena sativa | Cereals
(monocot) |
7 | 76.33 Mb | 130 | 25,425 |
| 7 | Foxtail millet | Setaria italica | Cereals
(monocot) |
9 | 400.91 Mb | 34,584 | 66,027 |
| 8 | Papayas | Carica papaya | Tree
(dicot) |
9 | 271.73 Mb | 20,333 | 77,343 |
| 9 | Corn | Zea mays | Cereals
(monocot) |
10 | 2103.64 Mb | 319 | 2,091,896 |
| 10 | Average genome size of monocot=651.46 | Average genome size of dicot=1352.475 Mb | |||||
∗Data resource: PlantGDB (ftp://ftp.plantgdb.org/download/Genomes/); National Center for biotechnology Information (https://www.ncbi.nlm.nih.gov); Phytozome (http://www.phytozome.net/); PLAZA for monocot and dicot (https://bioinformatics.psb.ugent.be/plaza/)
Retrieving the Most Similar Protein Sequences
The presence of PPO and POX genes in each plant species were identify using systematic BLASTP protocols (https://blast.ncbi.nlm.nih.gov/Blast.cgi). For this purpose, e-value and bit score was set <= e-20 and 100 respectively with default settings.
In Silico Physical Mapping of Genes and Genomes
The chromosomal place of POX genes was identified using CoGe BLAST20 (https://genomevolution.org/coge/CoGeBlast.pl). For PPO genes, we directly searched each gene in NCBI database by using accession number of individual plant using reciprocal BLAST search technique. To map the positions of the genes on chromosomes, the corporal maps was obtained using MapChart version 2.32 (https://www.wur.nl/en/show/Mapchart.htm), the locus of individually gene characterize on chromosomes in the form of base pair (bp).
Evolutionary Tree Analysis and Functional Classification of PPO and POX Gene Families
For phylogenetic tree, we used neighbour joining (NJ) method based on its realistic precision and cubic development reported by earlier researchers, which make this technique a superior opportunity to build confidential phylogenetic tree.21 For all identified PPO and POX genes, the multiple sequence alignment (MSA) were obtain by ClustalW version 2.1 downloaded from (http://www.clustal.org/clustal2/), and phylogenetic tree was built from T-Coffee web based software (https://www.ebi.ac.uk/Tools/msa/tcoffee/) that attempts to alleviate the pitfalls of progressive configuration methods using standard online tools from the European Bioinformatics Institute EMBL-EBI, (https://www.ebi.ac.uk/).22 For both PPO and POX genes, the neighbour-joining distance tree made evidently using default protocols and follows 1000 bootstrapping assessment replicates to confirm maximum declaration of interval and their precision23. Bootstrapping value considers to assessing the degree of support for each homologous group present in tree. The constructing of the first important tree for 135 PPO protein sequences was carried out by ClustalW 2.1, and then it was continued by MEME 5.0.4 suit (http://meme-suite.org/) results. For perceiving and penetration of motifs distribution we used a conventional tool MEME (Multiple EM for Motif Elicitation). The MEME tool stands actual beneficial for construction of evolutionary tree and clasp conserved nucleotide or amino acid structures, the earlier researcher used this contrivance to support the penetration of both phylogenies along with motifs. We used MEME tool (http://meme-suite.org/tools/meme) to find 10 subdomains having dynamically separated or heterogeneous motifs distribution between six and fifty residues. For observing the correlation of motif distribution we can use the phylogenetic tree construction with MEME motifs24. Therefore, we concluded that accepts of MEME are well supported and connected, as they interactively representation for creating and displaying phylogenetic tree we used online tool free available for public accesses named iTOL (interactive Tree of Life) (https://itol.embl.de/).25 Likewise, the phylogenetic tree for 1045 POX gene family is also constructed, by using the same method of POX gene families (ClustalW 2.1, Clustal Omega and iTOL server) but we do not check the motif distribution due to its maximum number of genes distribution on each plant species as like (1045).
In_Silico Investigation of PPO and POX Genes Expression
For in_silico expression analysis of PPO and POX genes, BLAST search accomplish among all corresponding plant species and EST sequences copied in the local database. The EST hits having 𝑒-20 E-value and <=100 bit score consider most significant hits. The in_silico expression of these identified genes depends upon the presence of significant hits and considered these genes as “not expressed”, if any significant hit not found. On the other hand, a gene is considered as less expressed if the significant hit range from 1-99. A gene cluster considers as moderate expressed if its hit ranges from 201 – 400 and more than 400 shows highly expresses gene.
Contraction of Protein- Protein Interaction Network
For protein-protein interaction, PPO and POX gene explored in STING database available at (https://string-db.org/).26 The STING database is a based on very diverse functionality correlating with wide range of properties such as data mining, gene expression analysis, literature and computational prediction etc., calculation of this database is completely dependent upon statistical measurement framework.27
Gene Ontology of PPO and POX Genes
A gene ontology (GO) of PPO and POX genes from O. sativa indica group was conceded using Cello2go (http://cello.life.nctu.edu.tw/cello2go/) online available tool for public accesses. The Cello2go uses built-in protocol of BLAST and search with default settings having 0.001 E-value to pursue amino acids and DNA sequence results that are marked as gene ontology in a house developed catalogue linked to derive from the knowledge based UniProt, SwissProt and TrEMBL database.28
Comparative Analysis of Synteny
The synteny analysis of PPO and POX genes in nine plant genomes was performed using orthologue evidence. For Orthologue pairs the hypothetical method best bidirectional hit (BBH) was used.29 Consequently, our evolutionary interval and approximation nature of BLAST outcome transformation in mutation rate so, the highest hit of blast some time miss some conceivable orthologue pairs. In addition to this, the BBH method gives the facility for orthologous pairs with one to one relation, after our object was to find one possible orthologue pair to form entire possible orthologous.30 Conversely, for every PPO and POX genes families we recognised a single clear orthologue, therefore the facts of possible changes acquiescently occur in monocot and dicot genome due to their divergence, however many genes may have one too many relation for orthologous combination; hence we used BBH method for getting this type of relations.
In InParanoid (INP) program is the best method for identification of orthologue formation to check the one to many relationships31. We hypothesized result based on both (BHH & INP) methodologies to find some favorable orthologue pairs and clearly defined the association of gene pairs which are important based on conservation of Sequence identity.32 We accomplished BLAST search for all genes against all plant genomes selected in this study and used three scaled criterion to filter the results with most significant hits. These parameters were the BLAST bit score ≤=100; E-value ≤ 𝑒 −20 as well as 20% uniqueness among amino acid sequences terminated at least 50% length of the protein. In the least two important BLAST hits which are clearly match the aforementioned criteria and ensure bidirectional hits through each other, and measured orthologues to counted as single orthologous pair.33
Results and Discussion
PPOs and POX Genes in Dicots and Monocots
For PPO and POX gene family we performed genome wide analysis in total nine plant species, which are identified in very diverse groups like legumes (P. sativum and V. angularis), fruits (M. notabilis), cereals (A. sativa, S. italica, and Z. mays), trees (C. papaya), monocot (O. sativa) and model dicot (S. moellendorffi) species. In this study we observed the well-known facts, on average genome size of monocot and dicots. The dicots genome size was larger than monocots. For their divergence, the inductions of variation in total amount of DNA and number of genes via shuffling of genome (entire set of genes) in monocot and dicot might have been important for different evolutionary paths34. In table 1, the statistical measurement of monocot and dicot genome sizes are given. On average, the genome sizes of dicots plant were found to have a higher in percentage as compared with monocot. The average percentage of dicot is 1101.82Mb or monocot is 738.21 Mb, then we also perceived that the five maximum number of dicot plants are used in this research possess a greater number of chromosomes with respect to monocots plant correspondingly. Even in previous research reports, the researcher show the negative correlation remained observed among basic number of chromosomes and genome sizes of monocot and dicot35. We detected the overall of 135 PPO and 1695 POX genes in nominated plant species given in material and methods regardless of their genome sizes in figure 1.
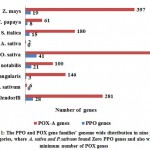 |
Figure 1: The PPO and POX gene families’ genome wide distribution |
For PPO and POX gene family, we identified the maximum number of PPO and POX genes in O. sativa. The minimum of POX gene was in A. sativa, and PPO in V. angularis, also absent in two plants A. sativa and P. sativum. The PPO concentration in monocots was founded maximum number of genes (75) as compard with dicots, because dicots were less number of PPO genes (62). There remained the 41 PPO genes in rice (O. sativa), highest in adzuki bean (V. angularis), not found in two plants one is dicot pea (P. sativum) and another one is monocot oat (A. sativa). In this study the number of POX gene families was also identified and they remain higher in gene concentrations on individual plant genome species such as S. moellendorffi (281), O. sativa (474), and further plants classes remains moderately comparable through the earlier research reports,36, 37, 38 although the diverse methodologies have been used for this type of study. After monocot divergence we scarcely explore to find out 135 PPO genes. On the other hand for POX gene family, we identify 1695 genes seem to be distributed in higher percentage among PPO genes.
PPO and POX genes Insilco Mapping and Genome-Wide Distribution
For discovery of physical place of genes on chromosomal position, we accomplished the BLAST consideration of the recognized PPO and POX gene families for the complete genomic sequences of nominated plants species. After that we see the PPO gene families are limited to explore on particular chromosomal site and POX families seem be to expended on complete genome of the individual species, the physical map of each chromosomes for PPO and POX gene families was created (Material are available in supplementary file 1). Many PPO and POX genes existed in main groups and S. moellendorffi and Z. mays chromosome measure has distributed in these genes, but we were observed the same trend for all selected plants used in current study (check supplementary figure 1).
The percentage of PPO genes on some chromosomes was founded to be less in dicots and on the other observation also funded that PPO genes are absent in two plant P. sativum dicot and A. sativa monocot. Whereas the reaming plant monocot PPOs gene differentially distributed on chromosomal positions. Hereafter, in respect of all PPOs genes, same as like monocot the S. moellendorffi plant seems to follow the distribution pattern. In dicots, excluding P. sativum, the greater ratio of PPOs genes were explored to proceeding on various chromosomes, while on monocots, many of the PPO genes are listed on chromosomal starting and ending positions. Many chromosomes of P. sativum (6) and A. sativa (2) were less in number PPOs or POX genes. Observation of physical mapping indicates the POX genes find in cluster of sub telomeric region of chromosomes and randomly distributed on entire genome of the selected plant species. Presence of gene clusters at sub-telomeric region on chromosomal position gives the significant value of proximal domain with whole possible occurrence of POX gene families as sub telomeric and exceptional higher ranks of sequence diversity between the genes members.39-40 For rapid evolution of these clusters the POX at telomere proximal region linked most probably for greater existence.41-42 The higher percentage of recognized genes and conceivable presence as a sub-telomeric gene family through collections of genes at telomeric proximal sections fact about evolution level of POX gene family with respect to PPOs gene family
PPO and POX Genes Phylogenetic Relation among Different Plant Species
Identification of PPO and POX gene families are highly divergent via belonging to individual plant species of monocot and dicots that also including with legumes, fruit and cereals. We have analysed the both PPO and POX gene families between monocot and dicot plant species for finding their evolutionary relation. First we was analysed 135 PPO genes via obtaining multiple sequence alignment (MSA) and then Neighbour joining (NJ) tree were constructed with the support of 1000 bootstrap replicates (BR) through default parameters, the 1000 BR used to find confidence of values for making a reliable phylogenetic tree.42 The NJ method has a cubic value to build a reliable tree for maximum length of sequences with very high-speed through original neighbour joining algorithm.43 For PPO and POX genes the motif distribution pattern recognised using online MEME software and exceptionally relation of phylogenetic tree and MEME results produced more with NJ tree as given in figure 2. We observed the clear correlation of NJ tree and motif pattern distribution, where many group and sub group of phylogenetic tree basically share the common motif pattern. We also perceive that many motifs are distributed in almost all groups and sub groups or some of them are more conserved, except the ones at the first end of the dicot and intermediate section of the phylogeny tree. Our identified preserved motifs are vital element to determine the molecular function of the PPO genes families among all individual plant species. The motif distribution of PPO genes 6 of 10 in P. sativum and two in A. sativa lacks lots of motifs and relationship with other groups might not show close evolutionary pedigree, we precede the novel method of motif via using MEME software to define the conserved region of gene distribution that are typically progress from the gene extension inside the similar clusters, where motif suitable into the developed and worse species.44 It can be clarified that the precursor genetic cause with many motif arrangement seem early in the progression, and then, same structure stood preserved by topical genes which are used give the information of evolution.
 |
Figure: 2 The PPO genes schematic demonstration of motif distribution |
In this study, we perceive configuration of motif sharing associated facts in the way of preservation of PPO genes in complete groups and sub groups, eliminating the two subgroups in the interior part of the phylogenetic hierarchy (Figure 3(a)). The interactively representation of phylogenetic tree established using iTOL and it found three main classes; monocot, dicot and mixed assigned with three different colour ranges with significant bootstrap replicates. The previous study also stated that, the particular selected plant having no significant hits and conserved domain with PPO genes and these plants made stress fortification due to exterior inheritance pattern45&46. The two selected plant P. sativum and A. sativa have zero POX genes and missing numerous motifs in sharing of MEME motif pattern, we were found in a distinct phylogenetic session or cluster among per two S. italica PPO genes (Figure 3(b)).
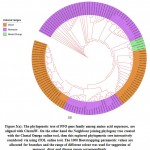 |
Figure 3(a): The phylogenetic tree of PPO gene family among amino |
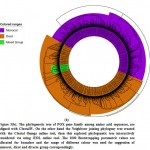 |
Figure 3(b): The phylogenetic tree of POX gene family among amino acid sequences |
The large group of monocots surveyed through flourishing conjugated dicot plant species group via those formed by the phylogenetic association among the pattern distributions and suggest the conservation of PPO genes, as well as their development of monocots to dicots. The current development study shows evidence of a phylogenetic relationship for lower plants based on genomic distribution and pattern distribution analysis.36-47 For POX genes families we established a close review via obtaining a lager neighbour joining tree show monocot and dicot distribution hits was not founded in single small group and subgroups, as like seem for PPO genes. In this study the four monocot species have largest group and subgroup given in colour range for phylogenetic tree are consist of many genes and same procedure was established for POX genes families to see group representation of dicots. Hereafter, not only any of the unique species can be measured equally basal one or descending one. The situation looks that throughout development of the POX protein sequence family; two foremost clusters have progressed, follow-on in extraordinary level of efficient divergence among the POX gene duplicates in both monocots and as well as dicots.
In Silico Expression Analysis
For PPO and POX genes families, we revealed the in-silico expression analysis in selected plant genomes, they all are expressed except two plant species P. stivum and A. sativa in which only PPO was absent and POX genes included with very low quantity. The earlier researcher analysed the negative correlation for PPO and POX genes families in these two plant species,45-46, they also observed the protection process mechanism that directly show positivity via depending upon external factors. For identified PPO (135) and POX (1695) genes families, we analysed and categorised these genes based on level of expression via considering significant hits of EST, then we distributed PPO and POX expressed genes; such as not expressed, low expressed, moderately expressed, or extremely expressed for all selected plant species. Among the known PPO genes (Figure 4 (a) and (b)), 91.15% existed to be either moderately or extremely expressed, where 85.21% POX genes remained perceived to be moderately or highly expressed (Figure5 (a) and (b)).
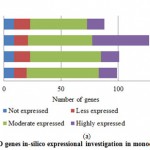 |
Figure 4(a): PPO genes in-silico expressional investigation in monocot plant species |
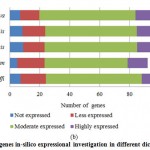 |
Figure 4(b): PPO genes in-silico expressional investigation in different dicot plant species. |
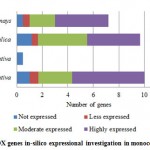 |
Figure 5(a): POX genes in-silico expressional investigation in monocot plant species |
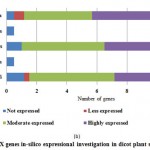 |
Figure 5(b): POX genes in-silico expressional investigation in dicot plant species |
PPO Protein-Protein Interaction Network Retrieval
The network analysis of PPO and POX gene families is retrieve from String database. Protein–protein interaction showed that PPO functional partner are mentioned in figure 6(a) and POX genes interacting with TRX-X, CAD4, OS10t0512400-01, CAD3, CAD1, CAD8D, CAD88, CAD8A, CAD6 and FC1 etc. The PPO genes confirming statistics of nodes is 21, total of edges is 100, averages nodes 9.52, average local cluster coefficient is 0.876, expected no of edges is 24 and PPI enrichment p-value was : < 1.0e-16 in figure 6(a). However, the POX genes having statistics of nodes is 21, number of edges is 97, the averages nodes 9.24, the average local cluster coefficient is 0.539, expected no of edges is 21 and PPI enrichment p-value was : < 1.0e-16 given in (supplementary file figure 2a).
To considered well interaction of network, all networks are distributed into 9 groups using K-mean cluster algorithum.48 The group was defined having close relations with each other; the balls mentioned in figure are charitable unspecified effects in the relationships among all genes. The arrows show positive relations among interacted partner genes. The networks are divided into different colour coded ranging used to indicate different groups and functional partner genes which we are predicted in our analysis, PPO genes in figure 6(B and C) and for POX genes figures are mentioned in supplementary file ((supplementary file figure 2b and c). The mechanism of cellular processing is applicable based on many functional partnerships and their interactions with systematic characterization which is important context for helping to explore molecular and system biology.25 We observed that positive correlation of PPO and POX protein among many other protein families.
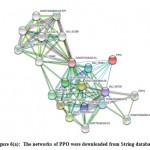 |
Figure 6(a): The networks of PPO were downloaded from String database |
 |
Figure 6(b): PPO protein-protein network divided into 9 k-means clusters |
 |
Figure 6(c): Predicted functional partner of PPO genes |
Gene ontology of PPO and POX Genes
The earlier researcher revolved that the PPO and POX genes in various plant species (apple and mango) maximum founded in region of chloroplast.49-50 The furthermost commonly recommended the starring role used for PPO in plants has been described for defence in paradox of pathogenic bacterium and herbivores, well-known chloroplast-contained enzyme physical separation from the vacuole-contained substrates.51
In this research we explore to use the CELLO2GO tool and this one revealed has a PPO protein sequence distribution is extreme nearly contained in the Chloroplast (43.1%) by total 2.156 in addition to GO investigation specified molecular function 47.4% , biological progression 53.7% as well as cellular constituent stayed 13.9% (Figure 7). On the other hand, the POX protein remains highly contained in the extracellular portion (62.2%) with score 3.108 and gene GO investigation specified molecular functionality 49.8% , biological progression 50.9% and cellular constituent 61.3% ( supplementary file figure 3 )
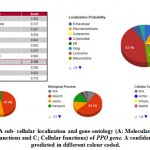 |
Figure 7(a): A sub- cellular localization and gene ontology |
Synteny Analysis and Orthology
For synteny analysis of PPO and POX genes we were count the orthologous pairs between each species having pairs as described in methods. The earlier researcher also founded that the negative relationship of PPO presence in other plant species both of monocot and dicot.37 For PPO genes families’ the orthologous pairs of A. sativa and P. sativum show lowest parentage orthology analysis as compared with other individual plant species (Figure 8(a)). The orthologous pair section analysis of targeted species was performed and analyzed the corresponding species, where individual ribbon come from diverse plant species in the form of different clades. Therefore the lowest arthology in V. angularis (bule clade) was found very clearly with other plant species. The approximation in C. papaya of PPO genes are identified with genomes of dicot plant and show the conserved links of orthologous 30% along with S. moellendorffi, 61% C. papaya, 63% Z. mays, as well as 55% M. notabilis, but with monocots, the rank of conservation of A. sativa remained minimum in percentage of 23% along with P. sativum, 22% S. italica, 21% O. sativa, and 18% with C. papaya. The comparatively less orthologous of PPO genes in A. sativa are founded among monocots is (75.6%) as compared to other monocot plant species as like Z. mays (87%), O. sativa (85.69%) in addition to S. italic is (83%). Interestingly, PPO genes from M. notabilis, a dicot species, indicate relatively further orthology direction near monocots (93.75%) as related by further dicots (85.81%). That is, once more, the situation is M. notabilis, a dicot species, looks to check location of monocot species and make sure an extra transference toward monocots as perceived through gene distribution and phylogeny investigation. For now a great ratio of orthological information, excepting A. sativa and P. sativum, facts leads toward the higher conservation of the PPO gene families between the individual plant species.
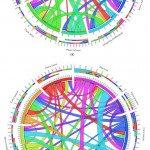 |
Figure 8: The individual plant species orthologous for (a) PPO and (b) POX gene families |
For conformation of the orthologues pairs of POX gene families, we determine the minimum ratio of orthology information among all species, but it was a slightly greater among all monocots. Based on comparison of orthological information of POX genes association indicated the conserved pairs for only 7 to 8% among dicots, whereas along with monocots, it is only from 11 to 16 % too can be perceived via the extensive ribbons (blue and purple) on behalf of monocot species (Figure 8(b)). We identified high percentage of POX genes families’ with least ratio of orthologue information among all individual plant species. It might be triggered through a higher percentage of progression as well as expansion, which taken a higher level of well-designed and functional divergence among the members.
Conclusion
We conduct research on comparative genomics, gene distribution, synthesis analysis, phylogeny exploration, network study between protein-protein interaction (PPI) and proportional analysis of gene ontology accomplished among individual plant species as stated in the material and methods; We explore a progressive protocol for genes from PPO and POX, to recognized the lowest number of PPO genes (135) and determined the relationship between different monocotyledons and dicotyledonous groups through analyzing the motif distribution patterns, in the direction of differentiate with the phylogenetic tree. The two main conclusions come from a higher percentage of orthology: (i) PPO gene family precedes the monocot / eudicot difference with hardly exploration of the monocot / eudicot divergence, and (ii) the other is an exceptional preservation of function. While the continuous random distribution of whole-genome clusters at the level of the sub-telomeric regions, POX genes have a very low proportion of orthology with many phylogenetic and special subgroups and subgroups of monocots and dicots may suggest the following: (i) possible existence of this family as a family of sub-telomeric genes (ii) possible expression of unusually high levels of sequence diversity (iii) higher development and expansion rates than the PPO gene family and (iv) the development of many groups and subgroups during family expansion, resulting in a high level of functional divergence.
Supplementary Information
Additional File: Additional Figure A – Figure 7(b)
Acknowledgments
This work was interest free with the support of Virtual University of Pakistan, Department of bioinformatics (VUPDB) Lahore, Pakistan. The authors are also thankful to NCBI, ENSEMBLE and other genomic biological databases and software’s for providing genomic resources in public domain.
Conflicts of Interest
The author of this study did not have any type of conflict of interest.
Funding
This research work did not have any grant from funding agencies in the government and private sector.
References
- Mordecai, E.A. Pathogen impacts on plant communities: Unifying theory, concepts, and empirical work. Ecol. Monogr. 2011, 81, 429–441.
- Tran LT, Constabel CP. The polyphenol oxidase gene family in poplar: phylogeny, differential expression and identification of a novel, vacuolar isoform. Planta. 2011 Oct 1;234(4):799.
- Wang J, Constabel CP. Biochemical characterization of two differentially expressed polyphenol oxidases from hybrid poplar. Phytochemistry. 2003 Sep 1;64(1):115-21.
- Afoakwa EO, Kongor JE, Takrama JF, Budu AS, Mensah-Brown H. Effects of pulp preconditioning on total polyphenols, o-diphenols and anthocyanin concentrations during fermentation and drying of cocoa (Theobroma cacao) beans. Journal of Food Science and Engineering. 2013 May 1;3(5):235.
- Queiroz C, Mendes Lopes ML, Fialho E, Valente-Mesquita VL. Polyphenol oxidase: characteristics and mechanisms of browning control. Food reviews international. 2008 Sep 16;24(4):361-75.
- Sun Y, He Z, Ma W, Xia X. Alternative splicing in the coding region of Ppo-A1 directly influences the polyphenol oxidase activity in common wheat (Triticum aestivum L.). Functional & integrative genomics. 2011 Mar 1;11(1):85-93.
- Taketa S, Matsuki K, Amano S, Saisho D, Himi E, Shitsukawa N, Yuo T, Noda K, Takeda K. Duplicate polyphenol oxidase genes on barley chromosome 2H and their functional differentiation in the phenol reaction of spikes and grains. Journal of experimental botany. 2010 Jul 8;61(14):3983-93.
- Tran LT, Taylor JS, Constabel CP. The polyphenol oxidase gene family in land plants: lineage-specific duplication and expansion. BMC genomics. 2012 Dec;13(1):395
- Constabel CP, Barbehenn R. Defensive roles of polyphenol oxidase in plants. InInduced plant resistance to herbivory 2008 (pp. 253-270). Springer, Dordrecht.
- Richter C, Dirks ME, Gronover CS, Prüfer D, Moerschbacher BM. Silencing and heterologous expression of ppo-2 indicate a specific function of a single polyphenol oxidase isoform in resistance of dandelion (Taraxacum officinale) against Pseudomonas syringae pv. tomato. Molecular plant-microbe interactions. 2012 Feb;25(2):200-10.
- War AR, Paulraj MG, Ahmad T, Buhroo AA, Hussain B, Ignacimuthu S, Sharma HC. Mechanisms of plant defense against insect herbivores. Plant signaling & behavior. 2012 Oct 1;7(10):1306-20.
- van Doorn WG, Ketsa S. Cross reactivity between ascorbate peroxidase and phenol (guaiacol) peroxidase. Postharvest biology and technology. 2014 Sep 1;95:64-9.
- Tudzynski B. Fungal phytohormones in pathogenic and mutualistic associations. InPlant Relationships 1997 (pp. 167-184). Springer, Berlin, Heidelberg.
- Gomes FB, Moraes JC, Santos CD, Goussain MM. Resistance induction in wheat plants by silicon and aphids. Scientia Agricola. 2005 Dec;62(6):547-51.
- Passardi F, Theiler G, Zamocky M, Cosio C, Rouhier N, Teixera F, Margis-Pinheiro M, Ioannidis V, Penel C, Falquet L, Dunand C. PeroxiBase: the peroxidase database. Phytochemistry. 2007 Jun 1;68(12):1605-11.
- Ito H, Hiraga S, Tsugawa H, Matsui H, Honma M, Otsuki Y, Murakami T, Ohashi Y. Xylem-specific expression of wound-inducible rice peroxidase genes in transgenic plants. Plant Science. 2000 Jun 12;155(1):85-100.
- Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet.. 2005 Dec 15;39:309-38.
- Fitch WM. Distinguishing homologous from analogous proteins. Systematic zoology. 1970 Jun 1;19(2):99-113.
- Remm M, Storm CE, Sonnhammer EL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. Journal of molecular biology. 2001 Dec 14;314(5):1041-52.
- Castillo AI, Nelson AD, Haug-Baltzell AK, Lyons E. A tutorial of diverse genome analysis tools found in the CoGe web-platform using Plasmodium spp. as a model. Database. 2018 Jan 1;2018.
- Telles GP, Araújo GS, Walter ME, Brigido MM, Almeida NF. Live neighbor-joining. BMC bioinformatics. 2018 Dec;19(1):172.
- Sievers F, Higgins DG. Clustal Omega for making accurate alignments of many protein sequences. Protein Science. 2018 Jan;27(1):135-45.
- Czech L, Huerta-Cepas J, Stamatakis A. A critical review on the use of support values in tree viewers and bioinformatics toolkits. Molecular biology and evolution. 2017 Mar 22;34(6):1535-42.
- Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic acids research. 2016 Apr 19;44(W1):W242-5.
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, Jensen LJ. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic acids research. 2010 Nov 2;39(suppl_1):D561-8.
- Macqueen JB. Some Methods for classification and Analysis of Multivariate Observations, Proceedings of 5-th Berkeley Symposium on Mathematical Statistics and Probability. Berkeley: University of California Press 1:281-297.
- Yu CS, Cheng CW, Su WC, Chang KC, Huang SW, Hwang JK, Lu CH. CELLO2GO: a web server for protein subcellular Localization prediction with functional gene ontology annotation. PloS one. 2014 Jun 9;9(6):e99368.
- Dalquen DA, Dessimoz C. Bidirectional best hits miss many orthologs in duplication-rich clades such as plants and animals. Genome biology and evolution. 2013 Sep 6;5(10):1800-6.
- Chen F, Mackey AJ, Vermunt JK, Roos DS. Assessing performance of orthology detection strategies applied to eukaryotic genomes. PloS one. 2007 Apr 18;2(4):e383.
- Kim K, Kim W, Kim S. ReMark: an automatic program for clustering orthologs flexibly combining a Recursive and a Markov clustering algorithms. Bioinformatics. 2011 May 5;27(12):1731-3.
- Del Castillo CS, Hikima JI, Jang HB, Nho SW, Jung TS, Wongtavatchai J, Kondo H, Hirono I, Takeyama H, Aoki T. Comparative sequence analysis of a multidrug-resistant plasmid from Aeromonas hydrophila. Antimicrobial agents and chemotherapy. 2013 Jan 1;57(1):120-9.
- Moreno-Hagelsieb G, Latimer K. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics. 2007 Nov 26;24(3):319-24.
- Trivers R, Burt A, Palestis BG. B chromosomes and genome size in flowering plants. Genome. 2004 Jan 1;47(1):1-8.
- Hulsen T, Huynen MA, de Vlieg J, Groenen PM. Benchmarking ortholog identification methods using functional genomics data. Genome biology. 2006 Feb;7(4):R31.
- Passardi F, Longet D, Penel C, Dunand C. The class III peroxidase multigenic family in rice and its evolution in land plants. Phytochemistry. 2004 Jul 1;65(13):1879-93.
- Rawal HC, Singh NK, Sharma TR. Conservation, divergence, and genome-wide distribution of PAL and POX A gene families in plants. International journal of genomics. 2013;2013.
- Tran LT, Taylor JS, Constabel CP. The polyphenol oxidase gene family in land plants: Lineage-specific duplication and expansion. BMC genomics. 2012 Dec;13(1):395.
- Tashiro S, Nishihara Y, Kugou K, Ohta K, Kanoh J. Subtelomeres constitute a safeguard for gene expression and chromosome homeostasis. Nucleic acids research. 2017 Sep 14;45(18):10333-49.
- Barry JD, Ginger ML, Burton P, McCulloch R. Why are parasite contingency genes often associated with telomeres?. International journal for parasitology. 2003 Jan 1;33(1):29-45.
- Moran GP, Coleman DC, Sullivan DJ. Comparative genomics and the evolution of pathogenicity in human pathogenic fungi. Eukaryotic cell. 2011 Jan 1;10(1):34-42.
- Kong H, Landherr LL, Frohlich MW, Leebens‐Mack J, Ma H, DePamphilis CW. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. The Plant Journal. 2007 Jun;50(5):873-85.
- Minh BQ, Nguyen MA, von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Molecular biology and evolution. 2013 Feb 15;30(5):1188-95.
- Mailund T, Pedersen CN. QuickJoin—Fast neighbour-joining tree reconstruction. Bioinformatics. 2004 Jun 16;20(17):3261-2.
- Di C, Xu W, Su Z, Yuan JS. Comparative genome analysis of PHB gene family reveals deep evolutionary origins and diverse gene function. InBMC bioinformatics 2010 Oct (Vol. 11, No. 6, p. S22). BioMed Central.
- Hernandez JA, Jimenez A, Mullineaux P, Sevilia F. Tolerance of pea (Pisum sativum L.) to long‐term salt stress is associated with induction of antioxidant defences. Plant, cell & environment. 2000 Aug;23(8):853-62.
- Levings CS, Stuber CW, Murphy CF. Inheritance of an Auxin Inducible Peroxidase in Oats (Avena sativa L.) 1. Crop science. 1971;11(2):271-2.
- Wang C, Wang Q, Zhu X, Cui M, Jia H, Zhang W, Tang W, Leng X, Shen W. Characterization on the conservation and diversification of miRNA156 gene family from lower to higher plant species based on phylogenetic analysis at the whole genomic level. Functional & integrative genomics. 2019 Jun 7:1-20.
- Jamil K, Jayaraman A, Rao R, Raju S. In silico evidence of signaling pathways of notch mediated networks in leukemia. Computational and structural biotechnology journal. 2012 Jul 1;1(2):e201207005.
- Robinson SP, Loveys BR, Chacko EK. Polyphenol oxidase enzymes in the sap and skin of mango fruit. Functional Plant Biology. 1993;20(1):99-107.
- Taranto F, Pasqualone A, Mangini G, Tripodi P, Miazzi M, Pavan S, Montemurro C. Polyphenol oxidases in crops: biochemical, physiological and genetic aspects. International journal of molecular sciences. 2017 Feb 10;18(2):377.
- Boeckx T, Winters AL, Webb KJ, Kingston-Smith AH. Polyphenol oxidase in leaves: is there any significance to the chloroplastic localization?. Journal of experimental botany. 2015 Apr 4;66(12):3571-9.
Abbreviation
| NCBI | National Centre For Biotechnology Information |
| PPO | Polyphenol Oxidase |
| POX | Peroxidase |
| ESTs | Expression Sequence Tags |
| BLAST | Basic Local Alignment Search |
| NG | Neighbour Joining Method |
| MSA | Multiple Sequence Alignment |
| EMBI – EBI | European Molecular Biology Laboratory – European Bioinformatics Institute |
| MEME | Expectation-Maximization Multiple For Motif Elicitation |
| ITOL | Interactive Tree Of Life |
| STRING | Search Tool For The Retrieval Of Interesting Genes |
| GO | Gene Ontology |
| BBH | Best Bidirectional Hit |
| INP | Inparanoid Program |
| Mb | Megabytes |
| PPI | Protein-Protein Interactions |
| aa | Amino Acids |
| PlantGDB | Plant Genome Databank |
| PDB | Protein Databank |

This work is licensed under a Creative Commons Attribution 4.0 International License.





