Published online Jun 26, 2022. doi: 10.12998/wjcc.v10.i18.5965
Peer-review started: February 17, 2022
First decision: March 29, 2022
Revised: March 30, 2022
Accepted: May 22, 2022
Article in press: May 22, 2022
Published online: June 26, 2022
Psoriasis is a chronic inflammatory skin disease, the pathogenesis of which is more complicated and often requires long-term treatment. In particular, moderate to severe psoriasis usually requires systemic treatment. Psoriasis is also associated with many diseases, such as cardiometabolic diseases, malignant tumors, infections, and mood disorders. Psoriasis can appear at any age, and lead to a substantial burden for individuals and society. At present, psoriasis is still a treatable, but incurable, disease. Previous studies have found that microRNAs (miRNAs) play an important regulatory role in the progression of various diseases. Currently, miRNAs studies in psoriasis and dermatology are relatively new. Therefore, the identification of key miRNAs in psoriasis is helpful to elucidate the molecular mechanism of psoriasis.
To identify key molecular markers and signaling pathways to provide potential basis for the treatment and management of psoriasis.
The miRNA and mRNA data were obtained from the Gene Expression Omnibus database. Then, differentially expressed mRNAs (DEmRNAs) and differentially expressed miRNAs (DEmiRNAs) were screened out by limma R package. Subsequently, DEmRNAs were analyzed for Gene Ontology and Kyoto Encyclopedia of Genes and Genomics functional enrichment. The “WGCNA” R package was used to analyze the co-expression network of all miRNAs. In addition, we constructed miRNA-mRNA regulatory networks based on identified hub miRNAs. Finally, in vitro validation was performed. All experimental procedures were approved by the ethics committee of Chinese PLA General Hospital (S2021-012-01).
A total of 639 DEmRNAs and 84 DEmiRNAs were identified. DEmRNAs screening criteria were adjusted P (adj. P) value < 0.01 and |logFoldChange| (|logFC|) > 1. DEmiRNAs screening criteria were adj. P value < 0.01 and |logFC| > 1.5. KEGG functional analysis demonstrated that DEmRNAs were significantly enriched in immune-related biological functions, for example, toll-like receptor signaling pathway, cytokine-cytokine receptor interaction, and chemokine signaling pathway. In weighted gene co-expression network analysis, turquoise module was the hub module. Moreover, 10 hub miRNAs were identified. Among these 10 hub miRNAs, only 8 hub miRNAs predicted the corresponding target mRNAs. 97 negatively regulated miRNA-mRNA pairs were involved in the miRNA-mRNA regulatory network, for example, hsa-miR-21-5p-claudin 8 (CLDN8), hsa-miR-30a-3p-interleukin-1B (IL-1B), and hsa-miR-181a-5p/hsa-miR-30c-2-3p-C-X-C motif chemokine ligand 9 (CXCL9). Real-time polymerase chain reaction results showed that IL-1B and CXCL9 were up-regulated and CLDN8 was down-regulated in psoriasis with statistically significant differences.
The identification of potential key molecular markers and signaling pathways provides potential research directions for further understanding the molecular mechanisms of psoriasis. This may also provide new research ideas for the prevention and treatment of psoriasis in the future.
Core Tip: Psoriasis is a common chronic, recurrent, immune-regulatory skin and joint disease. Although psoriasis is widespread and has significant negative impact on patients’ life quality, it has not yet been fully diagnosed and treated. Moreover, it is also associated with many other diseases. So far, psoriasis is still a treatable, but incurable, disease. We use weighted gene co-expression network analysis to identify key modules and microRNAs (miRNAs) related to psoriasis and explore potential key pathways related to psoriasis through the targeting relationship of miRNA-mRNA. This provides new research ideas for the prevention and treatment of psoriasis in the future.
- Citation: Shu X, Chen XX, Kang XD, Ran M, Wang YL, Zhao ZK, Li CX. Identification of potential key molecules and signaling pathways for psoriasis based on weighted gene co-expression network analysis. World J Clin Cases 2022; 10(18): 5965-5983
- URL: https://www.wjgnet.com/2307-8960/full/v10/i18/5965.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i18.5965
Psoriasis is a common chronic, recurrent, immune-regulatory skin and joint disease. It has a variety of clinical skin manifestations, but the most common clinical manifestations are erythematous, scaling papules, and plaques[1]. Previous studies have found that psoriasis is affected by family history and age[2]. Mechanical stress, air pollutants, sun exposure, infection, lifestyle, obesity, dyslipidemia, and mental stress are associated with the progression of psoriasis[3]. The pathogenesis of psoriasis is more complicated and often requires long-term treatment. Mild to moderate psoriasis can be treated topically with a combination of glucocorticoids, vitamin D analogues, and phototherapy. Moderate to severe psoriasis usually requires systemic treatment[4]. Psoriasis is also associated with many diseases, for example cardiometabolic diseases, malignant tumors, infections and mood disorders[5]. So far, psoriasis is still a treatable, but incurable, disease. Therefore, identification of key genes and signaling pathways are of great significance for understanding the molecular mechanism of psoriasis and provides potential basis for the treatment and management of psoriasis.
MicroRNAs (miRNAs) are a class of small non-coding single-stranded RNAs that regulate gene expression[6]. MiRNAs play a vital role in the progression of diseases, for example cancer, infectious diseases, and immune diseases[7]. MiRNAs can regulate cell proliferation, keratinocyte differentiation, apoptosis, and atypical immune activation in psoriasis[8]. Previous studies have found that miR-187 can inhibit the proliferation of keratinocytes by targeting B7 homolog 3 protein. In addition, in the mouse model of psoriasis, overexpression of miR-187 can reduce acanthosis and reduce the severity of the disease[9]. MiR-183-3p can inhibit the proliferation and migration of keratinocytes in psoriasis by inhibiting growth factor receptor binding 2-associated binding protein 1[10]. MiR-320b negatively regulates keratinocyte proliferation in psoriasis by targeting AKT serine/threonine kinase 3[11]. The low expression of miR-31 in dermal mesenchymal stem cells (DMSCs) in patients with psoriasis results in the increased expression of some of its target genes, which in turn promotes the activation of T lymphocyte by inhibiting the proliferation of DMSCs, thereby contributing to the pathogenesis of psoriasis[12]. Previous studies have found that miR-146 regulates inflammatory responses in keratinocytes and skin fibroblasts, which may affect the pathogenesis of psoriasis[13]. Furthermore, in human keratinocytes, the expression of miR-146a is induced by pro-inflammatory cytokines tumor necrosis factor-α (TNF-α), interleukin-1B (IL-1B), and IL-17a, and the expression of miR-146b is induced by interferon-γ (IFN-γ) and IL-22. MiR-203 promotes keratinocyte proliferation by targeting liver X receptor-α and peroxisome proliferator-activated receptor-γ in psoriasis[14]. MiR-125 regulates keratinocytes proliferation by regulating targeted genes[15,16]. MiR-197 over expression inhibits keratinocytes proliferation induced by IL-22 and keratinocytes migration[17]. In addition, miR-99 also plays an important role in regulating the abnormal proliferation and differentiation of keratinocytes in psoriasis[18]. These studies demonstrate that miRNAs play a vital role in the pathogenesis of psoriasis. Currently, miRNAs studies in psoriasis and dermatology are relatively new. Therefore, the identification of key miRNAs in psoriasis is helpful to elucidate the molecular mechanism of psoriasis.
Weighted gene co-expression network analysis (WGCNA) can be used for finding clusters (modules) of highly correlated genes[19]. WGCNA has been used to identify key genes for many diseases, including cancer[20], cardiovascular disease[21], and immunological diseases[22]. In addition, WGCNA has also been used to identify key mRNAs and long noncoding RNAs in psoriasis. So far, we have not found studies that use WGCNA to identify key miRNAs in psoriasis. Therefore, we use WGCNA to identify key modules and miRNAs related to psoriasis and explore potential key pathways related to psoriasis through the targeting relationship of miRNA-mRNA.
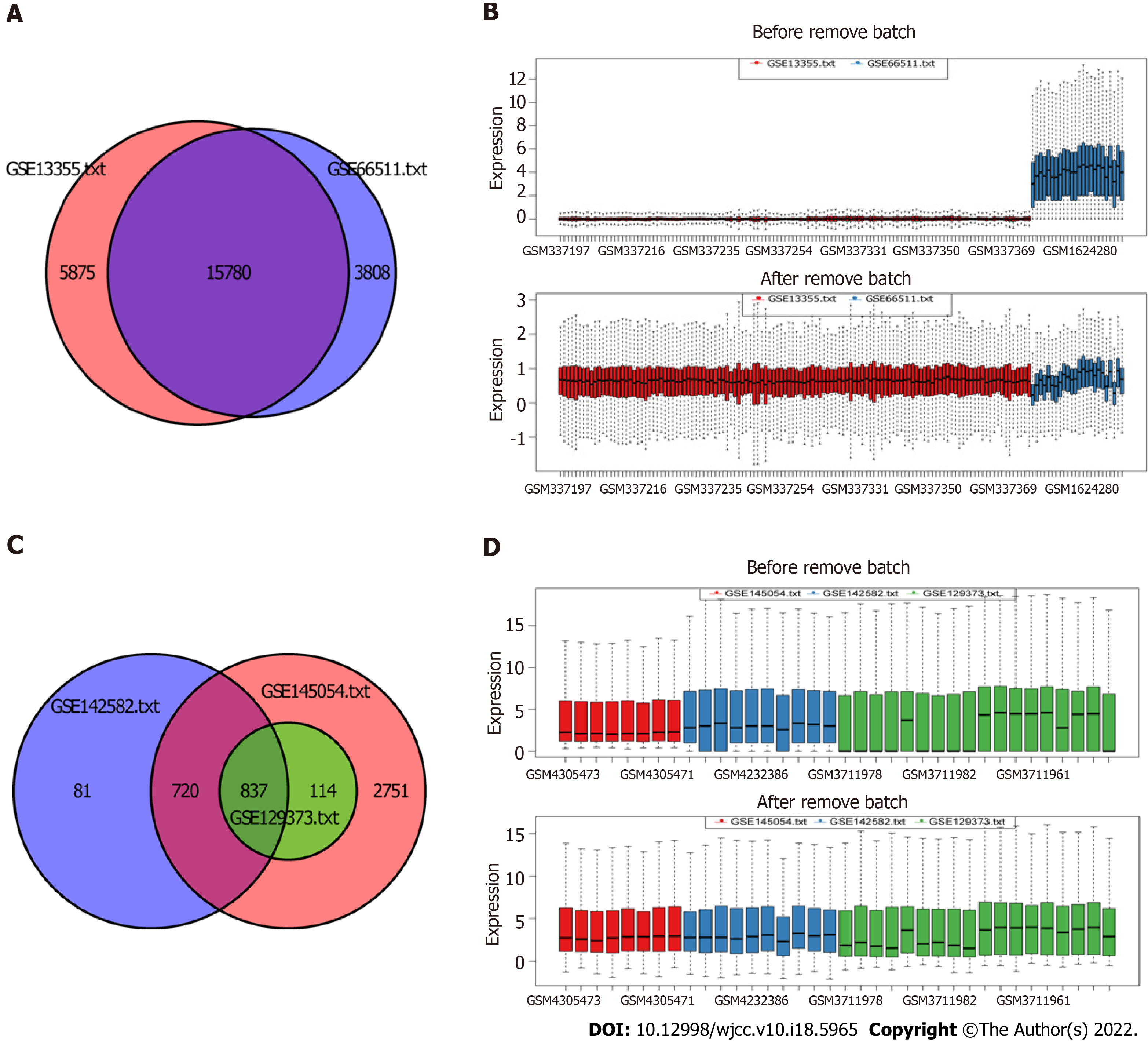
The miRNA and mRNA data were obtained from the Gene Expression Omnibus (GEO) database[23]. The keyword we searched in the GEO database was “psoriasis”. Cell line or animal level studies and single-sample studies were excluded. Five datasets GSE13355, GSE66511, GSE145054, GSE142582, and GSE129373 were involved in this study (Table 1). All data were derived from skin samples. GSE13355 and GSE66511 were mRNA data from GPL570 and GPL16288 platform, respectively. GSE145054, GSE142582, and GSE129373 were miRNA data from GPL19117, GPL20301, and GPL11154, respectively. The gene expression matrix files in GSE13355 and GSE145054 were downloaded. The GPL annotation file was used to annotate the expression matrix. The probe ID was converted to the gene symbol. Multiple probes correspond to the same gene, taking the average value. The probe that corresponds to multiple genes was removed. The original data of gene expression profile in GSE66511, GSE142582, and GSE129373 were downloaded, and performed by logarithm processing. Then, the combat function in “sva” R package was used to remove batch effects.
| GEO ID | Samples, Normal:Case | Platforms | Author | Type | Source |
| GSE13355 | 64:58 | GPL570 | Gudjonsson JE | mRNA | Skin |
| GSE66511 | 12:12 | GPL12688 | Keermann M | mRNA | Skin |
| GSE145054 | 4:4 | GPL19117 | Mildner M | miRNA | Skin |
| GSE142582 | 5:5 | GPL20301 | Yu Z | miRNA | Skin |
| GSE129373 | 9:9 | GPL11154 | Srivastava A | miRNA | Skin |
After the above pretreatment, differentially expressed mRNAs (DEmRNAs) and differentially expressed miRNAs (DEmiRNAs) were screened out by limma R package[24]. DEmRNAs screening criteria were adjusted P (adj. P) value < 0.01 and |logFoldChange| (|logFC|) > 1. DEmiRNAs screening criteria were adj. P value < 0.01 and |logFC| > 1.5.
To investigate the possible involvement of DEmRNAs in the biological processes related to psoriasis, we performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomics (KEGG) functional enrichment analyses. David database was used for the enrichment analysis of DEmRNAs[25], and the screening criterion was P < 0.05. GO enrichment analysis, which is comprised of biological processes (BP), cellular components (CC), and molecular functions (MF). The “GOplot” R package was used to visualize the enrichment results.
The “WGCNA” R package was used to analyze the co-expression network of all miRNAs. To detect outliers, the “hclust” function was used to cluster the sample data. Subsequently, to construct a scale-free topology, the “pickSoftThreshold” function was used to select an appropriate soft threshold power regulator (height 0.90, β value 2). Calculate the adjacency matrix based on the kernel value. The adjacency matrix was transformed into topological overlap matrix (TOM) and corresponding dissimilarity matrix (1-TOM). Genes with similar expression patterns were gathered together, and modules were divided according to the function of “cutreeDynamic” with default parameters. Since the modules identified by the dynamic tree cutting algorithm may be similar, they are combined with a truncation of 0.25 height[26].
To identify the important modules associated with psoriasis, the module eigengene (ME) of each module was calculated using the “moduleEigengenes” function. Then, the correlation between ME and psoriasis was analyzed using Pearson method. Subsequently, the module with the highest correlation with psoriasis was identified as the hub module. According to module connectivity (MM) and clinical trait relationship (GS) of each gene in the hub module, the candidate hub miRNAs were screened out[27]. The screening criteria were MM > 0.8 and GS > 0.5. Finally, the intersection of candidate hub miRNAs and DEmiRNAs were selected as real hub miRNAs.
To explore the correlation between miRNA and mRNA, we constructed a miRNA-mRNA regulatory network. Target mRNAs of miRNAs were predicted using miRDB (http://mirdb.org) database. MiRNA-mRNA pairs involved in common DEmRNA and negatively regulated were selected to construct the network. Cytoscape (www.cytoscape.org/) was used to visualize the miRNA-mRNA regulatory network.
We collected skin tissue samples from healthy control individuals and patients with psoriasis. Basic information (including age, sex, etc) of patients and healthy controls were recorded in detail during sample collection (Table 2). The specific inclusion criteria of patients with psoriasis: (1) Patients had at least one well-circumscribed erythematous and squamous lesion, which had been confirmed by at least two dermatologists; (2) Patient’s pathological tissues were confirmed by clinical histopathology; (3) Patients had no systemic anti-psoriatic treatment 2 wk before the skin biopsy; and (4) Patients had no topical anti-psoriatic treatment 1 wk before the skin biopsy. Patients with psoriasis who were treated with immune-related drugs before sampling and whose clinical information was incomplete were excluded. The individuals in the normal control group were sex and age matched with the psoriasis group, and without history of psoriasis or other autoimmune diseases.
| Group | Sample number | Sampling location | Sex | Age | Height, cm | Body weight, kg | BMI | Smoking | Drinking | History of ASCVD | History of hypertension | History of hyperlipidemia | History of diabetes | Psoriatic arthritis | Severity index, PASI | Whether to collect non-lesion parts |
| Normal control group | C1 | Right forearm extension side | Female | 47 | 158 | 54 | 21.6 | No | No | No | No | No | No | |||
| C2 | Right sole of the foot | Male | 60 | 178 | 70 | 22.1 | Yes | Yes | No | No | No | No | ||||
| C3 | Left sole of the foot | Male | 59 | 183 | 81 | 24.2 | No | No | No | No | No | No | ||||
| C4 | Left armpit | Female | 27 | 160 | 52 | 20.3 | No | Yes | No | No | No | No | ||||
| C7 | Left forearm extension side | Female | 18 | 160 | 50 | 19.5 | No | No | No | No | No | No | ||||
| C9 | Right shoulder | Female | 27 | 161 | 62 | 23.9 | No | No | No | No | No | No | ||||
| C11 | Right calf | Female | 60 | 156 | 55 | 22.6 | No | No | No | No | No | No | ||||
| Psoriasis group | E2-2 | Right forearm | Male | 19 | 177 | 58 | 18.5 | No | No | No | No | No | No | No | 10.5 | Yes |
| E3-2 | Back | Female | 58 | 157 | 58 | 23.5 | No | No | No | No | Yes | No | No | 8 | Yes | |
| E5-2 | Left calf stretched side | Male | 55 | 176 | 68 | 22 | No | No | No | Yes | No | Yes | No | 13.4 | Yes | |
| E6-2 | Outer left thigh | Female | 44 | 166 | 67 | 24.3 | No | No | No | No | No | No | No | 11.9 | Yes | |
| E7-2 | Right thigh internal test | Male | 62 | 170 | 66 | 22.8 | Yes | No | No | No | No | No | No | 8.6 | Yes | |
| E8-2 | Right forearm extension side | Male | 48 | 170 | 58 | 20.1 | Yes | No | No | No | No | No | No | 15.4 | Yes | |
| E10-2 | Back | Male | 18 | 172 | 60 | 20.3 | Yes | No | No | No | No | No | No | 22.2 | Yes |
According to screening criteria, 7 normal and 7 patient skin tissue samples were obtained. Firstly, total RNA of the samples was extracted using TRIzol. Then, fastKing cDNA first strand synthesis kit and miRNA first strand cDNA synthesis kit (Tailing Reaction) were used for mRNA and miRNA reverse transcription, respectively. Subsequently, SuperReal PreMix Plus (SYBR Green) and miRNA quantitative PCR kit (SYBR Green Method) were used to perform real-time polymerase chain reaction (RT-PCR) validation of mRNA and miRNA, respectively. GAPDH, ACTB, and hsa-U6 were internal reference. ABI7300 fluorescence quantitative PCR instrument was used for detection. Finally, the 2-ΔΔCt method was used for relative quantitative analysis of the data[28]. This study complied with the Declaration of Helsinki. Informed consent was obtained from the participants. All experimental procedures were approved by the ethics committee of Chinese PLA General Hospital, No. S2021-012-01.
All statistical analyses were performed using R software. Limma R package was used to screen for DEmRNAs and DEmiRNAs. Pearson’s method was used to analyze the correlation between ME and psoriasis. RT-PCR validation data were statistically analyzed by one-way ANOVA. P < 0.05 was statistically significant.
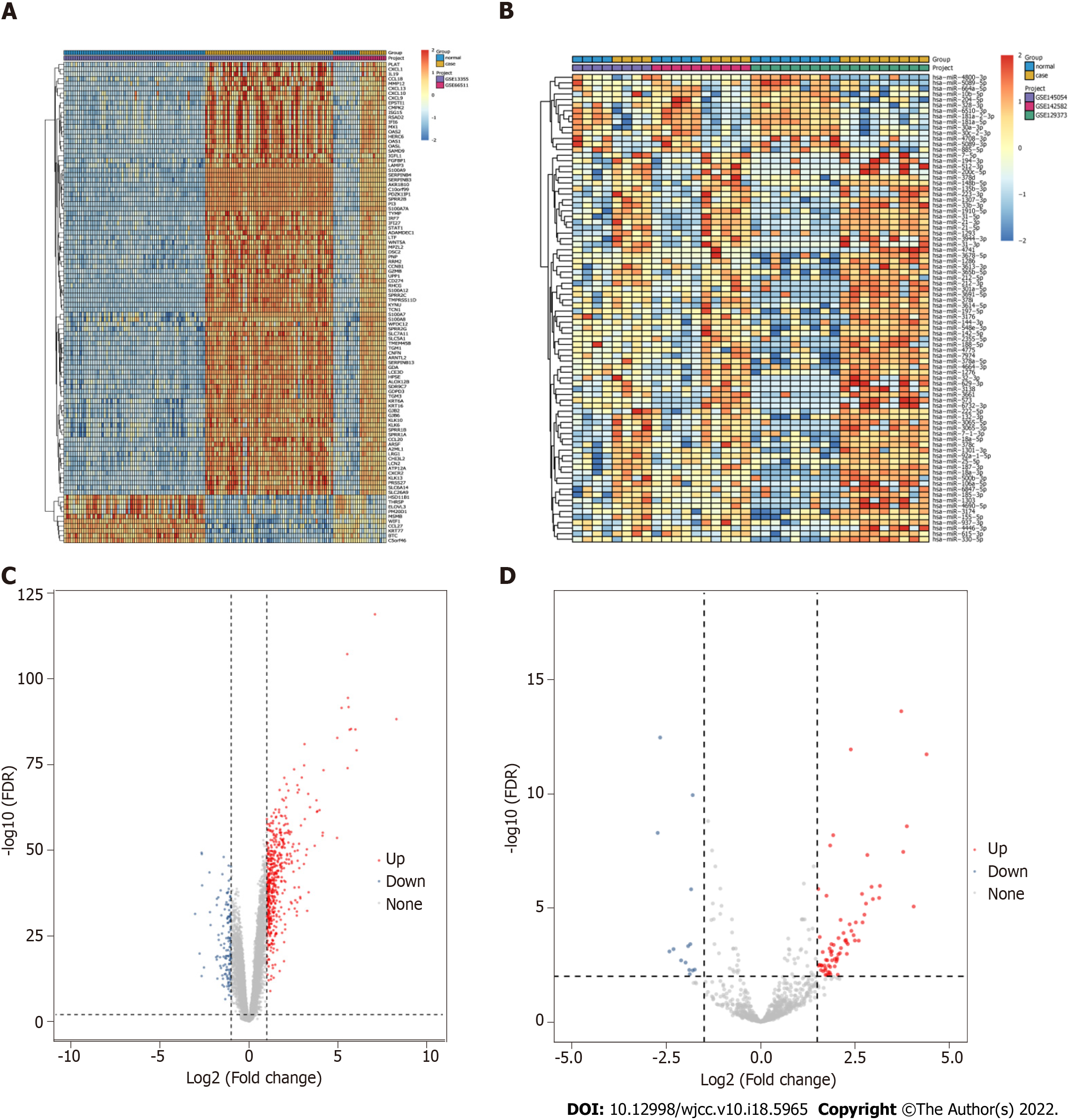
After pretreatment of the raw data, 15780 mRNAs and 837 miRNAs were screened out (Figure 1). Compared with the control group, 639 DEmRNAs were identified in the psoriasis group (adj. P value < 0.01 and |logFC| > 1). Among them, 497 were up-regulated and 142 were down-regulated. Compared with the control group, 84 DEmiRNAs were identified in the psoriasis group (adj. P value < 0.01 and |logFC| > 1.5). Among them, 70 were up-regulated and 14 were down-regulated. Heat maps showed that there were significant differences of mRNA (Figure 2A) and miRNA (Figure 2B) expression between psoriasis group and control group. The volcano maps of DEmRNAs and DEmiRNAs were shown in Figure 2C and D.
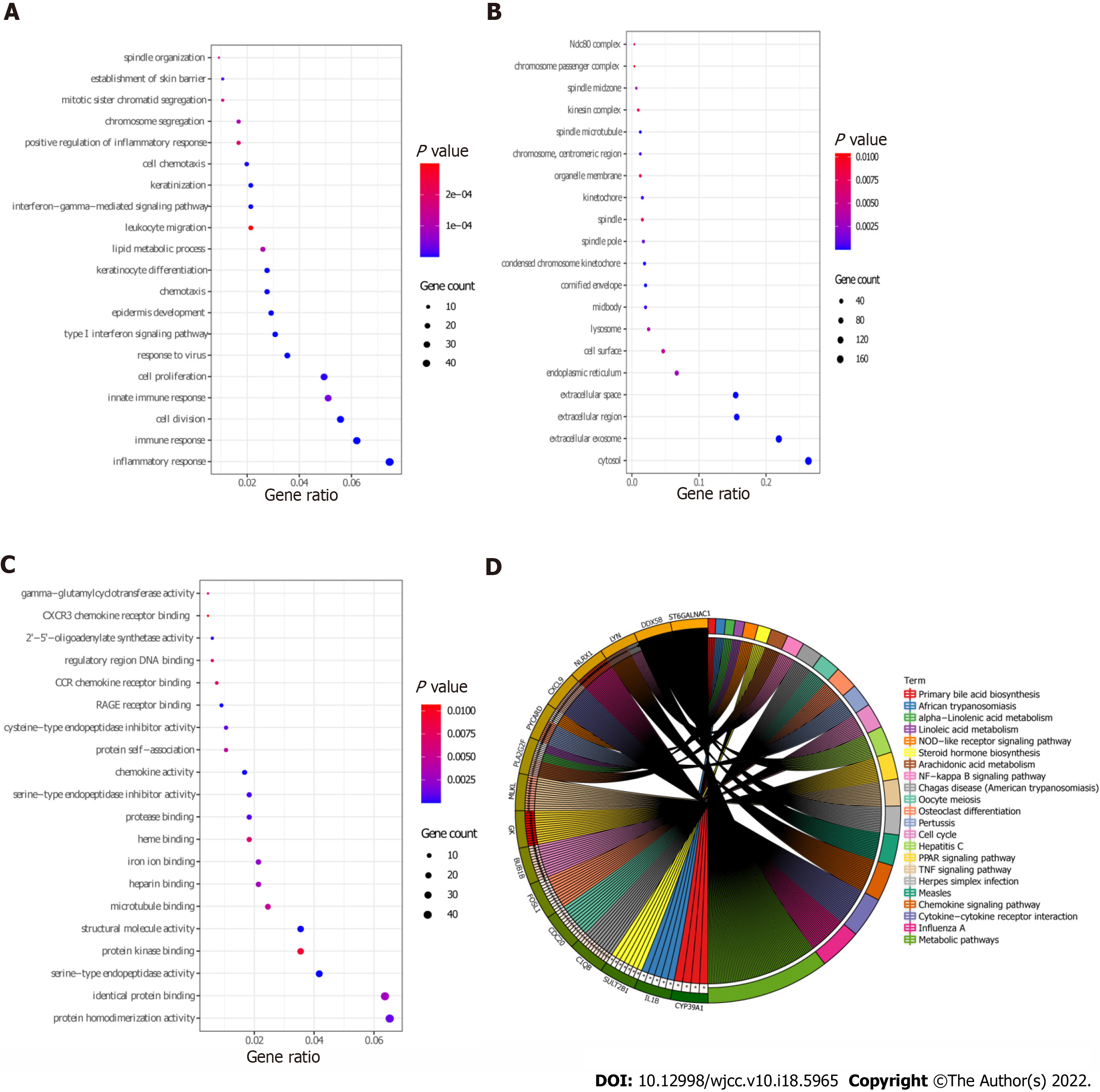
Functional analysis of 639 DEmRNAs was performed using David database (P < 0.05). GO enrichment results showed that most of the DEmRNAs were distributed in inflammatory response (GO:BP), immune response (GO:BP), cytosol (GO:CC), extracellular exosome (GO:CC), protein homodimerization activity (GO:MF), and other biological functions (Figure 3A-C). KEGG enrichment results demonstrated that DEmRNAs were significantly enriched in metabolic pathways, influenza A, chemokine signaling pathway, and cytokine-cytokine receptor interaction (Figure 3D and Table 3).
| Category | hsa | Term | Count | P value | Genes |
| KEGG_PATHWAY | hsa03320 | PPAR signaling pathway | 14 | 3.07E-06 | GK, MMP1, ADIPOQ, LPL, FADS2, ACADL, ACOX2, FABP5, FABP7, ACSBG1, PLIN1, ANGPTL4, SLC27A4, PPARD |
| KEGG_PATHWAY | hsa05164 | Influenza A | 22 | 1.05E-05 | NLRX1, RSAD2, DDX58, STAT1, TMPRSS4, MX1, MAPK13, PYCARD, CXCL10, SOCS3, OAS1, OAS2, IL-1B, OAS3, TNFSF10, IRF7, CCL2, PRSS3, KPNA2, PRSS2, MYD88, IRF9 |
| KEGG_PATHWAY | hsa05133 | Pertussis | 12 | 2.56E-04 | PYCARD, C1QB, CASP7, NOS2, CALML5, IL-1B, IRF1, IRF8, CALML3, FOS, MYD88, MAPK13 |
| KEGG_PATHWAY | hsa05162 | Measles | 16 | 4.14E-04 | DDX58, STAT1, MX1, CD3G, CD3D, OAS1, CCNE2, CCNE1, OAS2, IL-1B, OAS3, TNFSF10, IRF7, PRKCQ, MYD88, IRF9 |
| KEGG_PATHWAY | hsa04668 | TNF signaling pathway | 14 | 4.87E-04 | MLKL, CCL20, CXCL1, FOS, NOD2, SELE, MMP9, MAPK13, CXCL10, SOCS3, CASP7, IL-1B, CCL2, JUNB |
| KEGG_PATHWAY | hsa04062 | Chemokine signaling pathway | 19 | 7.44E-04 | LYN, CXCL9, CCL22, STAT1, CCL20, CXCR4, CXCL1, CXCL13, CXCL10, HCK, CCL8, PLCB4, CXCR2, RAC2, CCL2, CCR7, CCL19, CCL18, CCL27 |
| KEGG_PATHWAY | hsa04060 | Cytokine-cytokine receptor interaction | 21 | 0.002838 | CXCL9, IL20, IL4R, CCL22, CCL20, CXCR4, IL19, CXCL1, CXCL13, CXCL10, CCL8, IL-1B, CXCR2, TNFSF10, CCL2, CCR7, CCL19, IL7R, CCL18, CCL27, TNFRSF21 |
| KEGG_PATHWAY | hsa05160 | Hepatitis C | 14 | 0.003635 | DDX58, STAT1, IFIT1, CLDN1, MAPK13, SOCS3, OAS1, OAS2, IRF1, OAS3, CLDN8, IRF7, CLDN17, IRF9 |
| KEGG_PATHWAY | hsa00590 | Arachidonic acid metabolism | 9 | 0.003655 | PLA2G2F, HPGDS, GPX2, PLA2G4D, PLA2G2A, PLA2G3, ALOX12B, CYP2E1, CYP4F8 |
| KEGG_PATHWAY | hsa04110 | Cell cycle | 13 | 0.00555 | BUB1B, TTK, CDC6, CDC25B, CCNA2, CDC20, CCNB2, CCNB1, CCNE2, PTTG1, CCNE1, CDK1, MAD2L1 |
| KEGG_PATHWAY | hsa05142 | Chagas disease (American trypanosomiasis) | 11 | 0.011418 | C1QB, GNA15, PLCB4, NOS2, IL-1B, CCL2, CD3G, FOS, CD3D, MYD88, MAPK13 |
| KEGG_PATHWAY | hsa04114 | Oocyte meiosis | 11 | 0.017478 | CDC20, CCNB2, CCNB1, PTTG1, CALML5, CCNE2, CCNE1, CDK1, CALML3, MAD2L1, AURKA |
| KEGG_PATHWAY | hsa00592 | Alpha-Linolenic acid metabolism | 5 | 0.018996 | PLA2G2F, FADS2, PLA2G4D, PLA2G2A, PLA2G3 |
| KEGG_PATHWAY | hsa05168 | Herpes simplex infection | 15 | 0.020979 | DDX58, STAT1, TAP1, FOS, IFIT1, SOCS3, OAS1, OAS2, IL-1B, OAS3, IRF7, CDK1, CCL2, MYD88, IRF9 |
| KEGG_PATHWAY | hsa04380 | Osteoclast differentiation | 12 | 0.021031 | FOSL1, SOCS3, FCGR3B, STAT1, LCK, IL-1B, BLNK, ACP5, FOS, JUNB, IRF9, MAPK13 |
| KEGG_PATHWAY | hsa04064 | NF-kappa B signaling pathway | 9 | 0.028614 | LYN, BCL2A1, DDX58, LCK, IL-1B, BLNK, PRKCQ, CCL19, MYD88 |
| KEGG_PATHWAY | hsa04621 | NOD-like receptor signaling pathway | 7 | 0.028679 | PYCARD, IL-1B, CARD6, CCL2, CXCL1, NOD2, MAPK13 |
| KEGG_PATHWAY | hsa00591 | Linoleic acid metabolism | 5 | 0.031316 | PLA2G2F, PLA2G4D, PLA2G2A, PLA2G3, CYP2E1 |
| KEGG_PATHWAY | hsa00120 | Primary bile acid biosynthesis | 4 | 0.031945 | CYP39A1, CH25H, ACOX2, CYP7B1 |
| KEGG_PATHWAY | hsa00140 | Steroid hormone biosynthesis | 7 | 0.033377 | SULT2B1, HSD11B1, STS, HSD3B1, HSD17B2, CYP2E1, CYP7B1 |
| KEGG_PATHWAY | hsa01100 | Metabolic pathways | 64 | 0.037857 | ST6GALNAC1, GDA, GLDC, PIK3C2G, PYGL, HK2, SMPD3, IL4I1, HPGDS, PNP, ACADL, SPTLC2, KYNU, NAMPT, LIPG, HYAL4, PGM2, TK1, UPP1, AASS, PLA2G4D, ARG1, GPT2, AMPD3, NME1, ALDH3A1, PLCB4, ACOX2, CMPK2, ACSBG1, CYP2E1, IDO1, PLA2G3, ALOX12B, FUT2, CYP2C18, FUT1, FUT3, TYMP, HSD11B1, UGCG, RDH12, HSD17B2, RDH16, ATP6V0A4, XDH, HAO2, PLA2G2F, GALNT6, RRM2, GK, GCH1, NOS2, PLA2G2A, HSD3B1, CYP4F8, ALDH4A1, SQLE, HAL, AKR1B10, DHRS9, POLE2, POLR3G, HPSE |
| KEGG_PATHWAY | hsa05143 | African trypanosomiasis | 5 | 0.047416 | PLCB4, IL-1B, SELE, MYD88, IDO1 |
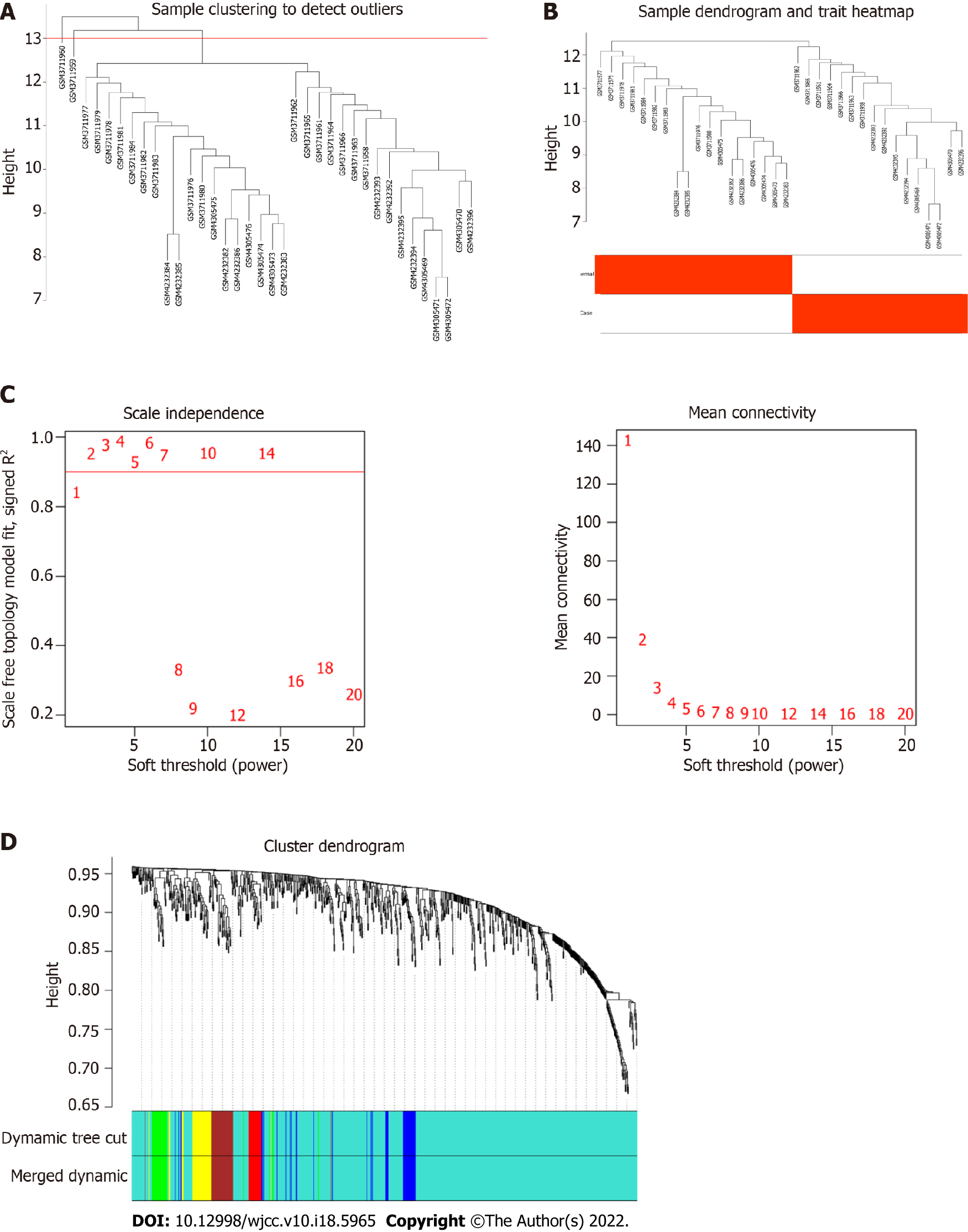
WGCNA was performed on 837 miRNAs. The samples were clustered to remove abnormal samples (GSM3711960 and GSM3711959) (Figure 4A and B). After calculation, when the soft threshold β was 2, it was approximately a scale-free topology (Figure 4C). After determining the soft threshold, the cluster tree graph was constructed. Subsequently, with the minimum number of genes for modules set to 20, the modules with dissimilarity < 25% merged using the dynamic tree cutting method. Finally, 6 modules were determined (Figure 4D).
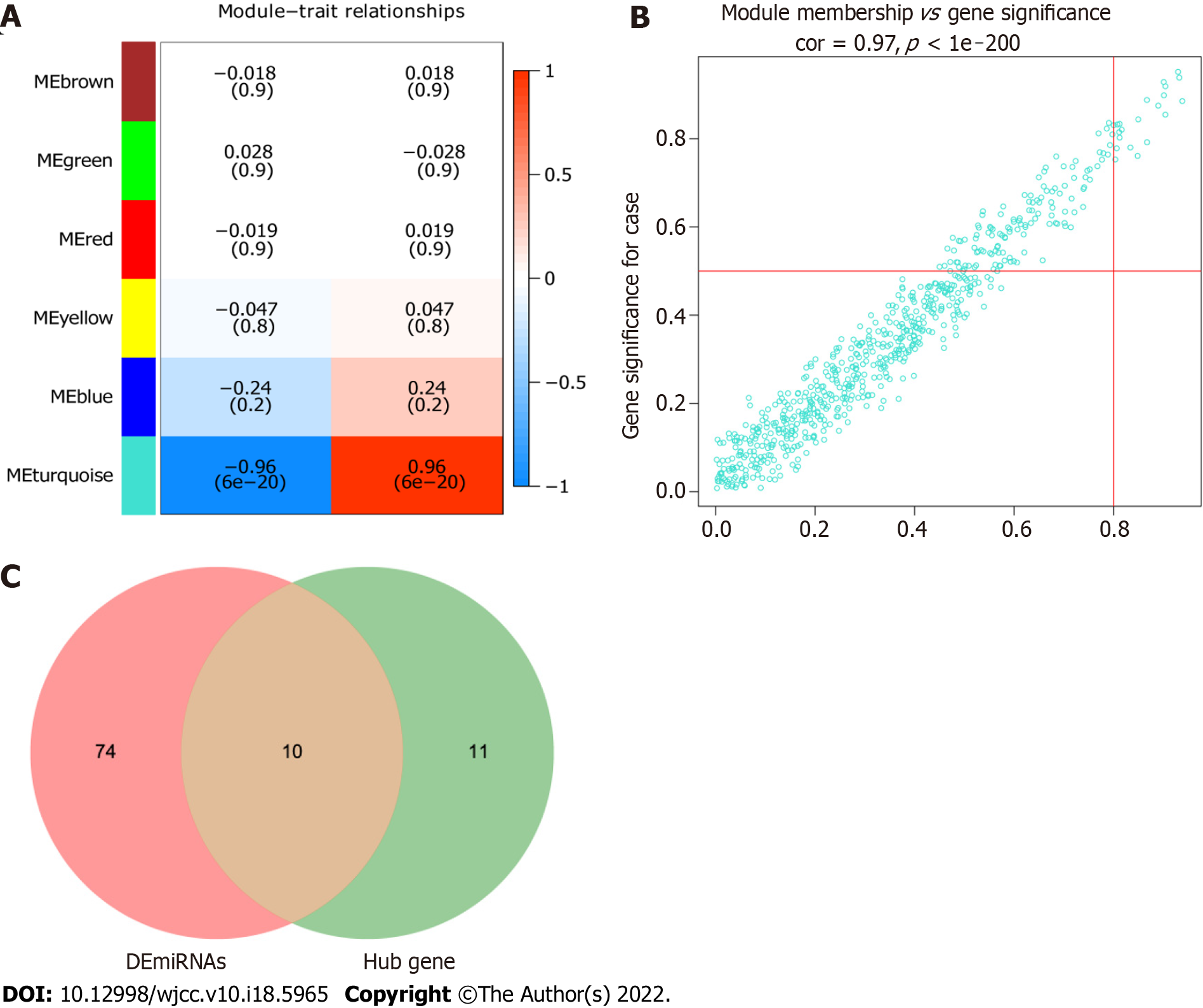
The correlation between modules and psoriasis was analyzed by Pearson’s method. The results showed that turquoise module had the highest correlation with psoriasis (r = 0.96) (Figure 5A). Therefore, turquoise module was considered the hub module. Subsequently, 21 miRNAs were screened out from the turquoise module as candidate hub miRNAs (MM > 0.8 and GS > 0.5) (Figure 5B). Intersection of DEmiRNAs and candidate hub miRNAs was obtained (Figure 5C). Ten intersecting miRNAs were identified as real hub miRNA. Among them, 5 hub miRNAs were up-regulated (hsa-miR-21-3p, hsa-miR-21-5p, hsa-miR-31-5p, hsa-miR-18a-5p, and hsa-miR-33b-3p) and 5 hub miRNAs were down-regulated (hsa-miR-181a-2-3p, hsa-miR-181a-5p, hsa-miR-6510-3p, hsa-miR-30a-3p, and hsa-miR-30c-2-3p).
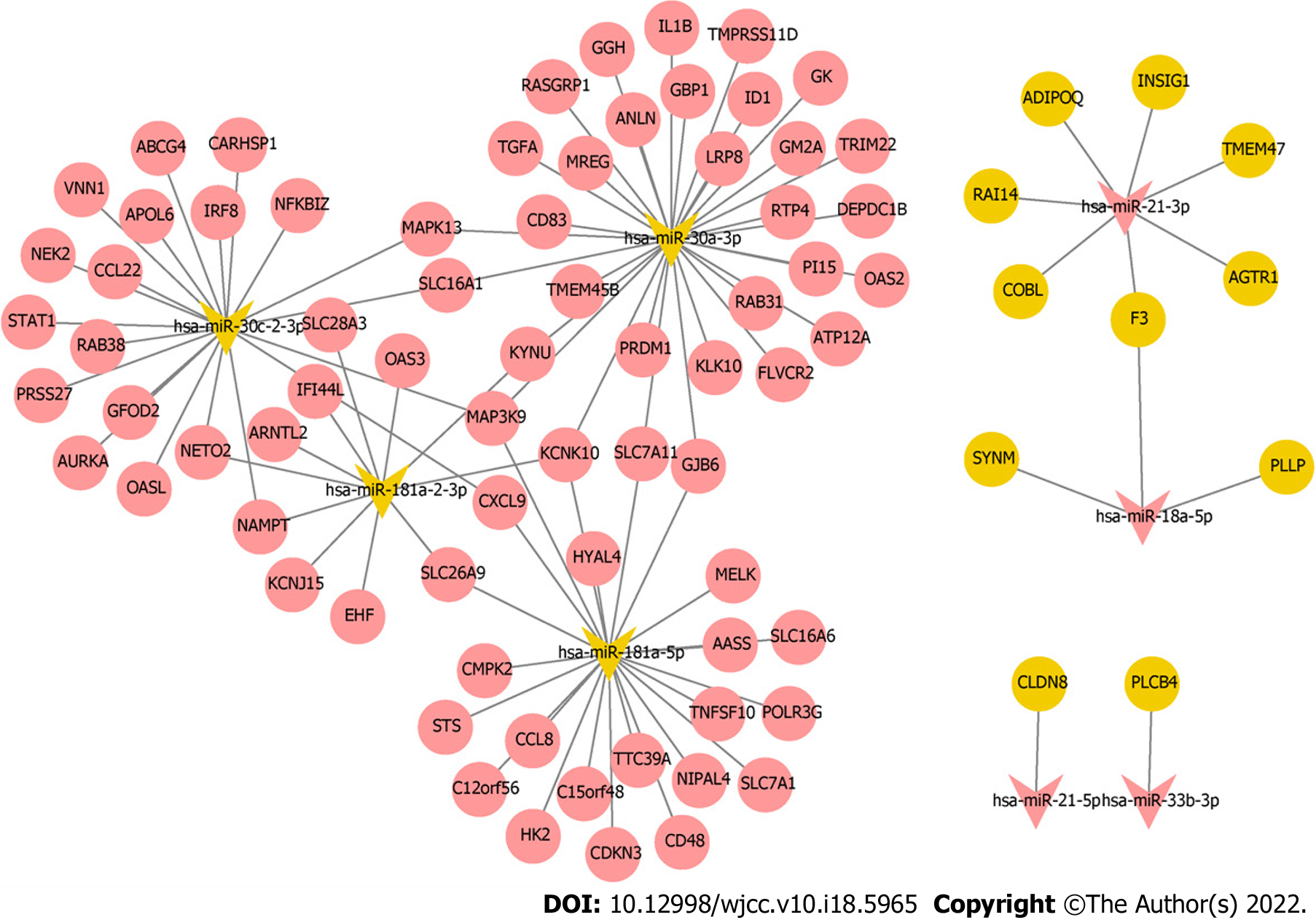
Target mRNAs of 10 hub miRNAs were predicted using miRDB database, but only 8 hub miRNAs (4 up-regulated and 4 down-regulated) were predicted to the corresponding target mRNAs. Eleven target mRNAs of up-regulated hub miRNAs (hsa-miR-21-3p, hsa-miR-21-5p, hsa-miR-18a-5p, and hsa-miR-33b-3p) were matched with DEmRNAs, and 72 target mRNAs of down-regulated hub miRNAs (hsa-miR-181a-2-3p, hsa-miR-181a-5p, hsa-miR-30a-3p, and hsa-miR-30c-2-3p) were matched with DEmRNAs. Ninety-seven negatively regulated miRNA-mRNAs were involved in the miRNA-mRNA regulatory network (Figure 6); for example, hsa-miR-21-3p/hsa-miR-18a-5p-F3, hsa-miR-21-5p-claudin 8 (CLDN8), hsa-miR-33b-3p-PLCB4, hsa-miR-30a-3p-IL-1B, hsa-miR-181a-5p-C-C motif chemokine ligand 8 (CCL8), hsa-miR-181a-5p/hsa-miR-30c-2-3p-C-X-C motif chemokine ligand 9 (CXCL9), and hsa-miR-30c-2-3p-KYNU.
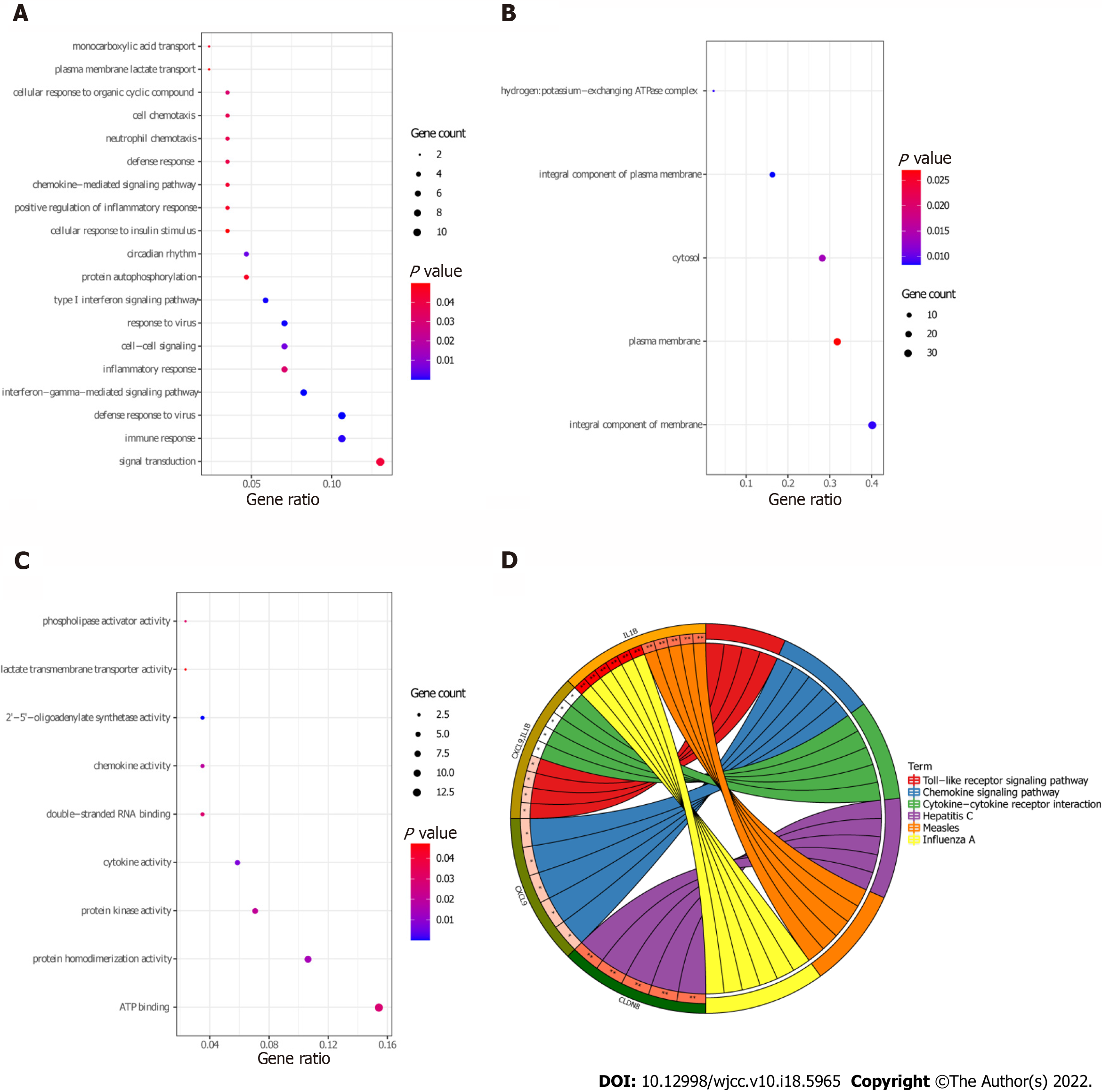
Functional analysis of 83 target DEmRNAs was performed using David database (P < 0.05). GO enrichment results demonstrated that most of the target DEmRNAs were distributed in signal transduction (GO:BP), immune response (GO:BP), integral component of membrane (GO:CC), plasma membrane (GO:CC), ATP binding (GO:MF), and other biological functions (Figure 7A-C). KEGG enrichment results demonstrated that the target DEmRNAs were significantly enriched in influenza A, hepatitis C, measles, and chemokine signaling pathway (Figure 7D and Table 4).
| Category | hsa | Term | Count | P value | Genes |
| KEGG_PATHWAY | hsa05164 | Influenza A | 6 | 0.002804 | STAT1, OAS2, IL-1B, OAS3, TNFSF10, MAPK13 |
| KEGG_PATHWAY | hsa05160 | Hepatitis C | 5 | 0.006506 | STAT1, OAS2, CLDN8, OAS3, MAPK13 |
| KEGG_PATHWAY | hsa05162 | Measles | 5 | 0.006506 | STAT1, OAS2, IL-1B, OAS3, TNFSF10 |
| KEGG_PATHWAY | hsa04062 | Chemokine signaling pathway | 5 | 0.020355 | CXCL9, CCL8, PLCB4, CCL22, STAT1 |
| KEGG_PATHWAY | hsa04620 | Toll-like receptor signaling pathway | 4 | 0.021751 | CXCL9, STAT1, IL-1B, MAPK13 |
| KEGG_PATHWAY | hsa04060 | Cytokine-cytokine receptor interaction | 5 | 0.047526 | CXCL9, CCL8, CCL22, IL-1B, TNFSF10 |
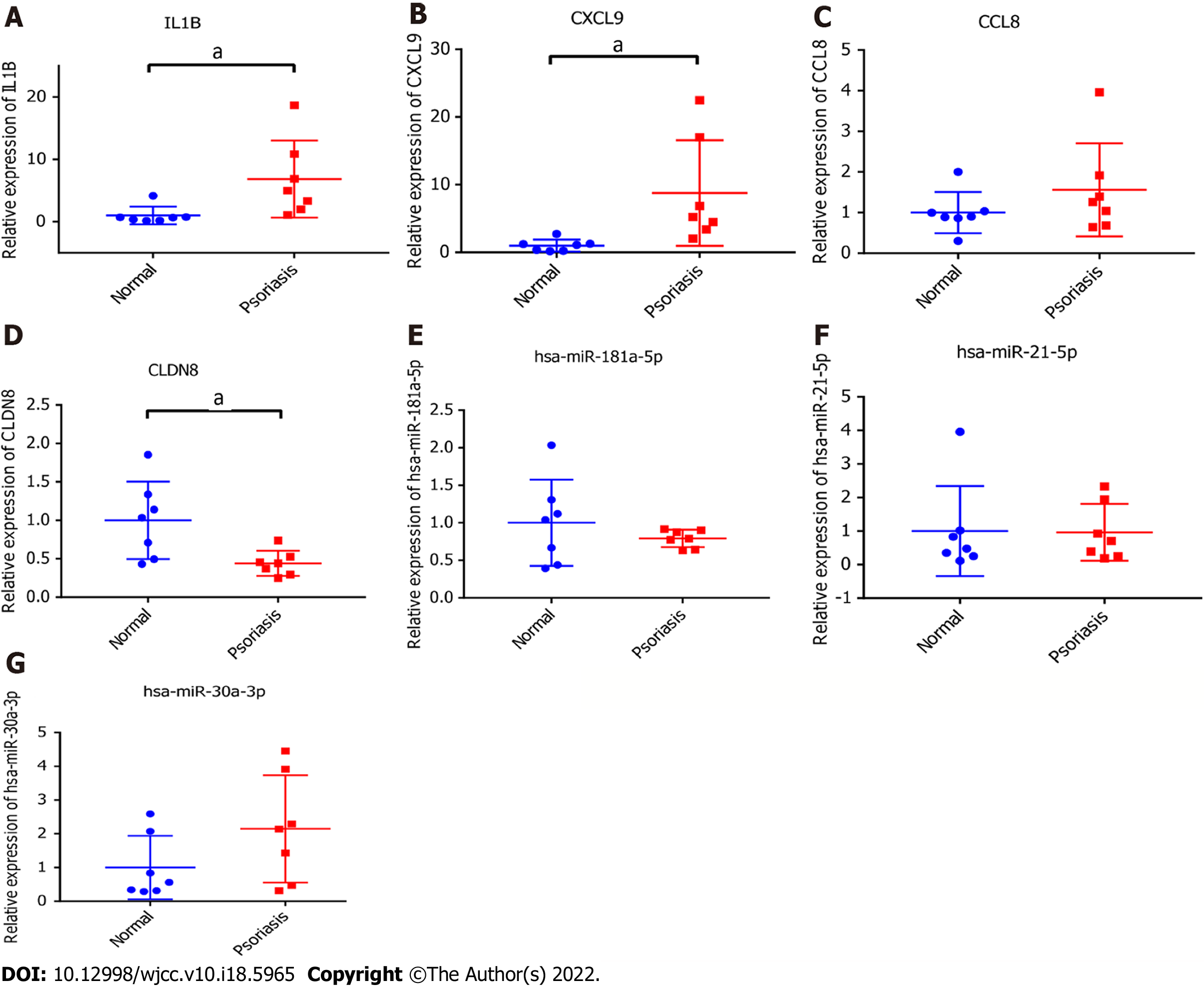
IL-1B, CXCL9, CLDN8, CCL8, hsa-miR-181a-5p, hsa-miR-30a-3p, and hsa-miR-21-5p related to psoriasis were selected from hub miRNAs and target DEmRNAs for in vitro validation. Primers for each mRNAs and miRNAs were shown in Table 5. IL-1B, CXCL9 and CCL8 were up-regulated and CLDN8 and hsa-miR-181a-5p were down-regulated in psoriasis tissues (Figure 8). Among them, IL-1B, CXCL9, and CLDN8 showed significant difference in expression levels. In addition, we found that expression trends of hsa-miR-30a-3p and hsa-miR-21-5p were contrary to the results of bioinformatics analysis. The reason for the inconsistency between RT-PCR and bioinformatics analysis results may be the small sample size. Further research is needed.
| Primer name | Primer sequence, 5’ to 3’ |
| GAPDH-F (internal reference) | 5-GGAGCGAGATCCCTCCAAAAT-3 |
| GAPDH-R (internal reference) | 5-GGCTGTTGTCATACTTCTCATGG-3 |
| ACTB-F (internal reference) | 5-CATGTACGTTGCTATCCAGGC-3 |
| ACTB-R (internal reference) | 5-CTCCTTAATGTCACGCACGAT-3 |
| IL-1B-F | 5-TTCGACACATGGGATAACGAGG-3 |
| IL-1B-R | 5-TTTTTGCTGTGAGTCCCGGAG-3 |
| CXCL9-F | 5-CCAGTAGTGAGAAAGGGTCGC-3 |
| CXCL9-R | 5-AGGGCTTGGGGCAAATTGTT-3 |
| CCL8-F | 5-TGGAGAGCTACACAAGAATCACC-3 |
| CCL8-R | 5-TGGTCCAGATGCTTCATGGAA-3 |
| CLDN8-F | 5-CTACAGGCAGCCAGAGGACT-3 |
| CLDN8-R | 5-ACAGGGATGAGCACCACCAT-3 |
| hsa-U6 (internal reference) | |
| hsa-miR-30a-3p | 5-CTTTCAGTCGGATGTTTGCAGC-3 |
| hsa-miR-181a-5p | 5-AACATTCAACGCTGTCGGTGAGT-3 |
| hsa-miR-21-5p | 5-TAGCTTATCAGACTGATGTTGA-3 |
Although psoriasis is widespread and has significant negative impact on patients’ quality of life, it has not yet been fully diagnosed and treated. In the research, we used the WGCNA method to identify 10 hub miRNAs that may be related to the pathogenesis of psoriasis. Then, target mRNAs of hub miRNAs were predicted by using miRDB database. Only 8 hub miRNAs were predicted to the corresponding target mRNAs. Subsequently, to understand the key biological functions involved in DEmRNAs and target DEmRNAs, we performed GO and KEGG functional analysis. Results demonstrated that they were significantly enriched in immune-related biological functions, for example, immune response, cell chemotaxis, and chemokine signaling pathway.
Hsa-miR-21-5p is abnormally expressed in psoriasis, but the specific molecular mechanism remains unclear[29]. In this study, results demonstrated that hsa-miR-21-5p expression was up-regulated and negatively regulated with CLDN8. CLDN8 is down-regulated in psoriasis and has an important molecular regulatory role[30]. CLDN8 has also been identified as a key downstream component of the IL-9 and IL-23 inflammatory cascade[31,32]. In addition, KEGG functional enrichment analysis showed that CLDN8 was enriched in hepatitis C. Cohen et al[33] found that psoriasis is associated with hepatitis C[33]. Cathelicidin, toll like receptor 9 (TLR9), IFN-γ, and TNF-α are inflammatory cytokines. Hepatitis C may increase susceptibility to psoriasis by up-regulating these inflammatory factors[34,35]. Thus, we hypothesized that hsa-miR-21-5p may play a vital regulatory role in hepatitis C through regulating CLDN8 and, thus, affect the pathogenesis of psoriasis.
Hsa-miR-30a-3p can affect the migration and proliferation of cancer cells by targeting related genes[36,37]. Hsa-miR-30a-3p is also contacted with platelet apoptosis and adhesion in immune thrombocytopenia[38]. However, we have not found any report about hsa-miR-30a-3p in psoriasis. In this study, we found that hsa-miR-30a-3p was up-regulated and negatively regulated with multiple DEmRNAs in psoriasis. It may play a vital regulatory role in psoriasis by targeting these DEmRNAs. We found the pro-inflammatory factor IL-1B in target DEmRNAs of hsa-miR-30a-3p. IL-1B has been found to play an important role in autoimmune or autoinflammatory conditions[39]. IL-1B is abundant in the tissue fluid of psoriasis and participates in the disease progression through dual effects[40]. First, it induces insulin resistance through p38 mitogen-activated protein kinase (p38MAPK), preventing insulin-dependent differentiation of keratinocytes, while IL-1B promotes keratinocyte proliferation, both hallmarks of psoriasis. Through KEGG functional enrichment analysis, we also found that IL-1B participates in multiple signal pathways, for example, measles, toll-like receptor signaling pathway, and cytokine-cytokine receptor interaction. Measles can relieve psoriasis through an immunosuppressive effect[41]. Toll-like receptor signaling pathway play an important role in psoriasis by affecting keratinocyte production[42]. The cytokine-cytokine receptor interaction is related to the occurrence and progression of psoriasis through combined transcriptomic analysis[43], which is consistent with our analysis. Thus, we hypothesized that hsa-miR-30a-3p may play a vital molecular role in the progression of psoriasis by targeting DEmRNAs to regulate multiple biological signaling pathways.
Hsa-miR-181a-5p is involved in the catabolic pathway of chondrocytes and oxidative stress in osteoarthritis[44]. Hsa-miR-181a-5p can also regulate the pathogenesis of sepsis-related inflammation through CRNDE/hsa-miR-181a-5p/TLR4 pathway[45]. In a case-control study, hsa-miR-181a-5p was significantly down-regulated in psoriasis[29]. The miRNA-mRNA regulatory network results demonstrated that hsa-miR-181a-5p was negatively regulated with multiple DEmRNAs. Among these DEmRNAs, we found inflammatory mediators CXCL9 and CCL8. CXCL9 is an important chemokine involved in T cell recruitment and is up-regulated in the plasma of patients with psoriasis[46,47]. Increased CXCL9 expression can aggravate the progression of psoriasis[48]. CXCL9 and hsa-miR-30c-2-3p were also negatively regulated in the miRNA-mRNA regulatory network. Oncology studies have shown that hsa-miR-30c-2-3p play a vital role in tumor pathogenesis by regulating the proliferation, apoptosis, migration, and invasion of cancer cells[49,50]. So far, we have not found any studies on hsa-miR-30c-2-3p in psoriasis. This article may be the first to report that hsa-miR-30c-2-3p plays a role in the pathogenesis of psoriasis. As a chemokine, CCL8 is involved in immune regulation and inflammatory processes in a variety of diseases[51-53]. Although no relevant studies on CCL8 have been found in psoriasis, the expression of CCL8 is up-regulated in atopic dermatitis[54]. In addition, CXCL9 and CCL8 were found to be enriched in chemokine signaling pathway and cytokine-cytokine receptor interaction by functional analysis. Therefore, this further suggests that hsa-miR-181a-5p and hsa-miR-30c-2-3p may play a regulatory role in psoriasis by targeting DEmRNAs to mediate multiple biological signaling pathways.
Current research results highlighted that silencing hsa-miR-181a-2-3p could enhance cadmium-induced inflammatory response and activation of inflammasome[55]. In the research, we found that hsa-miR-181a-2-3p was negatively regulated with multiple DEmRNAs. Among these DEmRNAs, OAS3 was found to be involved in hepatitis C and measles in KEGG functional enrichment. Some researchers have found that the OAS3 is concerned with the occurrence and progression of psoriasis through transcriptomic analysis, which is consistent with our analysis[56,57]. Thus, we hypothesized that hsa-miR-181a-2-3p may play a regulatory role in the progression of psoriasis by targeting OAS3 to mediate hepatitis C and measles. This provides potential molecular research directions for further research on the pathogenesis of psoriasis.
The expression of hsa-miR-21-3p in psoriasis was significantly increased[58]. Moreover, hsa-miR-21-3p plays a pro-inflammatory role in keratinocytes, and high expression in the skin is concerned with psoriasis[59]. So far, we have not found relevant reports about hsa-miR-18a-5p in psoriasis. However, hsa-miR-18a-5p promotes the proliferation and migration of pulmonary smooth muscle cells by targeting notch receptor 2[60]. Oncology studies have shown that hsa-miR-18a-5p promotes melanoma cell proliferation by targeting EPH receptor A7 signaling[61]. Hsa-miR-18a-5p can affect keratinocytes apoptosis by targeting B-cell lymphoma/leukemia-2-like protein 10 in patients with toxic epidermal necrolysis and is related to the skin erythema or erosion area of drug eruptions[62]. The miRNA-mRNA regulatory network regulatory network result demonstrates that hsa-miR-21-3p and hsa-miR-18a-5p jointly negatively regulate coagulation factor III, tissue factor (F3). This finding provides new research ideas for the pathogenesis of psoriasis in the future.
Results of previous studies demonstrate that down-regulation of interleukin 1 receptor associated kinase 3 by hsa-miR-33b-3p can alleviate the inflammation and apoptosis induced by IL-1B in CHON-001 cells[63]. As a key miRNA of major depression disorder and Kawasaki disease, hsa-miR-33b-3p play an important role in their pathogenesis[64,65]. Hsa-miR-33b-3p has also been reported in cancer[66]. In the miRNA-mRNA regulatory network, phospholipase C beta 4 (PLCB4) is the only negatively regulated target DEmRNA of hsa-miR-33b-3p. Neutrophils are an important part of the innate immune system and an early line of defense against microbial invasion. PLCB4 can regulate the number of neutrophils[67]. In addition, KEGG functional analysis result demonstrated that PLCB4 was enriched in chemokine signaling pathway. Thus, we hypothesized that hsa-miR-33b-3p may play a vital molecular role in psoriasis by targeting PLCB4 to regulate chemokine signaling pathway.
This study has some limitations. First, sample size of in vitro validation was small, leading to a certain degree of error between RT-PCR validation results and bioinformatics analysis results. Further studies with a larger sample size are needed. Secondly, the specific regulatory mechanism of the identified genes and signaling pathways in psoriasis remain unclear, so further research is needed.
In conclusion, identification of potential key molecular markers and signaling pathways provides potential molecular research directions for further understanding the pathological mechanisms of psoriasis. This may also provide new research ideas for the prevention and treatment of psoriasis in the future.
Previous studies have found that microRNAs (miRNAs) play an important regulatory role in the progression of various diseases. Currently, miRNAs studies in psoriasis and dermatology are relatively new.
Although psoriasis is widespread and has significant negative impact on patients’ life quality, it has not yet been fully diagnosed and treated.
Identification of key miRNAs in psoriasis is helpful to elucidate the molecular mechanism of psoriasis.
Differentially expressed mRNAs (DEmRNAs) and differentially expressed miRNAs were screened out by limma R package. DEmRNAs were analyzed for Gene Ontology and Kyoto Encyclopedia of Genes and Genomics functional enrichment. The “Weighted gene co-expression network analysis (WGCNA)” R package was used to analyze the co-expression network of all miRNAs. We constructed miRNA-mRNA regulatory networks based on identified hub miRNAs.
We identified a large number of DEmRNAs and screened possible signaling pathways related to psoriasis, for example, toll-like receptor signaling pathway, cytokine-cytokine receptor interaction, and chemokine signaling pathway. Ten hub miRNAs were identified by WGCNA. Eight hub miRNAs predicted the corresponding target mRNAs. Ninety-seven negatively regulated miRNA-mRNA pairs were involved in the miRNA-mRNA regulatory network, for example, hsa-miR-21-5p-CLDN8, hsa-miR-30a-3p-IL-1B and hsa-miR-181a-5p/hsa-miR-30c-2-3p-CXCL9.
The identification of potential key molecular markers and signaling pathways provides potential research directions for further understanding the molecular mechanisms of psoriasis.
This study provide new research ideas for the prevention and treatment of psoriasis in the future.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Bhargava S, India; Grigoras A, Romania A-Editor: Yao QG, China S-Editor: Fan JR L-Editor: Filipodia P-Editor: Qi WW
| 1. | Langley RG, Krueger GG, Griffiths CE. Psoriasis: epidemiology, clinical features, and quality of life. Ann Rheum Dis. 2005;64 Suppl 2:ii18-23; discussion ii24. [PubMed] [DOI] [Cited in This Article: ] |
| 2. | López-Estebaranz JL, Sánchez-Carazo JL, Sulleiro S. Effect of a family history of psoriasis and age on comorbidities and quality of life in patients with moderate to severe psoriasis: Results from the ARIZONA study. J Dermatol. 2016;43:395-401. [PubMed] [DOI] [Cited in This Article: ] |
| 3. | Kamiya K, Kishimoto M, Sugai J, Komine M, Ohtsuki M. Risk Factors for the Development of Psoriasis. Int J Mol Sci. 2019;20. [PubMed] [DOI] [Cited in This Article: ] |
| 4. | Rendon A, Schäkel K. Psoriasis Pathogenesis and Treatment. Int J Mol Sci. 2019;20. [PubMed] [DOI] [Cited in This Article: ] |
| 5. | Takeshita J, Grewal S, Langan SM, Mehta NN, Ogdie A, Van Voorhees AS, Gelfand JM. Psoriasis and comorbid diseases: Epidemiology. J Am Acad Dermatol. 2017;76:377-390. [PubMed] [DOI] [Cited in This Article: ] |
| 6. | Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, Ghaffari SH. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J Cell Physiol. 2019;234:5451-5465. [PubMed] [DOI] [Cited in This Article: ] |
| 7. | Bhaskaran M, Mohan M. MicroRNAs: history, biogenesis, and their evolving role in animal development and disease. Vet Pathol. 2014;51:759-774. [PubMed] [DOI] [Cited in This Article: ] |
| 8. | Timis TL, Orasan RI. Understanding psoriasis: Role of miRNAs. Biomed Rep. 2018;9:367-374. [PubMed] [DOI] [Cited in This Article: ] |
| 9. | Tang L, He S, Zhu Y, Feng B, Su Z, Liu B, Xu F, Wang X, Liu H, Li C, Zhao J, Zheng X, Sun C, Lu C, Zheng G. Downregulated miR-187 contributes to the keratinocytes hyperproliferation in psoriasis. J Cell Physiol. 2019;234:3661-3674. [PubMed] [DOI] [Cited in This Article: ] |
| 10. | Liu T, Zhang X, Wang Y. miR-183-3p suppresses proliferation and migration of keratinocyte in psoriasis by inhibiting GAB1. Hereditas. 2020;157:28. [PubMed] [DOI] [Cited in This Article: ] |
| 11. | Wang Y, Yu X, Wang L, Ma W, Sun Q. miR-320b Is Down-Regulated in Psoriasis and Modulates Keratinocyte Proliferation by Targeting AKT3. Inflammation. 2018;41:2160-2170. [PubMed] [DOI] [Cited in This Article: ] |
| 12. | Wang Q, Chang W, Yang X, Cheng Y, Zhao X, Zhou L, Li J, Zhang K. Levels of miR-31 and its target genes in dermal mesenchymal cells of patients with psoriasis. Int J Dermatol. 2019;58:198-204. [PubMed] [DOI] [Cited in This Article: ] |
| 13. | Hermann H, Runnel T, Aab A, Baurecht H, Rodriguez E, Magilnick N, Urgard E, Šahmatova L, Prans E, Maslovskaja J, Abram K, Karelson M, Kaldvee B, Reemann P, Haljasorg U, Rückert B, Wawrzyniak P, Weichenthal M, Mrowietz U, Franke A, Gieger C, Barker J, Trembath R, Tsoi LC, Elder JT, Tkaczyk ER, Kisand K, Peterson P, Kingo K, Boldin M, Weidinger S, Akdis CA, Rebane A. miR-146b Probably Assists miRNA-146a in the Suppression of Keratinocyte Proliferation and Inflammatory Responses in Psoriasis. J Invest Dermatol. 2017;137:1945-1954. [PubMed] [DOI] [Cited in This Article: ] |
| 14. | Xiao Y, Wang H, Wang C, Zeng B, Tang X, Zhang Y, Peng Y, Luo M, Huang P, Yang Z. miR-203 promotes HaCaT cell overproliferation through targeting LXR-α and PPAR-γ. Cell Cycle. 2020;19:1928-1940. [PubMed] [DOI] [Cited in This Article: ] |
| 15. | Xu N, Brodin P, Wei T, Meisgen F, Eidsmo L, Nagy N, Kemeny L, Ståhle M, Sonkoly E, Pivarcsi A. MiR-125b, a microRNA downregulated in psoriasis, modulates keratinocyte proliferation by targeting FGFR2. J Invest Dermatol. 2011;131:1521-1529. [PubMed] [DOI] [Cited in This Article: ] |
| 16. | Su F, Jin L, Liu W. MicroRNA-125a Correlates with Decreased Psoriasis Severity and Inflammation and Represses Keratinocyte Proliferation. Dermatology. 2021;237:568-578. [PubMed] [DOI] [Cited in This Article: ] |
| 17. | Lerman G, Sharon M, Leibowitz-Amit R, Sidi Y, Avni D. The crosstalk between IL-22 signaling and miR-197 in human keratinocytes. PLoS One. 2014;9:e107467. [PubMed] [DOI] [Cited in This Article: ] |
| 18. | Chen CL, Liu XM, Xiong-Ming PU, Wei-Dong WU. Research Advances of miRNA Differential Expression in Psoriasis. Medical Recapitulate 2016.. [Cited in This Article: ] |
| 19. | Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. [PubMed] [DOI] [Cited in This Article: ] |
| 20. | Song ZY, Chao F, Zhuo Z, Ma Z, Li W, Chen G. Identification of hub genes in prostate cancer using robust rank aggregation and weighted gene co-expression network analysis. Aging (Albany NY). 2019;11:4736-4756. [PubMed] [DOI] [Cited in This Article: ] |
| 21. | Niu X, Zhang J, Zhang L, Hou Y, Pu S, Chu A, Bai M, Zhang Z. Weighted Gene Co-Expression Network Analysis Identifies Critical Genes in the Development of Heart Failure After Acute Myocardial Infarction. Front Genet. 2019;10:1214. [PubMed] [DOI] [Cited in This Article: ] |
| 22. | Zhao X, Zhang L, Wang J, Zhang M, Song Z, Ni B, You Y. Identification of key biomarkers and immune infiltration in systemic lupus erythematosus by integrated bioinformatics analysis. J Transl Med. 2021;19:35. [PubMed] [DOI] [Cited in This Article: ] |
| 23. | Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207-210. [PubMed] [DOI] [Cited in This Article: ] |
| 24. | Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. [PubMed] [DOI] [Cited in This Article: ] |
| 25. | Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44-57. [PubMed] [DOI] [Cited in This Article: ] |
| 26. | Zhong S, Chen C, Liu N, Yang L, Hu Z, Duan P, Shuai D, Zhang Q, Wang Y. Overexpression Of hsa-miR-664a-3p Is Associated With Cigarette Smoke-Induced Chronic Obstructive Pulmonary Disease Via Targeting FHL1. Int J Chron Obstruct Pulmon Dis. 2019;14:2319-2329. [PubMed] [DOI] [Cited in This Article: ] |
| 27. | Lin J, Yu M, Xu X, Wang Y, Xing H, An J, Yang J, Tang C, Sun D, Zhu Y. Identification of biomarkers related to CD8+ T cell infiltration with gene co-expression network in clear cell renal cell carcinoma. Aging (Albany NY). 2020;12:3694-3712. [PubMed] [DOI] [Cited in This Article: ] |
| 28. | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408. [PubMed] [DOI] [Cited in This Article: ] |
| 29. | Alatas ET, Kara M, Dogan G, Akın Belli A. Blood microRNA expressions in patients with mild to moderate psoriasis and the relationship between microRNAs and psoriasis activity. An Bras Dermatol. 2020;95:702-707. [PubMed] [DOI] [Cited in This Article: ] |
| 30. | He H, Bissonnette R, Wu J, Diaz A, Saint-Cyr Proulx E, Maari C, Jack C, Louis M, Estrada Y, Krueger JG, Zhang N, Pavel AB, Guttman-Yassky E. Tape strips detect distinct immune and barrier profiles in atopic dermatitis and psoriasis. J Allergy Clin Immunol. 2021;147:199-212. [PubMed] [DOI] [Cited in This Article: ] |
| 31. | Li L, Huang S, Wang H, Chao K, Ding L, Feng R, Qiu Y, Feng T, Zhou G, Hu JF, Chen M, Zhang S. Cytokine IL9 Triggers the Pathogenesis of Inflammatory Bowel Disease Through the miR21-CLDN8 Pathway. Inflamm Bowel Dis. 2018;24:2211-2223. [PubMed] [DOI] [Cited in This Article: ] |
| 32. | Wang H, Chao K, Ng SC, Bai AH, Yu Q, Yu J, Li M, Cui Y, Chen M, Hu JF, Zhang S. Pro-inflammatory miR-223 mediates the cross-talk between the IL23 pathway and the intestinal barrier in inflammatory bowel disease. Genome Biol. 2016;17:58. [PubMed] [DOI] [Cited in This Article: ] |
| 33. | Cohen AD, Weitzman D, Birkenfeld S, Dreiher J. Psoriasis associated with hepatitis C but not with hepatitis B. Dermatology. 2010;220:218-222. [PubMed] [DOI] [Cited in This Article: ] |
| 34. | Chun K, Afshar M, Audish D, Kabigting F, Paik A, Gallo R, Hata T. Hepatitis C may enhance key amplifiers of psoriasis. J Eur Acad Dermatol Venereol. 2017;31:672-678. [PubMed] [DOI] [Cited in This Article: ] |
| 35. | Imafuku S, Naito R, Nakayama J. Possible association of hepatitis C virus infection with late-onset psoriasis: a hospital-based observational study. J Dermatol. 2013;40:813-818. [PubMed] [DOI] [Cited in This Article: ] |
| 36. | Zhou K, Cao D, Wang Y, Wang L, Meng X. Hsa-miR-30a-3p attenuates gastric adenocarcinoma proliferation and metastasis via APBB2. Aging (Albany NY). 2021;13:16763-16772. [PubMed] [DOI] [Cited in This Article: ] |
| 37. | Zhong B, Guo S, Zhang W, Zhang C, Wang Y. Bioinformatics prediction of miR-30a targets and its inhibition of cell proliferation of osteosarcoma by up-regulating the expression of PTEN. BMC Med Genomics. 2017;10:64. [PubMed] [DOI] [Cited in This Article: ] |
| 38. | Deng G, Yu S, He Y, Sun T, Liang W, Yu L, Xu D, Li Q, Zhang R. MicroRNA profiling of platelets from immune thrombocytopenia and target gene prediction. Mol Med Rep. 2017;16:2835-2843. [PubMed] [DOI] [Cited in This Article: ] |
| 39. | Swindell WR, Beamer MA, Sarkar MK, Loftus S, Fullmer J, Xing X, Ward NL, Tsoi LC, Kahlenberg MJ, Liang Y, Gudjonsson JE. RNA-Seq Analysis of IL-1B and IL-36 Responses in Epidermal Keratinocytes Identifies a Shared MyD88-Dependent Gene Signature. Front Immunol. 2018;9:80. [PubMed] [DOI] [Cited in This Article: ] |
| 40. | Buerger C, Richter B, Woth K, Salgo R, Malisiewicz B, Diehl S, Hardt K, Boehncke S, Boehncke WH. Interleukin-1β interferes with epidermal homeostasis through induction of insulin resistance: implications for psoriasis pathogenesis. J Invest Dermatol. 2012;132:2206-2214. [PubMed] [DOI] [Cited in This Article: ] |
| 41. | Chakravarti VS, Lingam S. Measles induced remission of psoriasis. Ann Trop Paediatr. 1986;6:293-294. [PubMed] [DOI] [Cited in This Article: ] |
| 42. | Chen JQ, Szodoray P, Zeher M. Toll-Like Receptor Pathways in Autoimmune Diseases. Clin Rev Allergy Immunol. 2016;50:1-17. [PubMed] [DOI] [Cited in This Article: ] |
| 43. | Gao Y, Yi X, Ding Y. Combined Transcriptomic Analysis Revealed AKR1B10 Played an Important Role in Psoriasis through the Dysregulated Lipid Pathway and Overproliferation of Keratinocyte. Biomed Res Int. 2017;2017:8717369. [PubMed] [DOI] [Cited in This Article: ] |
| 44. | Xue J, Min Z, Xia Z, Cheng B, Lan B, Zhang F, Han Y, Wang K, Sun J. The hsa-miR-181a-5p reduces oxidation resistance by controlling SECISBP2 in osteoarthritis. BMC Musculoskelet Disord. 2018;19:355. [PubMed] [DOI] [Cited in This Article: ] |
| 45. | Wang Y, Xu Z, Yue D, Zeng Z, Yuan W, Xu K. Linkage of lncRNA CRNDE sponging miR-181a-5p with aggravated inflammation underlying sepsis. Innate Immun. 2020;26:152-161. [PubMed] [DOI] [Cited in This Article: ] |
| 46. | Duarte GV, Boeira V, Correia T, Porto-Silva L, Cardoso T, Macedo MN, Oliveira MF, Carvalho E. Osteopontin, CCL5 and CXCL9 are independently associated with psoriasis, regardless of the presence of obesity. Cytokine. 2015;74:287-292. [PubMed] [DOI] [Cited in This Article: ] |
| 47. | Costa MC, Paixão CS, Viana DL, Rocha BO, Saldanha M, da Mota LMH, Machado PRL, Pagliari C, de Oliveira MF, Arruda S, Carvalho EM, Carvalho LP. Mononuclear Phagocyte Activation Is Associated With the Immunopathology of Psoriasis. Front Immunol. 2020;11:478. [PubMed] [DOI] [Cited in This Article: ] |
| 48. | Wang H, Chen W, He J, Xu W, Liu J. Network analysis of potential risk genes for psoriasis. Hereditas. 2021;158:21. [PubMed] [DOI] [Cited in This Article: ] |
| 49. | Tang CT, Liang Q, Yang L, Lin XL, Wu S, Chen Y, Zhang XT, Gao YJ, Ge ZZ. RAB31 Targeted by MiR-30c-2-3p Regulates the GLI1 Signaling Pathway, Affecting Gastric Cancer Cell Proliferation and Apoptosis. Front Oncol. 2018;8:554. [PubMed] [DOI] [Cited in This Article: ] |
| 50. | Zhang HD, Jiang LH, Hou JC, Zhou SY, Zhong SL, Zhu LP, Wang DD, Yang SJ, He YJ, Mao CF, Yang Y, Wang JY, Zhang Q, Xu HZ, Yu DD, Zhao JH, Tang JH, Ji ZL. Circular RNA hsa_circ_0072995 promotes breast cancer cell migration and invasion through sponge for miR-30c-2-3p. Epigenomics. 2018;10:1229-1242. [PubMed] [DOI] [Cited in This Article: ] |
| 51. | Abu El-Asrar AM, Berghmans N, Al-Obeidan SA, Gikandi PW, Opdenakker G, Van Damme J, Struyf S. The CC chemokines CCL8, CCL13 and CCL20 are local inflammatory biomarkers of HLA-B27-associated uveitis. Acta Ophthalmol. 2019;97:e122-e128. [PubMed] [DOI] [Cited in This Article: ] |
| 52. | Asano K, Takahashi N, Ushiki M, Monya M, Aihara F, Kuboki E, Moriyama S, Iida M, Kitamura H, Qiu CH, Watanabe T, Tanaka M. Intestinal CD169(+) macrophages initiate mucosal inflammation by secreting CCL8 that recruits inflammatory monocytes. Nat Commun. 2015;6:7802. [PubMed] [DOI] [Cited in This Article: ] |
| 53. | Zhang X, Chen L, Dang WQ, Cao MF, Xiao JF, Lv SQ, Jiang WJ, Yao XH, Lu HM, Miao JY, Wang Y, Yu SC, Ping YF, Liu XD, Cui YH, Zhang X, Bian XW. CCL8 secreted by tumor-associated macrophages promotes invasion and stemness of glioblastoma cells via ERK1/2 signaling. Lab Invest. 2020;100:619-629. [PubMed] [DOI] [Cited in This Article: ] |
| 54. | Brunner PM, Suárez-Fariñas M, He H, Malik K, Wen HC, Gonzalez J, Chan TC, Estrada Y, Zheng X, Khattri S, Dattola A, Krueger JG, Guttman-Yassky E. The atopic dermatitis blood signature is characterized by increases in inflammatory and cardiovascular risk proteins. Sci Rep. 2017;7:8707. [PubMed] [DOI] [Cited in This Article: ] |
| 55. | Kim J, Kim DY, Heo HR, Choi SS, Hong SH, Kim WJ. Role of miRNA-181a-2-3p in cadmium-induced inflammatory responses of human bronchial epithelial cells. J Thorac Dis. 2019;11:3055-3069. [PubMed] [DOI] [Cited in This Article: ] |
| 56. | Gao LJ, Shen J, Ren YN, Shi JY, Wang DP, Cao JM. Discovering novel hub genes and pathways associated with the pathogenesis of psoriasis. Dermatol Ther. 2020;33:e13993. [PubMed] [DOI] [Cited in This Article: ] |
| 57. | Keermann M, Kõks S, Reimann E, Prans E, Abram K, Kingo K. Transcriptional landscape of psoriasis identifies the involvement of IL36 and IL36RN. BMC Genomics. 2015;16:322. [PubMed] [DOI] [Cited in This Article: ] |
| 58. | Li X, Li J, Lu F, Cao Y, Xing J, Hou R, Yin G, Zhang K. Role of SPRED1 in keratinocyte proliferation in psoriasis. J Dermatol. 2020;47:735-742. [PubMed] [DOI] [Cited in This Article: ] |
| 59. | Degueurce G, D'Errico I, Pich C, Ibberson M, Schütz F, Montagner A, Sgandurra M, Mury L, Jafari P, Boda A, Meunier J, Rezzonico R, Brembilla NC, Hohl D, Kolios A, Hofbauer G, Xenarios I, Michalik L. Identification of a novel PPARβ/δ/miR-21-3p axis in UV-induced skin inflammation. EMBO Mol Med. 2016;8:919-936. [PubMed] [DOI] [Cited in This Article: ] |
| 60. | Miao R, Liu W, Qi C, Song Y, Zhang Y, Fu Y, Lang Y, Zhang Z. MiR-18a-5p contributes to enhanced proliferation and migration of PASMCs via targeting Notch2 in pulmonary arterial hypertension. Life Sci. 2020;257:117919. [PubMed] [DOI] [Cited in This Article: ] |
| 61. | Guo Y, Shi W, Fang R. miR18a5p promotes melanoma cell proliferation and inhibits apoptosis and autophagy by targeting EPHA7 signaling. Mol Med Rep. 2021;23. [PubMed] [DOI] [Cited in This Article: ] |
| 62. | Ichihara A, Wang Z, Jinnin M, Izuno Y, Shimozono N, Yamane K, Fujisawa A, Moriya C, Fukushima S, Inoue Y, Ihn H. Upregulation of miR-18a-5p contributes to epidermal necrolysis in severe drug eruptions. J Allergy Clin Immunol. 2014;133:1065-1074. [PubMed] [DOI] [Cited in This Article: ] |
| 63. | Tao T, Zhang Y, Wei H, Heng K. Downregulation of IRAK3 by miR-33b-3p relieves chondrocyte inflammation and apoptosis in an in vitro osteoarthritis model. Biosci Biotechnol Biochem. 2021;85:545-552. [PubMed] [DOI] [Cited in This Article: ] |
| 64. | Wu C, Zhao Y, Liu Y, Yang X, Yan M, Min Y, Pan Z, Qiu S, Xia S, Yu J, Yang P, Wan B, Shao Q. Identifying miRNA-mRNA regulation network of major depressive disorder in ovarian cancer patients. Oncol Lett. 2018;16:5375-5382. [PubMed] [DOI] [Cited in This Article: ] |
| 65. | Chen Y, Ding YY, Ren Y, Cao L, Xu QQ, Sun L, Xu MG, Lv HT. Identification of differentially expressed microRNAs in acute Kawasaki disease. Mol Med Rep. 2018;17:932-938. [PubMed] [DOI] [Cited in This Article: ] |
| 66. | Xu N, Li Z, Yu Z, Yan F, Liu Y, Lu X, Yang W. MicroRNA-33b suppresses migration and invasion by targeting c-Myc in osteosarcoma cells. PLoS One. 2014;9:e115300. [PubMed] [DOI] [Cited in This Article: ] |
| 67. | Okada Y, Kamatani Y, Takahashi A, Matsuda K, Hosono N, Ohmiya H, Daigo Y, Yamamoto K, Kubo M, Nakamura Y, Kamatani N. Common variations in PSMD3-CSF3 and PLCB4 are associated with neutrophil count. Hum Mol Genet. 2010;19:2079-2085. [PubMed] [DOI] [Cited in This Article: ] |