First Steps of Asthma Management with a Personalized Ontology Model
Abstract
:1. Introduction
2. State of the Art Systems
3. System Architecture
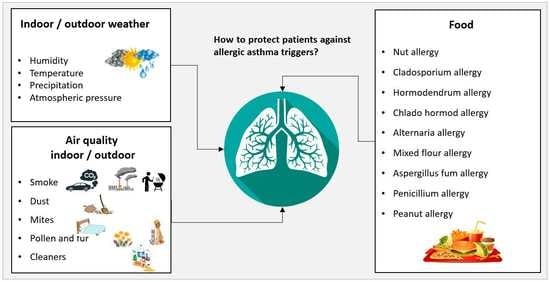
3.1. Data Acquisition
3.2. Representation Layer
- - Which information are necessary for having a detailed description of each context?
- - Which concepts are needed for supporting the design of medical rules allowing patient monitoring?
- - Which data have to be provided by patients to enable reasoning processes?
- -
- Asthma from BioPortal
- -
- Weather and environment ontologies from COPDology [30]
- -
- Food allergens from FoodOn
- -
- Symptoms and pollen concepts from SNOMED-CT
3.3. Reasoning Layer
Patient(?P)^Environment(?Outdoor)^LocatedAt(?P,?Outdoor)^Allergen(PM2.5)^hasCurrentValue(?PM2.5,?CV^hasMinNormalrange(?CV,?Min)^hasMaxNormalRange(CV,Max)^ swrlb:greaterThanOrEqual(?CV,Min)^swrlb:lessThanOrEqual(?CV,Max)->has_alarm_level(?P, No_Risk)
3.4. The Application Layer
4. Implementation
4.1. Design of Ontology Model
4.2. Applicability to Other Domains
- collecting data from asthmatic individual context parameters;
- collecting patient medical profile information;
- identifying and validate medical rules for risk factor control;
- identifying the risk factors of the disease;
- identifying the different recommendations;
- providing disease surveillance services.
5. Conclusions and Future Work
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Asthma. 2020. Available online: https://www.who.int/news-room/fact-sheets/detail/asthma (accessed on 25 November 2020).
- Government of Canada. Asthma and Chronic Obstructive Pulmonary Disease (COPD) in Canada. 2018. Available online: https://www.canada.ca/en/public-health/services/publications/diseases-conditions/asthma-chronic-obstructive-pulmonary-disease-canada-2018.html (accessed on 25 November 2020).
- Madore, A.M.; Laprise, C. Immunological and genetic aspects of asthma and allergy. J. Asthma Allergy 2010, 3, 107–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asthma Canada. Asthma Action Plan. 2019. Available online: https://asthma.ca/get-help/asthma-3/control/asthma-action-plan (accessed on 8 June 2022).
- Michael Schatz, Lanny Rosenwasser, The Allergic Asthma Phenotype. J. Allergy Clin. Immunol. Pract. 2014, 2, 645–648. [CrossRef] [PubMed]
- Emons, J.A.M.; van Wijk, R.G. Food Allergy and Asthma: Is There a Link? Curr. Treat. Options Allergy 2018, 5, 436–444. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, A.B.; Zhang, Z. Allergic Asthma: Influence of Genetic and Environmental Factors. J. Biol. Chem. 2011, 286, 32883–32889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toskala, E.; Kennedy, D.W. Asthma risk factors. Int. Forum Allergy Rhinol. 2015, 5, S11–S16. [Google Scholar] [CrossRef]
- Agache, I.; Akdis, C.; Jutel, M.; Virchow, J.C. Untangling asthma phenotypes and endotypes. Allergy 2012, 67, 835–846. [Google Scholar] [CrossRef]
- Froidure, A.; Mouthuy, J.; Durham, S.R.; Chanez, P.; Sibille, Y.; Pilette, C. Asthma phenotypes and IgE responses. Eur. Respir. J. 2016, 47, 304–319. [Google Scholar] [CrossRef] [Green Version]
- Quinde, M.; Khan, N.; Augusto, J.C. Personalisation of Context-Aware Solutions Supporting Asthma Management. In Computers Helping People with Special Needs; ICCHP 2018. Lecture Notes in Computer Science; Miesenberger, K., Kouroupetroglou, G., Eds.; Springer: Cham, Switzerland, 2018; Volume 10897. [Google Scholar] [CrossRef] [Green Version]
- Al-Dowaihi, D.; Al-Ajlan, M.; Al-Zahrani, N.; Al-Quwayfili, N.; Al-Jwiser, N.; Kanjo, E. MBreath: Asthma monitoring system on the go. In Proceedings of the 2013 International Conference on Computer Medical Applications (ICCMA), Sousse, Tunisia, 20–22 January 2013; pp. 1–4. [Google Scholar] [CrossRef]
- Kwan, A.M.; Fung, A.G.; Jansen, P.A.; Schivo, M.; Kenyon, N.J.; Delplanque, J.-P.; Davis, C.E. Personal Lung Function Monitoring Devices for Asthma Patients. IEEE Sens. J. 2014, 15, 2238–2247. [Google Scholar] [CrossRef]
- Anantharam, P.; Banerjee, T.; Sheth, A.; Thirunarayan, K.; Marupudi, S.; Sridharan, V.; Forbis, S.G. Knowledge-driven personalized contextual mHealth service for Asthma management in children. In Proceedings of the 2015 IEEE International Conference on Mobile Services, New York City, NY, USA, 27 June–2 July 2015; pp. 284–291. [Google Scholar] [CrossRef] [Green Version]
- Ra, H.K.; Salekin, A.; Yoon, H.J.; Kim, J.; Nirjon, S.; Stone, D.J.; Kim, S.; Lee, J.M.; Son, S.H.; Stankovic, J.A. AsthmaGuide: An Asthma monitoring and advice ecosystem. In Proceedings of the 2016 IEEE Wireless Health, Bethesda, MD, USA, 25–27 October 2016; pp. 128–135. [Google Scholar] [CrossRef]
- Dieffenderfer, J.; Goodell, H.; Mills, S.; McKnight, M.; Yao, S.; Lin, F.; Beppler, E.; Bent, B.; Lee, B.; Misra, V.; et al. Low-Power Wearable Systems for Continuous Monitoring of Environment and Health for Chronic Respiratory Disease. IEEE J. Biomed. Health Inform. 2016, 20, 1251–1264. [Google Scholar] [CrossRef]
- Gyrard, A.; Gaur, M.; Shekarpour, S.; Thirunarayan, K.; Sheth, A. (October 2018). Personalized Health Knowledge Graph. ISWC 2018 Contextualized Knowledge Graph Workshop. Available online: https://scholarcommons.sc.edu/aii_fac_pub/42/ (accessed on 8 June 2022).
- Quinde, M.; Khan, N. Juan Carlos Augusto: Case-Based Reasoning for Context-Aware Solutions Supporting Personalised Asthma Management. ICAISC 2019, 2, 260–270. [Google Scholar]
- Galante, C.M. Asthma management updates. Nursing 2022, 52, 25–34. [Google Scholar] [CrossRef]
- Singhal, S.; Sinha, A.; Singh, B. Context Awareness for Healthcare Service Delivery with Intelligent Sensors. In Frontiers of Data and Knowledge Management for Convergence of ICT, Healthcare, and Telecommunication Services; Springer: Cham, Switzerland, 2022; pp. 61–83. [Google Scholar]
- Sheehan, W.J.; Phipatanakul, W. Indoor allergen exposure and asthma outcomes. Curr. Opin. Pediatr. 2016, 28, 772–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baxi, S.N.; Phipatanakul, W. The role of allergen exposure and avoidance in asthma. Adolesc. Med. 2010, 21, 57–ix. [Google Scholar]
- Gautier, C.; Charpin, D. Environmental triggers and avoidance in the management of asthma. J. Asthma Allergy 2017, 10, 47–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orellano, P.; Quaranta, N.; Reynoso, J.; Balbi, B.; Vasquez, J. Effect of outdoor air pollution on asthma exacerbations in children and adults: Systematic review and multilevel meta-analysis. PLoS ONE 2017, 12, e0174050. [Google Scholar] [CrossRef]
- Hedman, L.; Bjerg, A.; Sundberg, S.; Forsberg, B.; Rönmark, E. Both environmental tobacco smoke and personal smoking is related to asthma and wheeze in teenagers. Thorax 2011, 66, 20–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandenplas, O. Occupational Asthma: Etiologies and Risk Factors. Allergy Asthma Immunol. Res. 2011, 3, 157–167. [Google Scholar] [CrossRef] [Green Version]
- Hyrkäs-Palmu, H.; Ikäheimo, T.M.; Laatikainen, T.; Jousilahti, P.; Jaakkola, M.S.; Jaakkola, J.J.K. Cold weather increases respiratory symptoms and functional disability especially among patients with asthma and allergic rhinitis. Sci. Rep. 2018, 8, 10131. [Google Scholar] [CrossRef]
- Sánchez, D.; Batet, M. Semantic similarity estimation in the biomedical domain: An ontology-based information-theoretic perspective. J. Biomed. Inform. 2011, 44, 749–759. [Google Scholar] [CrossRef] [Green Version]
- Cancer Biomedical Informatics Grid, Unified Medical Language System. Available online: http://bioportal.bioontology.org/ontologies/SNOMEDCT (accessed on 8 June 2022).
- Ajami, H.; Mcheick, H. Ontology-Based Model to Support Ubiquitous Healthcare Systems for COPD Patients. Electronics 2018, 7, 371. [Google Scholar] [CrossRef] [Green Version]
- Mcheick, H.; Saleh, L.; Ajami, H.; Mili, H. Context Relevant Prediction Model for COPD Domain Using Bayesian Belief Network. Sensors 2017, 17, 1486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, L.; Brochhausen, M. The CHRONIOUS Ontology Suite: Methodology and Design Principles. In Proceedings of the International Conference on Biomedical Ontologies (ICBO 2011), Buffalo, NY, USA, 26–30 July 2011; Smith, B., Ed.; SUNY University Buffalo: Buffalo, NY, USA, 2011; pp. 167–173. [Google Scholar]
- Paganelli, F.; Giuli, D. An Ontology-Based System for Context-Aware and Configurable Services to Support Home-Based Continuous Care. IEEE Trans. Inf. Technol. Biomed. 2011, 15, 324–333. [Google Scholar] [CrossRef]
- Lasierra, N.; Alesanco, A.; Guillén, S.; García, J. A three stage ontology-driven solution to provide personalized care to chronic patients at home. J. Biomed. Inform. 2013, 46, 516–529. [Google Scholar] [CrossRef]
- Wieringa, W.; Jones, V.M.; Hermens, H.J. Ontology-Based Generation of Dynamic Feedback on Physical Activity. In Artificial Intelligence in Medicine; AIME 2011. Lecture Notes in Computer Science; Peleg, M., Lavrač, N., Combi, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; Volume 6747. [Google Scholar]
- Riaño, D.; Real, F.; López-Vallverdú, J.A.; Campana, F.; Ercolani, S.; Mecocci, P.; Annicchiarico, R.; Caltagirone, C. An ontology-based personalization of health-care knowledge to support clinical decisions for chronically ill patients. J. Biomed. Inform. 2012, 45, 429–446. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.K.; Kim, J.H.; Chung, K.Y.; Rim, K.W.; Lee, J.H. Ontology Based Context Information Model for u-Healthcare Service. In Proceedings of the 2011 International Conference on Information Science and Applications, Jeju Island, Korea, 26–29 April 2011; pp. 1–6. [Google Scholar]
- Wolf, K.-H.; Gietzelt, M.; Al Scharaa, O.; Tegtbur, U.; Haux, R.; Marschollek, M.; Song, B. Decision Support for Teletraining of COPD Patients. Methods Inf. Med. 2010, 49, 96–102. [Google Scholar] [CrossRef]
- Kim, J.; Kim, J.; Lee, D.; Chung, K.-Y. Ontology driven interactive healthcare with wearable sensors. Multimed. Tools Appl. 2012, 71, 827–841. [Google Scholar] [CrossRef]
- Kazadi, Y.K.; Fonou-Dombeu, J.V. Analysis of Advanced Complexity Metrics of Biomedical Ontologies in the Bioportal Re-pository. Int. J. Biosci. Biochem. Bioinform. 2017, 7, 20–32. [Google Scholar]
- Ajami, H.; Mcheick, H.; Laprise, C. Ontology for Asthma: First Step. Available online: https://github.com/hmcheick/Asthma (accessed on 8 June 2022).
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef] [Green Version]






| Step | Approach |
|---|---|
| 1 | Identification of the domain and scope of the ontology, asthma domain, and alert management. |
| 2 | Ontology reuse and addressing poor ontological coverage of pulmonary diseases such as asthma. |
| 3 | Development of a conceptual model. |
| Rules | Description |
|---|---|
| Rule 1 | To set a maximum level for carbon monoxide (CO) |
| Rule 2 | To set a maximum level for formaldehyde (HCHO) |
| Rule 3 | To set a maximum level for volatile organic compounds (TVOC) |
| Rule 4 | To set a maximum level for carbon dioxide (CO2) |
| Rule 5 | To set a maximum level for particulate matter PM10 |
| Rule 6 | To set a maximum level for particulate matter PM2.5 |
| Rule 7 | To set a maximum level for ozone (O3) |
| Rule 8 | To set a maximum level for bacteria |
| Rule 9 | To set a maximum level for nitrogen dioxide (NO2) |
| Rule 10 | To set a maximum level for sulfur dioxide (SO2) |
| Rule 11 | To set a maximum level for hydrogen sulfide (H2S) |
| Rule 12 | To set a maximum level for nitric oxide (NO) |
| Rule 13 | To set a maximum level for nitrogen oxides (NOx) |
| Rule 14 | To set a maximum level for total reduced sulphur (TRS) |
| Rule 15 | To set a maximum level for cat dander |
| Rule 16 | To set a maximum level for dog dander |
| Rule 17 | To set a maximum level for horse dander |
| Rule 18 | To set a maximum level for D. farinae |
| Rule 19 | To set a maximum level for D. pteronisius |
| Rule 20 | To set a maximum level for feathers |
| Rule 21 | To set a maximum level for indoor dust |
| Rule 22 | To set a maximum level for grasses |
| Rule 23 | To set a maximum level for ragweed |
| Rule 24 | To set a maximum level for weeds |
| Rule 25 | To set a maximum level for phleola |
| Rule 26 | To set a maximum level for perennial ryegrass |
| Rule 27 | To set a maximum level for tree |
| Rule 28 | To set a maximum level for birch |
| Rule 29 | To set a maximum level for maple |
| Rule 30 | To set a maximum level for oak |
| Rule 31 | To set a maximum level for elm |
| Rule 32 | To set a maximum and minimum level for temperature |
| Rule 33 | To set a maximum and minimum level for pressure |
| Rule 34 | To set a maximum level for windchill |
| Rule 35 | To set a maximum and minimum level for humidity |
| Rule 36 | To set a maximum level for precipitation |
| Rules | Description |
|---|---|
| Rule 1 | It can be used to rule out an egg allergy |
| Rule 2 | It can be used to rule out a nut allergy |
| Rule 3 | It can be used to rule out a Cladosporium allergy |
| Rule 4 | It can be used to rule out a Hormodendrum allergy |
| Rule 5 | It can be used to rule out a chlado hormod allergy |
| Rule 6 | It can be used to rule out an Alternaria allergy |
| Rule 7 | It can be used to rule out a mixed flour allergy |
| Rule 8 | It can be used to rule out an Aspergillus fum allergy |
| Rule 9 | It can be used to rule out a penicillium allergy |
| Rule 10 | It can be used to rule out a peanut allergy |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ajami, H.; Mcheick, H.; Laprise, C. First Steps of Asthma Management with a Personalized Ontology Model. Future Internet 2022, 14, 190. https://doi.org/10.3390/fi14070190
Ajami H, Mcheick H, Laprise C. First Steps of Asthma Management with a Personalized Ontology Model. Future Internet. 2022; 14(7):190. https://doi.org/10.3390/fi14070190
Chicago/Turabian StyleAjami, Hicham, Hamid Mcheick, and Catherine Laprise. 2022. "First Steps of Asthma Management with a Personalized Ontology Model" Future Internet 14, no. 7: 190. https://doi.org/10.3390/fi14070190