Abstract
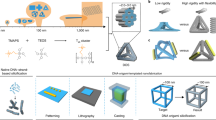
Genetically encoded protein scaffolds often serve as templates for the mineralization of biocomposite materials with complex yet highly controlled structural features that span from nanometres to the macroscopic scale1,2,3,4. Methods developed to mimic these fabrication capabilities can produce synthetic materials with well defined micro- and macro-sized features, but extending control to the nanoscale remains challenging5,6. DNA nanotechnology can deliver a wide range of customized nanoscale two- and three-dimensional assemblies with controlled sizes and shapes7,8,9,10,11. But although DNA has been used to modulate the morphology of inorganic materials12,13 and DNA nanostructures have served as moulds14,15 and templates16,17, it remains challenging to exploit the potential of DNA nanostructures fully because they require high-ionic-strength solutions to maintain their structure, and this in turn gives rise to surface charging that suppresses the material deposition. Here we report that the Stöber method, widely used for producing silica (silicon dioxide) nanostructures, can be adjusted to overcome this difficulty: when synthesis conditions are such that mineral precursor molecules do not deposit directly but first form clusters, DNA–silica hybrid materials that faithfully replicate the complex geometric information of a wide range of different DNA origami scaffolds are readily obtained. We illustrate this approach using frame-like, curved and porous DNA nanostructures, with one-, two- and three-dimensional complex hierarchical architectures that range in size from 10 to 1,000 nanometres. We also show that after coating with an amorphous silica layer, the thickness of which can be tuned by adjusting the growth time, hybrid structures can be up to ten times tougher than the DNA template while maintaining flexibility. These findings establish our approach as a general method for creating biomimetic silica nanostructures.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Berman, A. et al. Biological control of crystal texture: a widespread strategy for adapting crystal properties to function. Science 259, 776–779 (1993).
Belcher, A. M. et al. Control of crystal phase switching and orientation by soluble mollusc-shell proteins. Nature 381, 56–58 (1996).
Aizenberg, J. et al. Skeleton of Euplectella sp.: structural hierarchy from the nanoscale to the macroscale. Science 309, 275–278 (2005).
Kröger, N., Deutzmann, R. & Sumper, M. Polycationic peptides from diatom biosilica that direct silica nanosphere formation. Science 286, 1129–1132 (1999).
Sumper, M. Biomimetic patterning of silica by long-chain polyamines. Angew. Chem. Int. Ed. 43, 2251–2254 (2004).
Wegst, U. G., Bai, H., Saiz, E., Tomsia, A. P. & Ritchie, R. O. Bioinspired structural materials. Nat. Mater. 14, 23–36 (2015).
Jones, M. R., Seeman, N. C. & Mirkin, C. A. Programmable materials and the nature of the DNA bond. Science 347, 1260901 (2015).
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Kershner, R. J. et al. Placement and orientation of individual DNA shapes on lithographically patterned surfaces. Nat. Nanotechnol. 4, 557–561 (2009).
Zheng, J. et al. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature 461, 74–77 (2009).
Yang, F., Zuo, X., Fan, C. & Zhang, X. Biomacromolecular nanostructures-based interfacial engineering: from precise assembly to precision biosensing. Nat. Sci. Rev. nwx134 https://doi.org/10.1093/nsr/nwx134 (2018).
Numata, M., Sugiyasu, K., Hasegawa, T. & Shinkai, S. Sol-gel reaction using DNA as a template: an attempt toward transcription of DNA into inorganic materials. Angew. Chem. Int. Ed. 43, 3279–3283 (2004).
Jin, C., Qiu, H., Han, L., Shu, M. & Che, S. DNA transcription into diverse porous silicas by a co-structure directing route: chiral, ring and ordered nanochannel arrays. Chem. Commun. 3407–3409 (2009).
Peng, Z. & Liu, H. Bottom-up nanofabrication using DNA nanostructures. Chem. Mater. 28, 1012–1021 (2016).
Sun, W. et al. Casting inorganic structures with DNA molds. Science 346, 1258361 (2014).
Surwade, S. P. et al. Nanoscale growth and patterning of inorganic oxides using DNA nanostructure templates. J. Am. Chem. Soc. 135, 6778–6781 (2013).
Diagne, C. T., Brun, C., Gasparutto, D., Baillin, X. & Tiron, R. DNA origami mask for sub-ten-nanometer lithography. ACS Nano 10, 6458–6463 (2016).
Carcouët, C. C. et al. Nucleation and growth of monodisperse silica nanoparticles. Nano Lett. 14, 1433–1438 (2014).
Kim, H., Surwade, S. P., Powell, A., O’Donnell, C. & Liu, H. Stability of DNA origami nanostructure under diverse chemical environments. Chem. Mater. 26, 5265–5273 (2014).
Li, J. et al. Ion-beam sculpting at nanometre length scales. Nature 412, 166–169 (2001).
Agarwal, N. P., Matthies, M., Gur, F. N., Osada, K. & Schmidt, T. L. Block copolymer micellization as a protection strategy for DNA origami. Angew. Chem. Int. Ed. 56, 5460–5464 (2017).
Zhang, F., Nangreave, J., Liu, Y. & Yan, H. Structural DNA nanotechnology: state of the art and future perspective. J. Am. Chem. Soc. 136, 11198–11211 (2014).
Bustamante, C., Bryant, Z. & Smith, S. B. Ten years of tension: single-molecule DNA mechanics. Nature 421, 423–427 (2003).
Smith, S., Finzi, L. & Bustamante, C. Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads. Science 258, 1122–1126 (1992).
Zhou, X. F. et al. Radial compression elasticity of single DNA molecules studied by vibrating scanning polarization force microscopy. Phys. Rev. E 71, 062901 (2005).
Goodman, R. P. et al. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science 310, 1661–1665 (2005).
Fan, H. et al. Modulus-density scaling behaviour and framework architecture of nanoporous self-assembled silicas. Nat. Mater. 6, 418–423 (2007).
Meyers, M. A., McKittrick, J. & Chen, P. Y. Structural biological materials: critical mechanics-materials connections. Science 339, 773–779 (2013).
Wang, T. et al. A DNA crystal designed to contain two molecules per asymmetric unit. J. Am. Chem. Soc. 132, 15471–15473 (2010).
Sha, R. et al. Self-assembled DNA crystals: the impact on resolution of 5′-phosphates and the DNA source. Nano Lett. 13, 793–797 (2013).
Acknowledgements
This Letter is dedicated to the memory of Q. Huang, who initiated the project along with C.F. and X.L. We thank Z. Liu (Shanghai Technical Platform for Testing and Characterization on Inorganic Materials, CASSIC, China) for providing technical support, Y. Li (South China Normal University, China) for providing diatom specimens, and the BL16B1/BL19U2 beamline at Shanghai Synchrotron Radiation Facility (SSRF) for ongoing support in data collection and molecular dynamics simulation. This project was supported by National Science Foundation of China (grant numbers 21390414, 21329501, 21603262 and 21675167), National Key R&D Program of China (grant numbers 2016YFA0201200 and 2016YFA0400900) and the Key Research Program of Frontier Sciences, CAS (grant number QYZDJ-SSW-SLH031). L.W., C.F. and H.Y. thank the National Key R&D Program of China (grant number 2016YFA0400900). H.Y., F.Z. and Y.L. thank the US National Science Foundation, Office of Naval Research, Army Research Office, National Institutes of Health, and Department of Energy for financial support. W.L. and D.Z. thank the National Science Foundation of China (grant numbers U1463206 and 21733003), National Key R&D Program of China (grant number 2018YFA0209401) and Key Basic Research Program of the Science and Technology Commission of Shanghai Municipality (grant number 17JC1400100).
Author information
Authors and Affiliations
Contributions
C.F. and H.Y. supervised the research. X.L., C.F. and H.Y. conceived the research, designed the experiments. F.Z. designed the DNA nanostructures. X.J. and X.L. carried out silicification experiments and characterization. M.P. analysed EM and AFM data. P.L. carried out molecular dynamics simulations. W.L. and D.Z. provided silicification characterization and discussion. B.Z. carried out FTIR and Raman experiments. All authors analysed data. X.L., F.Z., X.J., M.P., D.Z., C.F. and H.Y. interpreted data and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, additional discussions, Supplementary Tables S1-S7, Supplementary Figures 1-80, DNA sequences and additional references.
Video 1 Rotation of a DNA origami silicification tetrahedron framework visualized by TEM.
This video is related to Figure 3b. The sample holder rotation angle is from -30° to +30°, 2.5° each frame.
Video 2 Rotation of a DNA origami silicification cube framework (~0°) visualized by TEM.
This video is related to Figure 3b. The sample holder rotation angle is from -30° to +30°, 2.5° each frame.
Video 3 Rotation of a DNA origami silicification cube framework (~45°) visualized by TEM.
This video is related to Figure 3b. The sample holder rotation angle is from -30° to +30°, 2.5° each frame.
Video 4 Rotation of a DOS tetrahedron framework loaded with two gold nanorods visualized by TEM.
This video is related to Figure 4f. The sample holder rotation angle is from -30° to +30°, 2.5° each frame.
Rights and permissions
About this article
Cite this article
Liu, X., Zhang, F., Jing, X. et al. Complex silica composite nanomaterials templated with DNA origami. Nature 559, 593–598 (2018). https://doi.org/10.1038/s41586-018-0332-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0332-7
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.