Abstract
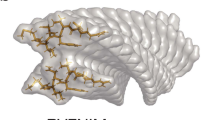
The aggregation of proteins is central to many aspects of daily life, including food processing, blood coagulation, eye cataract formation disease and prion-related neurodegenerative infections1,2,3,4,5. However, the physical mechanisms responsible for amyloidosis—the irreversible fibril formation of various proteins that is linked to disorders such as Alzheimer's, Creutzfeldt–Jakob and Huntington's diseases—have not yet been fully elucidated6,7,8,9. Here, we show that different stages of amyloid aggregation can be examined by performing a statistical polymer physics analysis of single-molecule atomic force microscopy images of heat-denatured β-lactoglobulin fibrils. The atomic force microscopy analysis, supported by theoretical arguments, reveals that the fibrils have a multistranded helical shape with twisted ribbon-like structures. Our results also indicate a possible general model for amyloid fibril assembly and illustrate the potential of this approach for investigating fibrillar systems.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Caughey, B. & Lansbury, P. T. Protofibrils, pores, fibrils and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu. Rev. Neurosci. 26, 267–298 (2003).
Stradner, A. et al. Equilibrium cluster formation in concentrated protein solutions and colloids. Nature 432, 492–495 (2004).
Mezzenga, R., Schurtenberger, P., Burbidge, A. & Michel, M. Understanding foods as soft materials. Nature Mater. 4, 729–740 (2005).
Selkoe, D. J. Folding proteins in fatal ways. Nature 426, 900–904 (2003).
Chiti, F. & Dobson, C. M. Amyloid formation by globular proteins under native conditions. Nature Chem. Biol. 5, 15–22 (2009).
Knowles, T. P. et al. Role of intermolecular forces in defining material properties of protein nanofibrils. Science 318, 1900–1903 (2007).
Nelson, R. et al. Structure of the cross-beta spine of amyloid-like fibrils. Nature 435, 773–778 (2005).
Chiti, F. & Dobson, C. M. Protein misfolding, functional amyloid and human disease. Annu. Rev. Biochem. 75, 333–366 (2006).
Dobson, C. M. Protein folding and misfolding. Nature 426, 884–890 (2003).
Graumann, P. L. Cytoskeletal elements in bacteria. Annu. Rev. Microbiol. 61, 589–618 (2007).
Kueh, H. Y. & Mitchison, T. J. Structural plasticity in actin and tubulin polymer dynamics. Science 325, 960–963 (2009).
Pearce, F. G., Mackintosh, S. H. & Gerrard, J. A. Formation of amyloid-like fibrils by ovalbumin and related proteins under conditions relevant to food processing. J. Agric. Food Chem. 55, 318–322 (2007).
Kavanagh, G. M., Clark, A. H. & Ross-Murphy, S. B. Heat-induced gelation of globular proteins: part 3. Molecular studies on low pH beta-lactoglobulin gels. Int. J. Biol. Macromol. 28, 41–50 (2000).
Bolder, S. G., Sagis, L. M., Venema, P. & van der Linden, E. Effect of stirring and seeding on whey protein fibril formation. J. Agric. Food Chem. 55, 5661–5669 (2007).
Gosal, W. S., Clark, A. H. & Ross-Murphy, S. B. Fibrillar beta-lactoglobulin gels: Part 1. Fibril formation and structure. Biomacromolecules 5, 2408–2419 (2004).
Jung, J. M., Savin, G., Pouzot, M., Schmitt, C. & Mezzenga, R. Structure of heat-induced beta-lactoglobulin aggregates and their complexes with sodium-dodecyl sulfate. Biomacromolecules 9, 2477–2486 (2008).
Gosal, W. S., Clark, A. H., Pudney, P. D. A. & Ross-Murphy, S. B. Novel amyloid fibrillar networks derived from a globular protein: beta-lactoglobulin. Langmuir 18, 7174–7181 (2002).
Veerman, C., Ruis, H., Sagis, L. M. & van der Linden, E. Effect of electrostatic interactions on the percolation concentration of fibrillar beta-lactoglobulin gels. Biomacromolecules 3, 869–873 (2002).
Arnaudov, L. N., de Vries, R., Ippel, H. & van Mierlo, C. P. Multiple steps during the formation of beta-lactoglobulin fibrils. Biomacromolecules 4, 1614–1622 (2003).
Bromley, E. H., Krebs, M. R. & Donald, A. M. Aggregation across the length-scales in beta-lactoglobulin. Faraday Discuss. 128, 13–27 (2005).
Sagis, L. M., Veerman, C. & van der Linden, E. Mesoscopic properties of semiflexible amyloid fibrils. Langmuir 20, 924–927 (2004).
Arnaudov, L. N. & de Vries, R. Strong impact of ionic strength on the kinetics of fibrilar aggregation of bovine beta-lactoglobulin. Biomacromolecules 7, 3490–3498 (2006).
Nilsson, M. R. Techniques to study amyloid fibril formation in vitro. Methods 34, 151–160 (2004).
Lashuel, H. A. & Wall, J. S. Molecular electron microscopy approaches to elucidating the mechanisms of protein fibrillogenesis. Methods Mol. Biol. 299, 81–101 (2005).
Ikeda, S. & Morris, V. J. Fine-stranded and particulate aggregates of heat-denatured whey proteins visualized by atomic force microscopy. Biomacromolecules 3, 382–389 (2002).
Chamberlain, A. K. et al. Ultrastructural organization of amyloid fibrils by atomic force microscopy. Biophys. J. 79, 3282–3293 (2000).
Khurana, R. et al. A general model for amyloid fibril assembly based on morphological studies using atomic force microscopy. Biophys. J. 85, 1135–1144 (2003).
Witz, G., Rechendorff, K., Adamcik, J. & Dietler, G. Conformation of circular DNA in two dimensions. Phys. Rev. Lett. 101, 148103 (2008).
Manning, G. S. Correlation of polymer persistence length with Euler buckling. Phys. Rev. A 34, 4467–4468 (1986).
Aggeli, A. et al. Hierarchical self-assembly of chiral rod-like molecules as a model for peptide beta-sheet tapes, ribbons, fibrils and fibers. Proc. Natl Acad. Sci. USA 98, 11857–11862 (2001).
Paravastu, A. K., Leapman, R. D., Yau, W. M. & Tycko, R. Molecular structural basis for polymorphism in Alzheimer's beta-amyloid fibrils. Proc. Natl Acad. Sci. USA 105, 18349–18354 (2008).
Jung, J. M. & Mezzenga, R. Liquid crystalline phase behavior of protein fibers in water: experiments versus theory. Langmuir 26, 504–514 (2010).
Marek, J. et al. Interactive measurement and characterization of DNA molecules by analysis of AFM images. Cytometry A 63, 87–93 (2005).
Acknowledgements
The authors thank G. Witz and J. Vieira for helpful discussions and assistance during the experiments.
Author information
Authors and Affiliations
Contributions
J.A. performed AFM imaging, analysed data and wrote the paper. J.M.J. prepared the fibrils. J.F. ran coarse-grain molecular dynamics simulations of the fibrils and analysed data. P.D.L.R and G.D. analysed data and wrote the paper. R.M designed the study, analysed the data and wrote the paper. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 565 kb)
Supplementary information
Supplementary movie 1 (MPG 3206 kb)
Supplementary information
Supplementary movie 2 (MPG 3188 kb)
Supplementary information
Supplementary movie 3 (MPG 2662 kb)
Supplementary information
Supplementary movie 4 (MPG 3522 kb)
Supplementary information
Supplementary movie 5 (MPG 5266 kb)
Rights and permissions
About this article
Cite this article
Adamcik, J., Jung, JM., Flakowski, J. et al. Understanding amyloid aggregation by statistical analysis of atomic force microscopy images. Nature Nanotech 5, 423–428 (2010). https://doi.org/10.1038/nnano.2010.59
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2010.59
This article is cited by
-
Heat treatment induced structural change and aggregation behavior of Moringa Oleifera seed salt-soluble protein
Journal of Food Measurement and Characterization (2024)
-
Data-mining unveils structure–property–activity correlation of viral infectivity enhancing self-assembling peptides
Nature Communications (2023)
-
Amyloid-polysaccharide interfacial coacervates as therapeutic materials
Nature Communications (2023)
-
Silk-derived peptide nanospirals assembled by self-propelled worm-like filaments
Nano Research (2023)
-
MIL-CELL: a tool for multi-scale simulation of yeast replication and prion transmission
European Biophysics Journal (2023)