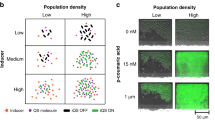
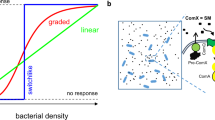
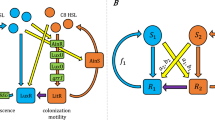
Abstract
Biological nanofactories, which are engineered to contain modules that can target, sense and synthesize molecules, can trigger communication between different bacterial populations. These communications influence biofilm formation1,2, virulence3,4, bioluminescence5,6 and many other bacterial functions7,8 in a process called quorum sensing9. Here, we show the assembly of a nanofactory that can trigger a bacterial quorum sensing response in the absence of native quorum molecules. The nanofactory comprises an antibody (for targeting) and a fusion protein that produces quorum molecules when bound to the targeted bacterium. Our nanofactory selectively targets the appropriate bacteria and triggers a quorum sensing response when added to two populations of bacteria. The nanofactories also trigger communication between two bacterial populations that are otherwise non-communicating. We envision the use of these nanofactories in generating new antimicrobial treatments that target the communication networks of bacteria rather than their viability.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Hardie, K. R. & Heurlier, K. Establishing bacterial communities by ‘word of mouth’: LuxS and autoinducer 2 in biofilm development. Nature Rev. Microbiol. 6, 635–643 (2008).
Irie, Y. & Parsek, M. R. Quorum sensing and microbial biofilms. Curr. Top. Microbiol. Immunol. 322, 67–84 (2008).
Higgins, D. A. et al. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature 450, 883–886 (2007).
Le Berre, R. et al. Quorum-sensing activity and related virulence factor expression in clinically pathogenic isolates of Pseudomonas aeruginosa. Clin. Microbiol. Infect. 14, 337–343 (2008).
Chen, X. et al. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415, 545–549 (2002).
Waters, C. M. & Bassler, B. L. The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes Dev. 20, 2754–2767 (2006).
DeLisa, M. P. & Bentley, W. E. Bacterial autoinduction: looking outside the cell for new metabolic engineering targets. Microb. Cell Fact. 1, 5 (2002).
Williamson, N. R., Fineran, P. C., Ogawa, W., Woodley, L. R. & Salmond, G. P. Integrated regulation involving quorum sensing, a two-component system, a GGDEF/EAL domain protein and a post-transcriptional regulator controls swarming and RhlA-dependent surfactant biosynthesis in Serratia. Environ. Microbiol. 10, 1202–1217 (2008).
Fuqua, W. C., Winans, S. C. & Greenberg, E. P. Quorum sensing in bacteria: the LuxR–LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176, 269–275 (1994).
Garcia, M. C. et al. Arachidonic acid stimulates cell adhesion through a novel p38 MAPK-RhoA signaling pathway that involves heat shock protein 27. J. Biol. Chem. 284, 20936–20945 (2009).
Sundrud, M. S. et al. Halofuginone inhibits TH17 cell differentiation by activating the amino acid starvation response. Science 324, 1334–1338 (2009).
Hung, D. T., Shakhnovich, E. A., Pierson, E. & Mekalanos, J. J. Small-molecule inhibitor of Vibrio cholerae virulence and intestinal colonization. Science 310, 670–674 (2005).
Rasko, D. A. et al. Targeting QseC signaling and virulence for antibiotic development. Science 321, 1078–1080 (2008).
LeDuc, P. R. et al. Towards an in vivo biologically inspired nanofactory. Nature Nanotech. 2, 3–7 (2007).
Keasling, J. D. Synthetic biology for synthetic chemistry. ACS Chem. Biol. 3, 64–76 (2008).
Chin, J. W. Modular approaches to expanding the functions of living matter. Nature Chem. Biol. 2, 304–311 (2006).
Wu, L. Q. & Payne, G. F. Biofabrication: using biological materials and biocatalysts to construct nanostructured assemblies. Trends Biotechnol. 22, 593–599 (2004).
Lowery, C. A., Dickerson, T. J. & Janda, K. D. Interspecies and interkingdom communication mediated by bacterial quorum sensing. Chem. Soc. Rev. 37, 1337–1346 (2008).
Vendeville, A., Winzer, K., Heurlier, K., Tang, C. M. & Hardie, K. R. Making ‘sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nature Rev. Microbiol. 3, 383–396 (2005).
Fernandes, R. & Bentley, W. E. AI-2 biosynthesis module in a magnetic nanofactory alters bacterial response via localized synthesis and delivery. Biotechnol. Bioeng. 102, 390–399 (2009).
Wang, L., Hashimoto, Y., Tsao, C. Y., Valdes, J. J. & Bentley, W. E. Cyclic AMP (cAMP) and cAMP receptor protein influence both synthesis and uptake of extracellular autoinducer 2 in Escherichia coli. J. Bacteriol. 187, 2066–2076 (2005).
Wu, C. F., Cha, H. J., Rao, G., Valdes, J. J. & Bentley, W. E. A green fluorescent protein fusion strategy for monitoring the expression, cellular location and separation of biologically active organophosphorus hydrolase. Appl. Microbiol. Biotechnol. 54, 78–83 (2000).
Taga, M. E., Miller, S. T. & Bassler, B. L. Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium. Mol. Microbiol. 50, 1411–1427 (2003).
Taga, M. E., Semmelhack, J. L. & Bassler, B. L. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol. 42, 777–793 (2001).
Xavier, K. B. & Bassler, B. L. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J. Bacteriol. 187, 238–248 (2005).
Duan, F. & March, J. C. Interrupting Vibrio cholerae infection of human epithelial cells with engineered commensal bacterial signaling. Biotechnol. Bioeng. 101, 128–134 (2008).
Auger, S., Krin, E., Aymerich, S. & Gohar, M. Autoinducer 2 affects biofilm formation by Bacillus cereus. Appl. Environ. Microbiol. 72, 937–941 (2006).
Frezza, M. et al. Ac2-DPD, the bis-(O)-acetylated derivative of 4,5-dihydroxy-2,3-pentanedione (DPD) is a convenient stable precursor of bacterial quorum sensing autoinducer AI-2. Bioorg. Med. Chem. Lett. 17, 1428–1431 (2007).
Miller, J. Experiments in Molecular Genetics (Cold Spring Harbor Laboratory Press, 1972).
Acknowledgements
The authors would like to thank B.L. Bassler for generously providing S. typhimurium MET715, Helim Aranda-Espinoza for providing access to microscopy facilities and T.A Dunn for his help in conducting the flow cytometry studies. Funding for this work was provided in part by the Defense Threat Reduction Agency (DTRA), the National Science Foundation and the R.W. Deutsch Foundation.
Author information
Authors and Affiliations
Contributions
R.F., V.R., H.- C.W. and W.E.B. all planned and designed the experiments. R.F., V.R. and H.-C.W. performed the experiments. R.F. and W.E.B. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 653 kb)
Rights and permissions
About this article
Cite this article
Fernandes, R., Roy, V., Wu, HC. et al. Engineered biological nanofactories trigger quorum sensing response in targeted bacteria. Nature Nanotech 5, 213–217 (2010). https://doi.org/10.1038/nnano.2009.457
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2009.457
This article is cited by
-
Regulation mechanism of metal ions towards magnetic properties in Mn1−xZnxFe2O4
Journal of Materials Science: Materials in Electronics (2020)
-
Intein-mediated assembly of tunable scaffoldins for facile synthesis of designer cellulosomes
Applied Microbiology and Biotechnology (2018)
-
A Facile Two-Step Enzymatic Approach for Conjugating Proteins to Polysaccharide Chitosan at an Electrode Interface
Cellular and Molecular Bioengineering (2017)
-
Directed assembly of a bacterial quorum
The ISME Journal (2016)
-
Nano-guided cell networks as conveyors of molecular communication
Nature Communications (2015)