Abstract
CD8+ effector T (TE) cell proliferation and cytokine production depends on enhanced glucose metabolism. However, circulating T cells continuously adapt to glucose fluctuations caused by diet and inter-organ metabolite exchange. Here we show that transient glucose restriction (TGR) in activated CD8+ TE cells metabolically primes effector functions and enhances tumour clearance in mice. Tumour-specific TGR CD8+ TE cells co-cultured with tumour spheroids in replete conditions display enhanced effector molecule expression, and adoptive transfer of these cells in a murine lymphoma model leads to greater numbers of immunologically functional circulating donor cells and complete tumour clearance. Mechanistically, TE cells treated with TGR undergo metabolic remodelling that, after glucose re-exposure, supports enhanced glucose uptake, increased carbon allocation to the pentose phosphate pathway (PPP) and a cellular redox shift towards a more reduced state—all indicators of a more anabolic programme to support their enhanced functionality. Thus, metabolic conditioning could be used to promote efficiency of T-cell products for adoptive cellular therapy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the findings of this study, as well as further information and requests for resources and reagents, will be made available on reasonable request by the corresponding author. scRNA-seq data have been deposited in the Gene Ontology Omnibus (accession no. GSE152018). Source data are provided with this paper.
References
Michalek, R. D. et al. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T-cell subsets. J. Immunol. 186, 3299–3303 (2011).
Maciver, N. J. et al. Glucose metabolism in lymphocytes is a regulated process with significant effects on immune cell function and survival. J. Leukoc. Biol. 84, 949–957 (2008).
Chang, C.-H. et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell 162, 1229–1241 (2015).
Chang, C.-H. et al. Post-transcriptional control of T-cell effector function by aerobic glycolysis. Cell 153, 1239–1251 (2013).
Blagih, J. et al. The energy sensor AMPK regulates T-cell metabolic adaptation and effector responses in vivo. Immunity 42, 41–54 (2015).
Rolf, J. et al. AMPKalpha1: a glucose sensor that controls CD8 T-cell memory. Eur. J. Immunol. 43, 889–896 (2013).
Klein Geltink, R. I. et al. Mitochondrial Priming by CD28. Cell 171, 385–397 (2017).
Buck, M. D. et al. Mitochondrial dynamics controls T-cell fate through metabolic programming. Cell 166, 63–76 (2016).
Sukumar, M. et al. Inhibiting glycolytic metabolism enhances CD8+ T-cell memory and anti-tumour function. J. Clin. Invest. 123, 4479–4488 (2013).
Sukumar, M. et al. Mitochondrial membrane potential identifies cells with enhanced stemness for cellular therapy. Cell Metab. 23, 63–76 (2016).
Scharping, N. E. et al. The tumor microenvironment represses T cell mitochondrial biogenesis to drive intratumoral T cell metabolic insufficiency and dysfunction. Immunity 45, 374–388 (2016).
Chowdhury, P. S., Chamoto, K., Kumar, A. & Honjo, T. PPAR-induced fatty acid oxidation in T cells increases the number of tumour-reactive CD8. Cancer Immunol. Res. 6, 1375–1387 (2018).
Buck, M. D., Sowell, R. T., Kaech, S. M. & Pearce, E. L. Metabolic instruction of immunity. Cell 169, 570–586 (2017).
Qiu, J. et al. Acetate promotes T-cell effector function during glucose restriction. Cell Rep. 27, 2063–2074 (2019).
Crompton, J. G., Sukumar, M. & Restifo, N. P. Targeting Akt in cell-transfer immunotherapy for cancer. Oncoimmunology 5, e1014776 (2016).
Nabe, S. et al. Reinforce the anti-tumour activity of CD8+ T cells via glutamine restriction. Cancer Sci. 109, 3737–3750 (2018).
Hosios, A. M. et al. Amino acids rather than glucose account for the majority of cell mass in proliferating mammalian cells. Dev. Cell 36, 540–549 (2016).
Garcia, D. & Shaw, R. J. AMPK: mechanisms of cellular energy sensing and restoration of metabolic balance. Mol. Cell 66, 789–800 (2017).
Saxton, R. A. & Sabatini, D. M. mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 (2017).
Jacobs, S. R. et al. Glucose uptake is limiting in T-cell activation and requires CD28-mediated Akt-dependent and independent pathways. J. Immunol. 180, 4476–4486 (2008).
Macintyre, A. N. et al. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab. 20, 61–72 (2014).
Siska, P. J. et al. Suppression of Glut1 and glucose metabolism by decreased Akt/mTORC1 signaling drives T-cell impairment in B cell leukemia. J. Immunol. 197, 2532–2540 (2016).
Waypa, G. B. et al. Hypoxia triggers subcellular compartmental redox signaling in vascular smooth muscle cells. Circ. Res. 106, 526–535 (2010).
Schieber, M. & Chandel, N. S. ROS function in redox signaling and oxidative stress. Curr. Biol. 24, R453–R462 (2014).
Hosios, A. M. & Vander Heiden, M. G. The redox requirements of proliferating mammalian cells. J. Biol. Chem. 293, 7490–7498 (2018).
Kong, H. & Chandel, N. S. Regulation of redox balance in cancer and T cells. J. Biol. Chem. 293, 7499–7507 (2018).
Mak, T. W. et al. Glutathione primes T-cell metabolism for inflammation. Immunity 46, 675–689 (2017).
Smith, C. A., Want, E. J., O’Maille, G., Abagyan, R. & Siuzdak, G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching and identification. Anal. Chem. 78, 779–787 (2006).
Huang, X. et al. X13CMS: global tracking of isotopic labels in untargeted metabolomics. Anal. Chem. 86, 1632–1639 (2014).
Ma, E. H. et al. Serine is an essential metabolite for effector T-cell expansion. Cell Metab. 25, 345–357 (2017).
Crompton, J. G. et al. Akt inhibition enhances expansion of potent tumour-specific lymphocytes with memory-cell characteristics. Cancer Res. 75, 296–305 (2015).
Vander Heiden, M. G., Cantley, L. C. & Thompson, C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009).
Warburg, O. The metabolism of carcinoma cells. J. Cancer Res. 9, 148–163 (1925).
Liberti, M. V. & Locasale, J. W. The Warburg effect: how does it benefit cancer cells?. Trends Biochem. Sci. 41, 211–218 (2016).
Zhao, J., Song, Y. & Liu, D. Clinical trials of dual-target CAR T cells, donor-derived CAR T cells, and universal CAR T cells for acute lymphoid leukemia. J. Hematol. Oncol. 12, 17 (2019).
Sena, L. A. et al. Mitochondria are required for antigen-specific T-cell activation through reactive oxygen species signaling. Immunity 38, 225–236 (2013).
Nathan, C. & Cunningham-Bussel, A. Beyond oxidative stress: an immunologist’s guide to reactive oxygen species. Nat. Rev. Immunol. 13, 349–361 (2013).
Chatterjee, S. et al. CD38−NAD+ axis regulates immunotherapeutic anti-tumour T-cell response. Cell Metab. 27, 85–100 (2018).
Johnson, M. O. et al. Distinct regulation of Th17 and Th1 cell differentiation by glutaminase-dependent metabolism. Cell 175, 1780–1795 (2018).
Ghergurovich, J. M. et al. A small-molecule G6PD inhibitor reveals immune dependence on pentose phosphate pathway. Nat. Chem. Biol. 16, 731–739 (2020).
Ma, E. H. et al. Metabolic profiling using stable isotope tracing reveals distinct patterns of glucose utilization by physiologically activated CD8. Immunity 51, 856–870 (2019).
Yang, J. C. & Rosenberg, S. A. Adoptive T-cell therapy for cancer. Adv. Immunol. 130, 279–294 (2016).
Chamoto, K. et al. Mitochondrial activation chemicals synergize with surface receptor PD-1 blockade for T-cell-dependent anti-tumour activity. Proc. Natl Acad. Sci. USA 114, E761–E770 (2017).
Kumar, A., Chamoto, K., Chowdhury, P. S. & Honjo, T. Tumours attenuating the mitochondrial activity in T cells escape from PD-1 blockade therapy. eLife 9, e52330 (2020).
Wills, J., Edwards-Hicks, J. & Finch, A. J. AssayR: a simple mass spectrometry software tool for targeted metabolic and stable isotope tracer analyses. Anal. Chem. 89, 9616–9619 (2017).
van der Windt, G. J. W., Chang, C. H. & Pearce, E. L. Measuring bioenergetics in T cells using a Seahorse extracellular flux analyser. Curr. Protoc. Immunol. 113, 3.16B.1–3.16B.14 (2016).
Pearce, E. L. et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature 460, 103–107 (2009).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Diaz-Papkovich, A., Anderson-Trocmé, L., Ben-Eghan, C. & Gravel, S. UMAP reveals cryptic population structure and phenotype heterogeneity in large genomic cohorts. PLoS Genet. 15, e1008432 (2019).
Aoki-Kinoshita, K. F. & Kanehisa, M. Gene annotation and pathway mapping in KEGG. Methods Mol. Biol. 396, 71–91 (2007).
Szklarczyk, D. et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019).
Acknowledgements
We thank members of the Pearce laboratories for support and helpful discussions and A. Quintana and J. Sutherland mouse colony management. This work was funded by the National Institutes of Health (NIH; CA181125 to E.L.P. and AI110481 to E.J.P.) and the Max Planck Society. R.Z. was supported by the Deutsche Forschungsgemeinschaft (DFG; SFB1160, B09; TRR167, B06) and the European Research Council (ERC; GvHDCure no. 681012).
Author information
Authors and Affiliations
Contributions
R.I.K.G., J.E.-H., P.A., D.O., D.E.S., D.J.P., J.M.B., K.M.G., A.M.K., M.S., F.M.U., M.F., R.Z., E.J.P. and E.L.P. designed the research, analysed data and provided conceptual input. A.E.P., N.A.M.L., J.D.C. and F.H. analysed data and performed the experiments. R.I.K.G., J.E.H. and E.L.P. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
E.L.P. is a SAB member of ImmunoMet Therapeutics, and E.L.P. and E.J.P. are founders of Rheos Medicines. The other authors declare no competing interests.
Additional information
Peer review information Primary Handling Editor: Christoph Schmitt.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 TGR CD8+ TE do not sustain mitochondrial metabolism by incorporating more glutamine-derived carbons into the TCA cycle.
WT CD8+ T cells isolated from spleens of C7Bl/6 mice were activated with anti-CD3 (5 μg/mL), anti-CD28 (0.5 μg/mL), IL-2 (100 U/mL), expanded for a total of 72 h, and exposed to 10 mM, 3 mM, or 1 mM glucose as indicated in cultures set to 1 million per ml. a, Polar metabolites were extracted from 500 µl media supernatant from 20 h cultures of 10 mM and 1 mM cells. Bar graphs represent glutamine concentration in the indicated conditions compared to complete culture media for n = 3 biological replicates. Significance was calculated using 2-tailed Student’s t tests, no significant changes (ns) were observed. b, TE were generated as above, but during the final 20 h all groups were cultured in 4 mM heavy labelled (U-13C) glutamine (100% U-13C glutamine). Polar metabolites were extracted and isotopologue distribution assessed by targeted mass spectrometry. TCA intermediates are plotted as percent label from newly metabolized U-13C (open bars) or remaining U-12C (black bars) glutamine carbons. Data are from n = 3 biological replicates, representative of 2 independent experiments. Significance was calculated using 2-tailed Student’s t tests, no significant changes were observed. c, TE were generated as above, but during the final 6 h 2 mM U-13C-lactate was added to the culture. Polar metabolites were extracted and isotopologue distribution assessed by targeted mass spectrometry. Significance was calculated using 2-tailed Student’s t tests comparing the predominant m + 2 isotopologue group in both conditions. Data shown for n = 3 biological replicates. Significance was calculated using 2-tailed Student’s t tests, No significant changes were observed. All error bars show SEM.
Extended Data Fig. 2 Limited transcriptional reprogramming during TGR.
WT CD8+ T cells isolated from spleens of C57BL/6 mice were activated with anti-CD3 (5 μg/mL), anti-CD28 (0.5 μg/mL), IL-2 (100 U/mL), expanded for a total of 72 h, and exposed to 10 mM (black) or 1 mM (orange) glucose in cultures set to 1 million per ml. RNA isolated from single cells was sequenced using the 10X genomics platform and analysed to explore transcriptional changes in each population of cells. a, A Uniform Manifold Approximation and Projection (UMAP) of the overlapped cells from each treatment (control, black; TGR, orange), clustered on the basis of transcriptional similarity is shown highlighting clusters and treatment distribution. Bar graphs depicting cluster distribution for each condition are also shown. b, Clusters 0 and 1, where least overlap between treatments was observed, were contrasted to look for differentially expressed genes (> 1.2-fold change and < 0.1 adjusted p-value); a heatmap of top up and down regulated genes is shown, along with UMAPs of example genes. c, The top 5 most enriched pathways in differentially regulated genes between cluster 0 and 1. d, Module scores based on average expression levels of gene programs were calculated for 18 differentially expressed OXPHOS genes (Atp5g1, Atp5g3, Cox5a, Cox6a1, Cyc1, Ndufa11, Ndufa12, Ndufa4, Ndufa8, Ndufab1, Ndufb7, Ndufb8, Ndufc1, Ndufc2, Ndufs6, Sdhb, Uqcr10, Uqcr11). Violin plots depict the global OXPHOS module score per condition (left) or per cluster and condition (right). e, Module scores based on average expression levels of gene programs were also calculated for enzyme-coding genes involved in glycolysis (mmu00010) or the pentose phosphate pathway (PPP, mmu00030) based on Kegg annotation. UMAP and Violin plots illustrate the OXPHOS, glycolysis and PPP modules, with the violin plots divided per cluster.
Extended Data Fig. 3 Signalling timecourse during TGR.
Protein isolates were prepared as described in (Fig. 3f), taking samples every 2 h over the 20 h exposure to limiting glucose concentration. Immunoblot analysis of protein extracts from equal cell numbers probed for phosphorylated acetyl-coA carboxylase at Ser79 (p-ACC1Thr79), total ACC1, phosphorylated AMPK at Thr172 (p-AMPkThr172), total AMPK, phosphorylated ribosomal protein S6 at Ser235/236 (p-S6Ser235/236), total S6, phosphorylated 4E-binding protein 1 at Thr37/46 (p-4E-BP1Thr37/46), total 4E-BP1, and Glut1. Tubulin was used as a loading control. Representative of 3 biological independent samples. Biological replicate data is shown in Fig. 3f.
Extended Data Fig. 4 TGR TE have altered glucose reallocation upon re-exposure to drive anabolic metabolism.
WT CD8+ T cells isolated from spleens of C57BL/6 mice were activated with anti-CD3 (5 μg/mL), anti-CD28 (0.5 μg/mL), IL-2 (100 U/mL), expanded for a total of 72 h, and exposed to 10 mM (control) or 1 mM (TGR) glucose during the final 5 min cells were re-exposed to normal (U-12C) glucose or heavy labelled (U-13C) glucose. 20 h cultures were started at 1 × 106 (control) and 1.5 × 106 (TGR) cells per/ml to generate similar end concentrations for glucose pulse experiments. a, Table showing the results of a Kegg-pathway analysis of the significantly increased and decreased metabolite pools in TGR TE after normal (U-12C) glucose re-feeding when compared to re-fed control TE. b, Table shows the results of a Kegg pathway analysis of the metabolites that exhibited increased glucose-derived U-13C assimilation (as determined by X13 CMS software) after re-feeding TGR TE with heavy labelled (U-13C) glucose compared to re-fed control TE. Data are from 3 biological replicates, representative of 3 independent experiments. Kegg Pathways were ranked according to the number of metabolite hits in that pathway (from highest to lowest). Metabolites in the highest ranked pathways were further validated by targeted analysis. c, Polar metabolites were extracted and untargeted metabolomic analysis was performed using XCMS on cells re-exposed to normal (U-12C) glucose. Volcano plot depicts relative metabolite pools compared between control and TGR cells. Orange circle represents a pentose phosphate pathway metabolite Pentose-phosphate, significantly increased in glucose re-fed TGR TE. Statistical significance was calculated using a Welch’s t-test, comparing relative intensities of each isotopologue in labelled samples of control TE versus those of TGR TE.
Extended Data Fig. 5 6h U-13C-glucose re-exposure following TGR.
WT CD8+ T cells isolated from spleens of C57BL/6 mice were activated with anti-CD3 (5 μg/mL), anti-CD28 (0.5 μg/mL), IL-2 (100 U/mL), expanded for a total of 72 h, and exposed to 10 mM (black) or 1 mM (orange) glucose. Cells were re-exposed to 10 mM U-13C-glucose for 6 h prior to polar metabolite extraction and analysis by LC-MS. 20 h cultures were started at 1 × 106 (control) and 1.5 × 106 (TGR) cells per/ml to generate similar end concentrations for glucose pulse experiments. a, Percent 13C label incorporation into nucleotide monophosphates was analysed. Data are from n = 3 biological replicates. Significance was calculated using 2-tailed Student’s t tests. * p < 0.05. b, Relative 13C label incorporation into serine was analysed. Data are from n = 3 biological replicates. Significance was calculated using 2way ANOVA. Significance is indicated for the 13C portion (open bars). *** p < 0.001. The 12C portion (filled bars) was also significant. ** p < 0.01. c, The GSH/GSSG ratio was calculated from summed isotopologues for each metabolite. Data are from n = 3 biological replicates. Significance was calculated using 2-tailed Student’s t tests, no significant difference was found. d, Relative 13C label incorporation into lactate was analysed. Data are from n = 3 biological replicates. Significance was calculated using 2way ANOVA. Significance is indicated for the 13C portion (open bars), which was not significant. The 12C portion (filled bars) was significant. *** p < 0.001. All error bars show SEM.
Extended Data Fig. 6 TGR enhances CD8+ tumour-specific antitumour function in vivo.
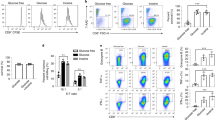
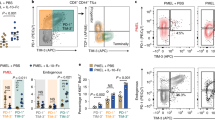
As depicted in Fig. 6f, congenically marked (CD45.1) C57BL/6 female recipient mice were injected subcutaneously with 1×106 B16-OVA melanoma cells and tumours established for 5 d. 5 ×106 control (black) or TGR (orange) OT-I+ TE were injected intravenously per tumour bearing mouse. Mice receiving no TE were used as controls. 3, 6 and 9 d after tumour inoculation, mice received 200 µg anti-PD-1 antibody or IgG2a isotype control in 100 µl PBS intraperitoneally. a, 21 d after tumour inoculation mice were humanely euthanized, blood was collected, red cells were lysed, and the white blood cell fraction stained for congenic markers. Data are presented as % of donor-derived (CD45.2+) CD8+ T cells as a fraction to total circulating CD8+ T cells and each dot represents an individual mouse (n = 5). Statistical significance was calculated by 2-tailed Student’s t test. * p < 0.05; ** p < 0.01. b, 21 d after tumour inoculation mice were humanely euthanized and the tumour mass was excised. Single cell suspensions were analysed by flow cytometry. The number of host (CD45.1) CD8+ T cells per mg tissue was quantified and each dot represents an individual mouse (n = 5). Statistical significance was calculated by 2-tailed Student’s t test. ns not significant. All error bars show SEM.
Supplementary information
Supplementary Information
Supplementary Note
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data and western blot optical density values.
Source Data Fig. 3
Uncropped western blots.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 3
Western blot optical density values.
Source Data Extended Data Fig. 3
Uncropped western blots.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Rights and permissions
About this article
Cite this article
Klein Geltink, R.I., Edwards-Hicks, J., Apostolova, P. et al. Metabolic conditioning of CD8+ effector T cells for adoptive cell therapy. Nat Metab 2, 703–716 (2020). https://doi.org/10.1038/s42255-020-0256-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-020-0256-z
This article is cited by
-
Cancer immunometabolism: advent, challenges, and perspective
Molecular Cancer (2024)
-
Metabolic engineering for optimized CAR-T cell therapy
Nature Metabolism (2024)
-
Defective mitochondria remodelling in B cells leads to an aged immune response
Nature Communications (2024)
-
The screening, identification, design and clinical application of tumor-specific neoantigens for TCR-T cells
Molecular Cancer (2023)
-
Extracellular acidosis restricts one-carbon metabolism and preserves T cell stemness
Nature Metabolism (2023)