Forensic characteristics and population genetics of Chinese Kazakh ethnic minority with an efficient STR panel
- Published
- Accepted
- Received
- Academic Editor
- Antonio Amorim
- Subject Areas
- Anthropology, Genetics, Genomics, Medical Genetics
- Keywords
- Non-CODIS STRs, Chinese Kazakh, Forensic population genetics, Geneticpolymorphism
- Copyright
- © 2019 Chen et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2019. Forensic characteristics and population genetics of Chinese Kazakh ethnic minority with an efficient STR panel. PeerJ 7:e6802 https://doi.org/10.7717/peerj.6802
Abstract
On the purpose of enhancing the forensic efficiency of CODIS STR loci, new STR loci have been gradually discovered and developed into some commercial multiplex systems. Recently, 22 STR loci including 18 non-CODIS STR loci and four CODIS STR loci were investigated in 501 unrelated healthy individuals of Kazakh ethnic group. Seven to 20 alleles at the different loci were identified and altogether 276 alleles for 22 selected loci were detected with the corresponding allelic frequencies ranging from 0.0010 to 0.3623. No significant deviation was observed from the Hardy–Weinberg equilibrium test for any of the 22 STRs. The value of cumulative power of discrimination in Kazakh group was 1-1.00E−28. Analyses of population differentiations and genetic distances between Kazakh and other Chinese groups presented that the Kazakh group with the Uygur group. These 22 STR loci evenly distributed on 22 different autosomal chromosomes were characterized by high genetic diversities and therefore could be utilized in the forensic cases to further increase the discrimination performance.
Introduction
Since the Federal Bureau of Investigation laboratory selected 13 autosomal STR loci as core loci of Combined DNA Index System (CODIS) in 1997, STR loci from CODIS have been applied into the forensic applications and commonly contained in many commercial kits (Gonçalves et al., 2002; Lan et al., 2018; Wang et al., 2012; Xiao et al., 2018). It is worth noting that seven new markers were added up to these 13 core STR loci, creating an expanded panel with 20 CODIS core loci in 2015 (Hares, 2015). However, in forensic applications, it is indispensable to combine the non-CODIS STRs with the previously commercial CODIS STR kits in order to increase the discrimination power (Inokuchi et al., 2018; Kuzniar, Jastrzebska & Ploski, 2006; Tsai et al., 2013) in some cases, like missing person investigations, complicated parentage testing cases and those with mutation events. In this study, 22 selected loci (Li et al., 2017) were introduced containing 18 non-CODIS STR loci and four CODIS STR loci in which three (D1S1656, D2S1338 and D12S391) were recognized as CODIS core loci in 2015 (Hares, 2015) and one (D16S539) belonged to 13 CODIS core loci.
The Kazakh group is one of the minority nationalities of China, with a population exceeding 1.46 million people (data derived from the sixth National Population Census of China, 2010) dwelling in the Xinjiang Uygur Autonomous Region, Qinghai and Gansu provinces. There have been some research focused on the Kazakh group from Xinjiang Uygur Autonomous Region (Liu et al., 2017; Mei et al., 2016; Yuan et al., 2014; Zhang et al., 2016), but very few studies were available for non-CODIS STRs in the Kazakh group. To enrich the database of population genetics and probe into the genetic background of Kazakh group, we calculated the allelic frequency distributions as well as forensic parameters of these 22 autosomal STRs for the first time in Kazakh group, and conducted comparative analysis with other five Chinese reference groups.
Materials and Methods
The collection of population samples
Whole blood samples were withdrawn from 501 unrelated healthy individuals of Chinese Xinjiang Kazakh group. During the process of sample collection, we ensured the genetic relationships by inquiring and no blood relationship was existed between any two individuals at least in three generations. Moreover, all individuals should meet the requirements of being the aboriginal inhabitants. The research was officially authorized by the ethics committee of Xi’an Jiaotong University Health Science Center, China (Ethical Application No: 2018-518) before the study, and was performed as per the human and ethical research principles of Xi’an Jiaotong University Health Science Center. All the participators signed informed consent statements prior to the specimen collections.
Multiplex amplification and STR genotyping
DNA was directly amplified without extraction based on a multiplex PCR system that simultaneously amplified 22 STR loci from Microreader™ 23sp ID kit. PCR amplification was implemented on the GeneAmp® PCR 9700 thermocycler (Applied Biosystems, Foster City, CA, USA) with 25 µL reaction volume, and amplified products were subsequently separated and analyzed on the ABI PRISM® 3130XL Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) with reference to internal lane standard Org500 (including different length fragments: 50, 75, 100, 139, 150, 160, 200, 300, 340, 350, 400, 450, 490 and 500 bp). The specific reaction system and conditions for electrophoresis were referred to the previously published research (Li et al., 2017). The analyzation of electrophoresis results were revealed by GeneMapper® ID-X 1.3 software (Applied Biosystems, Foster City, CA, USA).
Population genetic and forensic statistical analyses
For the studied Kazakh group, the distributions of allelic frequencies and forensic statistical parameters containing the observed heterozygosity (Ho), power of exclusion (PE), power of discrimination (PD), polymorphism information content (PIC) and Hardy–Weinberg equilibrium (HWE) of 22 STR loci were calculated by the modified Powerstats software v.1.2 (Tereba, 1999). Expected heterozygosity (He) was calculated as described by Nei (1978). The estimates of linkage disequilibrium (LD) (Slatkin, 2008) of 22 STR loci were performed by Genepop v.4.0.10 (http://genepop.curtin.edu.au/). Correlation coefficients (r2) for all allele combinations were ascertained by SHEsis online tool (Yongyong & Lin, 2005). The locus-by-locus fixation index (Fst values) and corresponding probability (p) values of population genetic differentiations were estimated by Arlequin software v.3.5 (Excoffier & Lischer, 2010) using the analysis of molecular variance (AMOVA) method. The DISPAN program (Ota, 1993) was implemented to calculate Nei’s genetic distance (DA) values (Nei, 1972) utilizing the raw STR genotyping data of Kazakh group. The population pairwise genetic differentiation Fst values and p values were carried out by Arlequin software v.3.5 (Excoffier & Lischer, 2010) based on raw STR genotyping data. The heat maps of population pairwise DA and Fst values were conducted using R statistical software v3.0.2 (Dean & Nielsen, 2007). The description of population genetic structure was presented by the STRUCTURE software v.2.3.4 (Pritchard, Stephens & Donnelly, 2000) and the optimum K value, the number of hypothetical ancestral populations, was estimated by Structure Harvester v.0.6.94 (Pritchard, Stephens & Donnelly, 2000).
Results
Hardy–Weinberg equilibrium and linkage disequilibrium analyses
HWE tests were performed and the results showed no significant deviations from HWE at these 22 STR loci in Kazakh group after Bonferroni correction (Curtin & Schulz, 1998) (p = 0.05∕22 = 0.00227273). In Fig. S1, the locations of STR loci were indicated on each autosomal chromosome. Furthermore, as shown in Table S1, significant p-values for LD analyses were detected in D10S1435-D15S659 and D13S325-D17S1290 after applying Bonferroni correction (Curtin & Schulz, 1998) (p = 0.05∕231 = 0.00021645). The r2 values of 22 STR loci were displayed in the Fig. S2. The result transpired that the r2 values of STR pairs were all less than 0.004.
| Alleles | D1S1656 | D2S1338 | D3S3045 | D4S2366 | D5S2500 | D6S477 | D7S3048 | D8S1132 | D9S925 | D10S1435 | D11S2368 | D12S391 | D13S325 | D14S608 | D15S659 | D16S539 | D17S1290 | D18S535 | D19S253 | D20S470 | D21S1270 | D22-GATA198B05 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 0.0070 | |||||||||||||||||||||
| 5 | 0.0010 | |||||||||||||||||||||
| 6 | 0.0230 | 0.0329 | ||||||||||||||||||||
| 7 | 0.0010 | 0.2116 | 0.0010 | 0.1936 | ||||||||||||||||||
| 8 | 0.0010 | 0.0050 | 0.0080 | 0.0240 | 0.0190 | 0.0020 | 0.0250 | 0.0050 | ||||||||||||||
| 9 | 0.2974 | 0.2804 | 0.0030 | 0.0020 | 0.0858 | 0.0090 | 0.2166 | 0.0030 | 0.1996 | 0.0130 | 0.0060 | 0.0030 | ||||||||||
| 9.2 | 0.0080 | |||||||||||||||||||||
| 10 | 0.0030 | 0.0369 | 0.0649 | 0.0778 | 0.0020 | 0.0040 | 0.0150 | 0.2265 | 0.0549 | 0.1397 | 0.0190 | 0.0200 | 0.0230 | 0.1377 | 0.2515 | |||||||
| 10.2 | 0.0220 | |||||||||||||||||||||
| 10.3 | 0.0020 | 0.0010 | ||||||||||||||||||||
| 11 | 0.0689 | 0.0519 | 0.2295 | 0.2784 | 0.0140 | 0.0349 | 0.1257 | 0.2315 | 0.1367 | 0.2196 | 0.0269 | 0.0210 | 0.1068 | 0.0209 | 0.0579 | |||||||
| 11.2 | 0.0050 | |||||||||||||||||||||
| 11.3 | 0.0010 | |||||||||||||||||||||
| 12 | 0.0698 | 0.1487 | 0.1637 | 0.1976 | 0.0489 | 0.0210 | 0.3623 | 0.1327 | 0.1826 | 0.2485 | 0.0060 | 0.1317 | 0.3243 | 0.0469 | 0.0499 | 0.0020 | ||||||
| 12.1 | 0.0010 | |||||||||||||||||||||
| 12.2 | 0.0070 | 0.0080 | ||||||||||||||||||||
| 12.3 | 0.0070 | 0.0539 | ||||||||||||||||||||
| 13 | 0.0629 | 0.2395 | 0.1317 | 0.0649 | 0.1776 | 0.0050 | 0.2325 | 0.0010 | 0.0559 | 0.1228 | 0.1287 | 0.0060 | 0.2794 | 0.2265 | 0.1507 | 0.1237 | ||||||
| 13.2 | 0.0020 | |||||||||||||||||||||
| 13.3 | 0.0090 | 0.0529 | ||||||||||||||||||||
| 14 | 0.0858 | 0.1647 | 0.1118 | 0.0629 | 0.2116 | 0.1088 | 0.2156 | 0.0010 | 0.0339 | 0.0269 | 0.0140 | 0.2675 | 0.0788 | 0.1617 | 0.2645 | 0.0050 | ||||||
| 14.2 | 0.0010 | |||||||||||||||||||||
| 14.3 | 0.0010 | 0.0030 | 0.0309 | |||||||||||||||||||
| 15 | 0.2665 | 0.0529 | 0.0180 | 0.2305 | 0.2754 | 0.2335 | 0.0209 | 0.0010 | 0.0100 | 0.0040 | 0.1996 | 0.2515 | 0.0698 | 0.0070 | 0.1756 | 0.0948 | 0.0240 | |||||
| 15.3 | 0.0190 | 0.0020 | 0.0030 | 0.0030 | ||||||||||||||||||
| 16 | 0.2076 | 0.0100 | 0.0030 | 0.0659 | 0.1826 | 0.0020 | 0.0030 | 0.3343 | 0.0050 | 0.0349 | 0.0160 | 0.0060 | 0.1427 | 0.3223 | 0.0090 | 0.0020 | 0.1527 | 0.0140 | 0.0778 | |||
| 16.1 | 0.0010 | |||||||||||||||||||||
| 16.3 | 0.0249 | 0.0060 | 0.0010 | |||||||||||||||||||
| 17 | 0.0768 | 0.0938 | 0.0180 | 0.0289 | 0.0140 | 0.0479 | 0.1896 | 0.1417 | 0.1157 | 0.0130 | 0.1008 | 0.1567 | 0.0589 | 0.0978 | ||||||||
| 17.3 | 0.0559 | 0.0130 | ||||||||||||||||||||
| 18 | 0.0060 | 0.1068 | 0.0010 | 0.0070 | 0.0998 | 0.1717 | 0.0589 | 0.0908 | 0.2086 | 0.0749 | 0.0160 | 0.1337 | 0.0170 | 0.0938 | ||||||||
| 18.1 | 0.0010 | |||||||||||||||||||||
| 18.2 | 0.0020 | |||||||||||||||||||||
| 18.3 | 0.0369 | 0.0100 | ||||||||||||||||||||
| 19 | 0.1946 | 0.0050 | 0.0738 | 0.1926 | 0.0010 | 0.1787 | 0.1916 | 0.2864 | 0.0010 | 0.0429 | 0.0100 | 0.1068 | ||||||||||
| 19.3 | 0.0130 | 0.0030 | ||||||||||||||||||||
| 20 | 0.1237 | 0.0020 | 0.1267 | 0.1307 | 0.0010 | 0.1467 | 0.1647 | 0.2395 | 0.0110 | 0.1298 | ||||||||||||
| 20.3 | 0.0010 | |||||||||||||||||||||
| 21 | 0.0239 | 0.1218 | 0.1208 | 0.2525 | 0.0778 | 0.1896 | 0.0050 | 0.2355 | ||||||||||||||
| 21.3 | 0.0010 | |||||||||||||||||||||
| 22 | 0.0479 | 0.0828 | 0.1647 | 0.0998 | 0.0968 | 0.1357 | 0.0020 | 0.1896 | ||||||||||||||
| 23 | 0.1707 | 0.1637 | 0.1257 | 0.0469 | 0.0409 | 0.0249 | 0.0359 | |||||||||||||||
| 24 | 0.1148 | 0.1557 | 0.0359 | 0.0050 | 0.0279 | 0.0190 | 0.0020 | |||||||||||||||
| 25 | 0.0898 | 0.1098 | 0.0040 | 0.0150 | 0.0030 | |||||||||||||||||
| 26 | 0.0150 | 0.0399 | 0.0010 | 0.0020 | 0.0030 | 0.0010 | ||||||||||||||||
| 27 | 0.0040 | 0.0100 | 0.0010 | 0.0010 | 0.0020 | |||||||||||||||||
| 28 | 0.0050 | 0.0010 |
Allelic frequencies and forensic parameters of Kazakh group
Raw genotyping data were presented in the Table S2. Allelic frequencies of the 22 STRs in the Kazakh group were enumerated in Table 1. Totally 276 alleles for 22 loci were found with the corresponding allelic frequencies varied from 0.001 to 0.3623. The least number of alleles (seven) was observed at D4S2366 locus, while the maximum (20) was at D12S391 locus. As presented in Fig. S3, the values of PE and Ho were in the range of 0.4903 to 0.7877, and 0.7385 to 0.8962, respectively, with the minimum values detected at D17S1290 locus and maximum at D20S470 locus. Additionally, the minimum values of PD, He and PIC emerged at D10S1435 locus with 0.8993, 0.7523 and 0.7124, separately. Conversely, the maximum values were all observed at D7S3048 locus with 0.9732, 0.8827 and 0.8700, respectively. The cumulative power of discrimination value for all 22 STR loci in the Kazakh group was 1-1.00E−28.
Interpopulation differentiation analyses between Kazakh and reference groups
In order to explore the population genetic differentiations between Kazakh group and the five previously reported reference groups, we calculated the locus-by-locus Fst values with p values of the 22 loci. As shown in Table 2, significant differences were observed between Kazakh and Xinjiang Uygur (Song et al., 2017) at nine loci, Xinjiang Hui (Fang et al., 2018) at 14 loci, Guangdong Han (Chen et al., 2016) at nine loci, Northern Han (Xie et al., 2015) (Hebei, Henan, Shaanxi) at 18 loci and Hainan Li (Chen et al., 2016) at 17 loci. However, after Bonferroni correction (p = 0.05∕110 = 0.00045455), Xinjiang Kazakh had significant differences with Xinjiang Uygur at one loci, Xinjiang Hui at three loci, Northern Han and Hainan Li at ten loci.
| Loci | Xinjiang Uygur | Xinjiang Hui | Northern Han | Guangdong Han | Hainan Li | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fst | p-value | Fst | p-value | Fst | p-value | Fst | p-value | Fst | p-value | |
| D1S1656 | 0.0008 | 0.2082 | 0.0018 | 0.0352 | 0.0020 | 0.0068 | 0.0002 | 0.8006 | 0.0069 | 0.0020 |
| D2S1338 | −0.0009 | 1.0000 | 0.0025 | 0.0029 | 0.0032 | 0.0000 | 0.0035 | 0.0811 | 0.0025 | 0.0694 |
| D3S3045 | 0.0021 | 0.0313 | 0.0009 | 0.1975 | 0.0020 | 0.0108 | 0.0001 | 0.7449 | 0.0089 | 0.0000 |
| D4S2366 | 0.0009 | 0.2151 | 0.0124 | 0.0000 | 0.0122 | 0.0000 | 0.0054 | 0.0264 | 0.0294 | 0.0000 |
| D5S2500 | 0.0018 | 0.0391 | 0.0044 | 0.0010 | 0.0039 | 0.0000 | 0.0023 | 0.2590 | 0.0056 | 0.0049 |
| D6S477 | 0.0045 | 0.0000 | 0.0028 | 0.0029 | 0.0009 | 0.1193 | 0.0007 | 0.5748 | 0.0055 | 0.0029 |
| D7S3048 | 0.0006 | 0.3656 | 0.0009 | 0.2209 | 0.0011 | 0.0489 | −0.0004 | 0.9580 | 0.0233 | 0.0000 |
| D8S1132 | 0.0005 | 0.3842 | 0.0040 | 0.0000 | 0.0057 | 0.0000 | 0.0081 | 0.0049 | 0.0204 | 0.0000 |
| D9S925 | 0.0002 | 0.5904 | 0.0032 | 0.0098 | 0.0039 | 0.0000 | 0.0038 | 0.1251 | 0.0027 | 0.0899 |
| D10S1435 | 0.0044 | 0.0039 | 0.0046 | 0.0020 | 0.0046 | 0.0000 | 0.0042 | 0.0929 | 0.0011 | 0.3969 |
| D11S2368 | 0.0013 | 0.0870 | 0.0021 | 0.0078 | 0.0006 | 0.2297 | 0.0051 | 0.0362 | 0.0046 | 0.0020 |
| D12S391 | 0.0017 | 0.0323 | 0.0000 | 0.8475 | 0.0006 | 0.2248 | 0.0046 | 0.0557 | 0.0051 | 0.0039 |
| D13S325 | 0.0006 | 0.3011 | 0.0001 | 0.6862 | 0.0044 | 0.0000 | 0.0047 | 0.0577 | 0.0167 | 0.0000 |
| D14S608 | 0.0030 | 0.0049 | 0.0021 | 0.0244 | 0.0022 | 0.0049 | 0.0109 | 0.0010 | 0.0044 | 0.0117 |
| D15S659 | 0.0015 | 0.0557 | 0.0018 | 0.0244 | 0.0028 | 0.0000 | 0.0073 | 0.0020 | 0.0141 | 0.0000 |
| D16S539 | 0.0002 | 0.6246 | 0.0067 | 0.0000 | 0.0060 | 0.0000 | 0.0056 | 0.0440 | 0.0042 | 0.0352 |
| D17S1290 | 0.0006 | 0.3519 | 0.0000 | 0.7341 | 0.0009 | 0.1095 | 0.0035 | 0.1486 | 0.0153 | 0.0000 |
| D18S535 | 0.0037 | 0.0029 | 0.0016 | 0.0528 | 0.0022 | 0.0049 | 0.0105 | 0.0010 | 0.0344 | 0.0000 |
| D19S253 | −0.0005 | 1.0000 | 0.0008 | 0.2815 | 0.0021 | 0.0088 | −0.0008 | 0.9638 | 0.0024 | 0.1202 |
| D20S470 | 0.0007 | 0.2590 | 0.0018 | 0.0205 | 0.0018 | 0.0059 | 0.0098 | 0.0010 | 0.0090 | 0.0000 |
| D21S1270 | 0.0018 | 0.0332 | 0.0016 | 0.0596 | 0.0021 | 0.0029 | 0.0057 | 0.0313 | 0.0019 | 0.1574 |
| D22-GATA198B05 | 0.0043 | 0.0010 | 0.0038 | 0.0010 | 0.0052 | 0.0000 | 0.0038 | 0.0704 | 0.0138 | 0.0000 |
Notes:
The numbers in bold indicated the loci showed significant differences between studied Kazakh group and the reference populations after the Bonferroni correction (the significant level =0.05∕110 = 0.00045455).
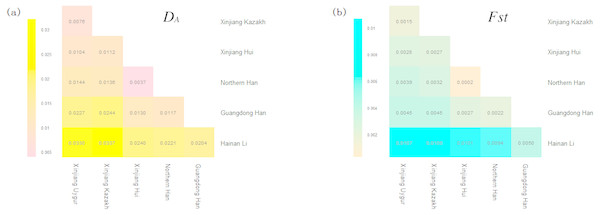
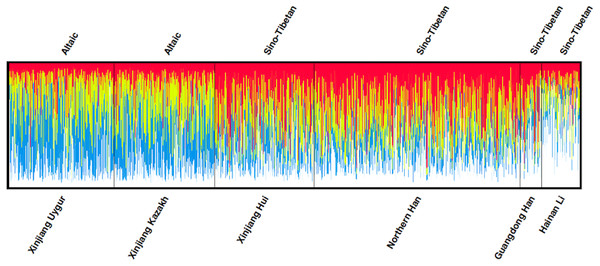
The DA and Fst (p) values of pairwise populations were listed in Tables S3 and S4, respectively. Kazakh group had the shortest distance with Uygur group (DA = 0.0076), followed by Xinjiang Hui (DA = 0.0112), Northern Han (DA = 0.0136), Guangdong Han (DA = 0.0244) and Hainan Li (DA = 0.0337) groups. The pairwise Fst values were also similar to the above DA results: the studied Kazakh group had the lowest Fst value with Xinjiang Uygur group and the highest with Hainan Li group. To visualize the results of population pairwise DA and Fst values more straightforwardly, we performed two heat maps of DA and Fst values between Kazakh and other reference groups as presented in Figs. 1A and 1B. The population structure analyses of the Kazakh and five reference groups were performed using the STRUCTURE software with the result depicted in Fig. 2. K = 4 was the most suitable configuration relying on the output posterior probability results (Evanno, Regnaut & Goudet, 2010). Individuals were represented by a vertical line and the colors stood for estimated membership fractions (Rosenberg et al., 2002).
Figure 1: (A) A heat map of pairwise DA values of Xinjiang Kazakh and five previously published populations based on R software (R Core Team, 2013). (B) A heat map of pairwise Fst values of STR loci among Xinjiang Kazakh and five previously published populations conducted by R software.
Figure 2: Clustering structure for the full-loci dataset assuming K = 4 of the six populations.
Structure analysis of six groups, assuming K = 4, was presented by the STRUCTURE software v.2.3.4. K = 4 was the most suitable number of the estimated ancestral components, which was represented by four colors. The genetic components of each individual were represented by a vertical line divided by colors.Discussion
STR loci from CODIS are commonly contained in present commercial kits. However, non-CODIS STRs can be used to enhance discrimination efficiency in some cases (He et al., 2018; Inokuchi et al., 2018). In our study, 22 autosomal STR loci, including 18 non-CODIS loci and four CODIS loci, were utilized to assess their genetic distributions in the Kazakh group living in Xinjiang Uygur Autonomous Region. Significant deviations of LD among these 22 STR loci were detected in D10S1435-D15S659 and D13S325-D17S1290, which were not found in other similar studies based on Microreader™ 23sp ID kit (Fang et al., 2018; Liu et al., 2016). Moreover, the fairly small r2 values of 22 STR pairs were conductive to determine the low correlations between these pairwise STRs. Hence, these 22 STR loci can be treated as independent markers in Xinjiang Kazakh group. Different ethnic groups in China might have various genetic backgrounds with different allelic frequency distributions, like Han, Xinjiang Kyrgyz and Xibe groups (Guo et al., 2018). Thus, allelic frequency data of these 22 STRs presented here is of great importance for forensic applications in Kazakh ethnic group. In our study, all loci were at a high degree of heterozygosity (He > 0.7, Ho > 0.7) (Akhteruzzaman et al., 2013; Nakamura et al., 1987). PD values of all the loci were greater than 0.9 except for D10S1435 locus (PD = 0.8993), and the PIC values were in the range of 0.7124 to 0.8700. Thus, STR loci in our study could obtain effective information in STR analyses for forensic cases. Besides, this study also enriched the Chinese non-CODIS STRs reference databases.
Various ethnic groups, in China, are deemed to have their special ethnic origins or different genetic backgrounds to some extent (He et al., 2018). In this research, we studied Kazakh group and previously published five Chinese groups with 2,345 individuals to elucidate the population genetic affiliations. The DA and Fst heat maps demonstrated that Kazakh group had closer genetic distances with Xinjiang Uygur group. This result was also supported by the structure analyses. As depicted in Fig. 2, the component of blue color found in Kazakh and Uygur was much higher than that in the other four groups at K = 4, the most suitable number of ancestry components.
Our findings were consistent with the previously published studies, such as Y-STR loci described by Shan et al. (2014) and autosomal STR loci depicted by Feng et al. (2018). In Chinese history, although Kazakh and Uygur groups had distinct ethnic origins, both groups all partially encompassed the genetic contributions of the Mongol and Turkic groups (http://www.khazaria.com/genetics/kazakhs.html). Furthermore, Kazakh and Uygur were the main ethnic groups in the ancient Silk Road and they had the similar religious faith, custom and culture (Kong et al., 2017). These historical and cultural backgrounds might explain, at least in part, the minor genetic differentiations between Xinjiang Kazakh and Uygur groups.
Conclusion
Our results illustrated that these 22 STR loci were highly polymorphic in the Xinjiang Kazakh group and, hence, can be utilized in forensic cases. In addition, close genetic distances and a similar genetic structure demonstrated the intimate genetic relationships between Kazakh and Uygur groups.