Comparative analysis of the complete plastomes of nine Pimpinella species (Apiaceae) from China
- Published
- Accepted
- Received
- Academic Editor
- Alison Nazareno
- Subject Areas
- Genomics, Molecular Biology, Plant Science, Taxonomy
- Keywords
- Apiaceae, Chloroplast genome, Pimpinella, Phylogenetic relationship
- Copyright
- © 2023 Wang et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2023. Comparative analysis of the complete plastomes of nine Pimpinella species (Apiaceae) from China. PeerJ 11:e14773 https://doi.org/10.7717/peerj.14773
Abstract
Pimpinella L. is one of the large genera in the Apiaceae family. In a previous study, the molecular phylogenies of Pimpinella were explored using nuclear ribosomal DNA internal transcribed spacers (ITS) and several chloroplast DNA segments. There have been few studies conducted on chloroplast genomes in Pimpinella, which has limited systematic understanding of this genus. We assembled the complete chloroplast genomes of nine Pimpinella species from China using data generated from next generation sequencing (NGS). The chloroplast (cp) DNA used were standard double-stranded molecules, ranging from 146,432 base pairs (bp) (P. valleculosa) to 165,666 bp (P. purpurea) in length. The circular DNA contained a large single-copy (LSC) region, small single-copy (SSC) region, and pair of inverted repeats (IRs). The cp DNA of the nine species contained 82–93 protein-coding genes, 36–37 transfer RNA (tRNA) genes, and eight ribosomal RNA (rRNA) genes, respectively. Four species (P. smithii, P. valleculosa, P. rhomboidea, and P. purpurea) exhibited striking distinctions in genome size, gene number, IR boundary, and sequence identity. We confirmed the non-monophyly of the Pimpinella species on the basis of the nine newly identified plastomes. The distant relationship between the above-mentioned four Pimpinella species and Pimpinelleae was indicated with high support values. Our study provides a foundation for future in-depth phylogenetic and taxonomic studies of genus Pimpinella.
Introduction
The genus Pimpinella L. (Apiaceae, Apioideae) is comprised of approximately 150 species that are mainly distributed throughout Europe, Africa, and Asia, with 39 species and two varieties found in China (Pu, 1985). This total was recently revised to include 44 species from the flora of China (Pu & Watson, 2005). Its members are characterized by ordinary ovate and laterally compressed mericarps without wings or spines on the ribs. The boundaries between the species within this genus are obscure. Moreover, the generic borders between Pimpinella and its related genera (such as Carum, Apium, Aegopodium, and Spuriopimpinella) are frequently blurred. The published molecular phylogenies of Pimpinella were inferred using the nuclear ribosomal internal transcribed spacer (ITS) region and a few plastid markers (rps16 intron, rpl16 intron, rps16 exon, trnL intron, and trnL-F spacer) (Magee et al., 2010; Wang et al., 2014; Fereidounfar, Ghahremaninejad & Khajehpiri, 2016; Fernández Prieto et al., 2018; Mousavi et al., 2022). The genus Pimpinella is not monophyletic and its members have been included among the Selineae, Echinophoreae, Pyramidoptereae, and Acronema clades and the East Asian clade (Wang et al., 2014; Mousavi et al., 2022).
A study of 26 Pimpinella species from China was conducted using data from the ITS and two cpDNA intron sequences (Wang et al., 2014), many parallel branches and lower support values were identified. There is a need to find more suitable polymorphic regions. In recent years, many comparative analyses of chloroplast genomes have enriched the molecular phylogenetic study of Apiaceae (Kang et al., 2019; Guo et al., 2020; Li et al., 2020; Ren et al., 2020; Liu et al., 2022). Few chloroplast genomes of the Pimpinella species have been reported (Tan & Yu, 2018; Wang, Cao & Liu, 2020) and the current comparative analysis of Pimpinella from China at the whole chloroplast genome level is still lacking. We sought to further investigate the plastome features of the Pimpinella species and enrich the phylogeny of the complicated Pimpinella genus. The nine complete plastid genome sequences of the newly sequenced Pimpinella will lay the foundation for understanding the relationships among these common Chinese Pimpinella members.
Materials & Methods
Sample collection, DNA extraction, and genome sequencing
In this study, we investigated nine Pimpinella species distributed throughout China (Table 1). Voucher specimens were preserved in the herbarium of Hengyang Normal University (HYNU). Fresh leaves collected in the field were dried and stored in silica. Total genomic DNA were then extracted using a modified cetyltrimethylammonium bromide protocol (Doyle, 1987), which was conducted by Novogene (Tianjin, China). DNA library prep and sequencing were completed at Novogene (Tianjin, China) on the Illumina Novaseq platform using the PE150 sequencing strategy.
Plastome sequence assembly and annotation
Utilizing NOVOPlasty 3.7 (Dierckxsens, Mardulyn & Smits, 2017) with the ribulose-1,5-bisphosphate carboxylase/oxygenase (rbc L) gene from Pimpinella diversifolia DC. (GenBank accession number MT561033) as the seed file, the raw reads of genome-related chloroplasts were assembled. After aligning with congeneric species in pairs, the plastome sequences were annotated using Geneious Prime 2019.2.3 (Kearse et al., 2012), supplemented with necessary manual adjustment. Nine plastome sequences accompanied with their annotations were submitted to NCBI (ON321877 –ON321885).
Chloroplast genome comparative analyses
In each of the nine plastome sequences, two inverted repeat (IR) regions were determined using the ‘Repeat Finder’ plugin in Geneious Prime (Kearse et al., 2012). Utilizing ‘Extract Annotations,’ all protein-coding genes (CDS) were extracted in Geneious Prime (Kearse et al., 2012) and subsequently concatenated into one sequence for each accession. We removed genes with lengths shorter than 300 bp, as well as repeated gene sequences. Using CodonW (https://sourceforge.net/projects/codonw/), the relative synonymous codon usage (RSCU) values of each species were determined. These RSCU values were presented on a heatmap created using TBtools v1.086 (Chen et al., 2020). With the help of an online software tool named PREP (Mower, 2009), RNA editing sites were forecasted.
REPuter (Kurtz et al., 2001) was utilized to search for the short-dispersed repeats (SDRs) of nine plastome sequences. All four options (forward, reverse, complement, and palindromic repeats) were chosen for ‘Match Direction’. Moreover, the Hamming distance value was set to 3 and minimal repeat size was set to 30. Using the MIcroSAtellite identification tool (MISA) (https://webblast.ipk-gatersleben.de/misa/) (Beier et al., 2017), simple sequence repeats (SSRs) were identified. Before that, 10, five, four, three, three, and three were selected as the minimum number of SSRs for mono-, di-, tri-, tetra-, penta-, and hexanucleotides, respectively.
| Accession no./taxon name | Sample site | Location | Voucher no. |
|---|---|---|---|
| Pimpinella candolleana Wight et Arn. | Kunming, Yunnan | 25°8′38″N, 102°44′34″E | w190804 |
| P. diversifolia DC. | Hangzhou, Zhejiang | 30°20′43″N, 119°26′43″E | w2101 |
| P. purpurea (Franch.) de Boiss. | Lijiang, Yunnan | 27°2′42″N, 100°11′47″E | w19080103 |
| P. rhomboidea Diels | Wushan, Chongqing | 31°17′11″N, 110°4′26″E | w20082201 |
| P. rubescens (Franch.) Wolff ex Hand.-Mazz. | Lijiang, Yunnan | 27°3′17″N, 100°11′46″E | w19080102 |
| P. scaberula (Franch.) Wolff | Deqin, Yunnan | 28°1′9″N, 98°54′1″E | w20081301 |
| P. smithii Wolff | Wushan, Chongqing | 31°17′11″N, 110°4′26″E | w20082202 |
| P. thellungiana H. Wolff | Lvliang, Shanxi | 37°14′21″N, 111°12′42″E | w190825 |
| P. valleculosa K.T.Fu | Wenxian, Gansu | 32°49′52″N, 105°23′39″E | H92903 |
We used the online tool IRSCOPE (https://irscope.shinyapps.io/irapp/) to display the structure of the nine plastomes, and adjusted manually if necessary. Utilizing the MAFFT (Katoh & Standley, 2013) plugin (alignment algorithm of FFT-NS-2 selected) within Geneious Prime (Kearse et al., 2012), nine Pimpinella plastomes were aligned. After that, the genetic distance of these nine Pimpinella plastomes was estimated using MEGA-X (https://www.megasoftware.net/) and the Kimura 2-parameter model (Kumar et al., 2018).
Phylogenetic analysis
In order to investigate the relationship between the nine Pimpinella species and allied taxa within Apioideae, we conducted phylogenetic analysis on the chloroplast genome sequences. Before that, the complete plastomes of the 13 other species within Apioideae of related genera were downloaded from the NCBI: Angelica sylvestris, Bupleurum boissieuanum, Bupleurum falcatum, Bupleurum latissimum, Cnidium officinale, Crithmum maritimum, Hansenia forbesii, Hansenia weberbaueriana, Haplosphaera phaea, Peucedanum praeruptorum, Pleurospermum camtshaticum, Pterygopleurum neurophyllum, and Tongoloa silaifolia (see GenBank accession numbers in the Supplemental File). The dataset was subsequently aligned using MAFFT (with algorithm of FFT-NS-2) in Geneious Prime (Kearse et al., 2012). Maximum likelihood (ML) analysis was performed using raxmlGUI (Silvestro & Michalak, 2012) with rapid bootstrap (1,000 replicates) and the GTRGAMMA model.
Results
The total characteristics of cp DNA of nine Pimpinella species
The complete cp genome of the nine Pimpinella species was a double-stranded molecule that ranged from 146,432 bp in P. valleculosa to 165,666 bp in P. purpurea. The cp genome had a conserved structure, identical to the majority of angiosperm plants, that contained a pair of IR regions (IRa and IRb), one LSC region, and one SSC region (Table 2). The plastid genomes of P. candolleana, P. diversifolia, P. rubescens, P. scaberula, and P. thellungiana were comprised of 130 genes: 84 protein-coding genes, 37 tRNA genes, and eight rRNA genes. Among these, 96 genes were unique, and 17 genes were duplicated in the IR region, including six protein-coding genes (ndhB, rpl23, rps7, rps12, ycf1, and ycf2), seven tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC), and four rRNA genes (rrn4.5, rrn5, rrn16, and rrn23).
Moreover, the plastid genomes of P. smithii and P. valleculosa contained 127 genes: 82 protein-coding genes, 36 tRNA genes, and eight rRNA genes. Among the 127 genes, 99 were unique and 14 genes were duplicated in the IR region, including four protein-coding genes (ndhB, rps7, rps12, and ycf1), six tRNA genes (trnA-UGC, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC) and four rRNA genes (rrn4.5, rrn5, rrn16, and rrn23).
The plastid genomes of P. rhomboidea consisted of 131 genes, including 85 protein-coding genes, 37 tRNA genes, and eight rRNA genes. Among these, 95 genes were unique and 18 genes were duplicated in the IR region, including seven protein-coding genes (ndhB, rpl2, rpl23, rps7, rps12, ycf1, and ycf2), seven tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC) and four rRNA genes (rrn4.5, rrn5, rrn16, and rrn23).
The plastid genomes of P. purpurea consisted of 139 genes, including 93 protein-coding genes, 37 tRNA genes and eight rRNA genes. Among these, 87 genes were unique and 26 genes were duplicated in the IR region, including 15 protein-coding genes (infA, ndhB, rpl2, rpl14, rpl16, rpl22, rpl23, rpl36, rps3, rps7, rps8, rps12, rps19, ycf1, and ycf2), seven tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC), and four rRNA genes (rrn4.5, rrn5, rrn16, and rrn23).
| Species | Size (bp) | LSC (bp) | SSC (bp) | IR (bp) | Number of protein-coding genes | Number of tRNA genes | Number of rRNA genes | GC content (%) |
|---|---|---|---|---|---|---|---|---|
| P. candolleana | 153,199 | 86,109 | 17,137 | 24,913 | 84 | 37 | 8 | 37.7 |
| P. diversifolia | 153,135 | 86,219 | 17,114 | 24,901 | 84 | 37 | 8 | 37.7 |
| P. purpurea | 165,666 | 86,110 | 17,458 | 31,049 | 93 | 37 | 8 | 37.9 |
| P. rhomboidea | 153,298 | 85,662 | 16,754 | 25,441 | 85 | 37 | 8 | 37.7 |
| P. rubescens | 152,236 | 85,209 | 17,113 | 24,957 | 84 | 37 | 8 | 37.6 |
| P. scaberula | 152,499 | 85,645 | 17,120 | 24,867 | 84 | 37 | 8 | 37.6 |
| P. smithii | 147,183 | 93,487 | 17,486 | 18,105 | 82 | 36 | 8 | 37.5 |
| P. thellungiana | 152,558 | 85,271 | 17,063 | 25,112 | 84 | 37 | 8 | 37.8 |
| P. valleculosa | 146,432 | 93,553 | 17,473 | 17,703 | 82 | 36 | 8 | 37.5 |
Codon usage and RNA editing site prediction
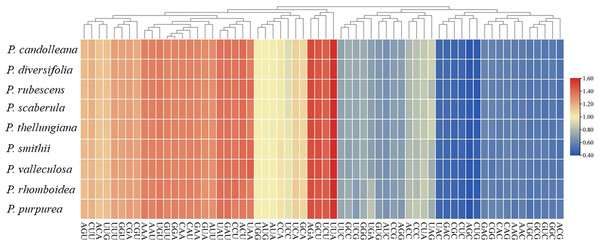
A total of 53 protein-coding genes were extracted from the nine Pimpinella plastomes. The total sequence sizes of these genes for codon analysis ranged from 63,423–63,516 bp. These protein sequences encoded 21,141–21,172 codons (Table 3). The GC content of the 1st, 2nd, and 3rd position of codons were 46.1, 38.3, and 29.8%, respectively. The RSCU values of the 30 codons were more than 1 (Table S1), and almost all of these ended with A/U. Approximately half of the codons were used more frequently, as shown in Fig. 1. These plastomes had an obvious bias in the use of codons towards the third position containing A/U. The amino acid leucine (Leu) had the largest number of codons (2,215–2,244), while cysteine (Cys) had the lowest number of codons (218–223).
| Index\Taxon | P. candolleana | P. diversifolia | P. rubescens | P. scaberula | P. thellungiana | P. smithii | P. valleculosa | P. rhomboidea | P. purpurea |
|---|---|---|---|---|---|---|---|---|---|
| Length (bp) | 63,516 | 63,513 | 63,447 | 63,516 | 63,516 | 63,432 | 63,483 | 63,435 | 63,423 |
| Codon No. | 21,172 | 21,171 | 21,149 | 21,172 | 21,172 | 21,144 | 21,161 | 21,145 | 21,141 |
| Amino acid No. | 21,119 | 21,118 | 21,096 | 21,119 | 21,119 | 21,091 | 21,108 | 21,092 | 21,088 |
| SC No. | 20,259 | 20,258 | 20,235 | 20,261 | 20,262 | 20,230 | 20,252 | 20,223 | 20,222 |
| ENC | 49.80 | 49.84 | 49.73 | 49.73 | 49.76 | 49.74 | 49.68 | 49.72 | 49.97 |
| CAI | 0.165 | 0.165 | 0.165 | 0.165 | 0.166 | 0.167 | 0.166 | 0.166 | 0.167 |
| CBI | −0.103 | −0.103 | −0.101 | −0.102 | −0.102 | −0.098 | −0.099 | −0.101 | −0.099 |
| FOP | 0.352 | 0.352 | 0.353 | 0.353 | 0.353 | 0.355 | 0.355 | 0.354 | 0.355 |
| GC content (%) | 0.381 | 0.382 | 0.381 | 0.381 | 0.382 | 0.380 | 0.380 | 0.381 | 0.383 |
In an analysis of the RNA editing sites, a total of 516 RNA editing sites were identified. The smallest number of editing sites was 56 for P. purpurea and the greatest was 58 in P. rhomboidea, P. smithii, P. thellungiana, and P. valleculosa (Table S2). In all of the nine Pimpinella plastomes, the ndhB gene contained the largest RNA editing sites (10 or 11). All of the RNA editing sites belonged to the same conversion type (cytosine to uracil (C-U)). Among these, the majority (41–45) were located in the 2nd codon position, and the remainder (13–17) were in the 1st position.
Repeats analysis
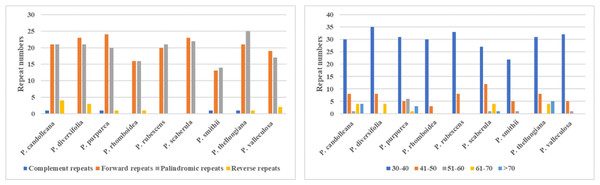
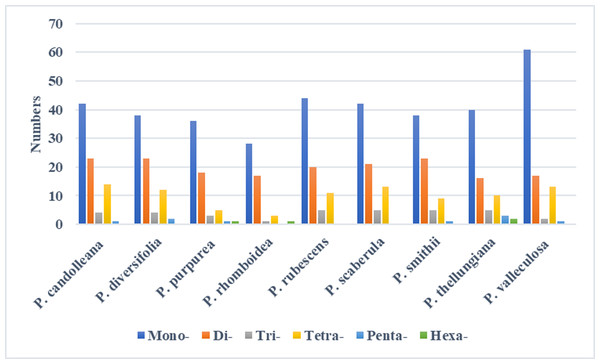
In the SDRs analysis, four types of repeats were determined: forward, reverse, complement, and palindromic repeats. Forward and palindromic repeats occurred in a large proportion in the nine Pimpinella plastomes. Moreover, the repeats with lengths of 30-40 bp accounted for the greatest proportion (Fig. 2). In the SSRs analysis, 50-94 SSRs were identified in the nine Pimpinella plastomes. The number of single nucleotide repeats was the largest (Fig. 3), with the majority being A/T repeats.
Figure 1: The RSCU values of codons in the nine Pimpinella plastomes.
Figure 2: The comparison of SDRs analysis in the nine Pimpinella plastomes.
Figure 3: The identification of SSRs in the nine Pimpinella plastomes.
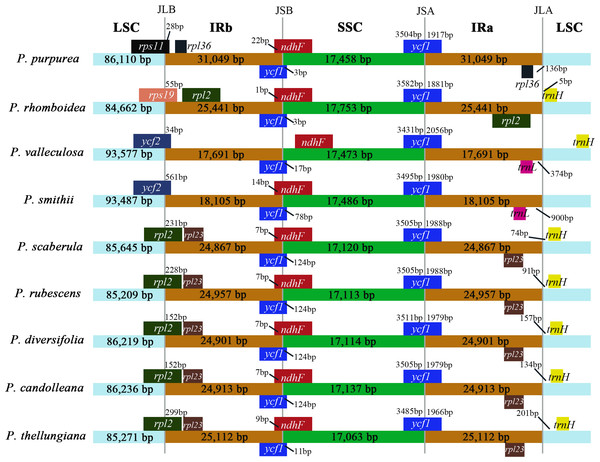
IR boundary comparison
The comparison of IR/SC borders among the nine plastomes are shown in Fig. 4. The gene arrangements and contents in the IR/SC borders of the plastomes of these five species (P. candolleana, P. diversifolia, P. rubescens, P. scaberula, and P. thellungiana) coincided. The LSC/IRb border of P. smithii and P. valleculosa, located in the ycf2 gene, and the trnL gene were 900 bp and 374 bp away from the IRa/LSC border, respectively. The LSC/IRb border of P. rhomboidea, located in the rps19 gene, and the trnH gene was five bp away from the IRa/LSC border. The LSC/IRb border of P. purpurea, located in the rps11 gene, and the rpl36 gene had a distance of 136 bp from the IRa/LSC border.
Figure 4: The IR boundary comparison in the nine Pimpinella plastomes.
Genetic distance analysis
Genetic distance analysis results are shown in Table 4. The highest value of pairwise genetic distance was 0.0365 between P. candolleana and P. purpurea, while the lowest was 0.0007 between P. candolleana and P. diversifolia.
| P. rubescens | P. scaberula | P. diversifolia | P. candolleana | P. thellungiana | P. valleculosa | P. smithii | P. rhomboidea | |
|---|---|---|---|---|---|---|---|---|
| P. scaberula | 0.0019 | |||||||
| P. diversifolia | 0.0048 | 0.0046 | ||||||
| P. candolleana | 0.0048 | 0.0047 | 0.0007 | |||||
| P. thellungiana | 0.0116 | 0.0116 | 0.0115 | 0.0115 | ||||
| P. valleculosa | 0.0211 | 0.0211 | 0.0208 | 0.0209 | 0.0201 | |||
| P. smithii | 0.0206 | 0.0205 | 0.0203 | 0.0204 | 0.0197 | 0.0070 | ||
| P. rhomboidea | 0.0350 | 0.0352 | 0.0353 | 0.0353 | 0.0347 | 0.0311 | 0.0308 | |
| P. purpurea | 0.0362 | 0.0364 | 0.0365 | 0.0365 | 0.0358 | 0.0319 | 0.0318 | 0.012 |
Phylogeny
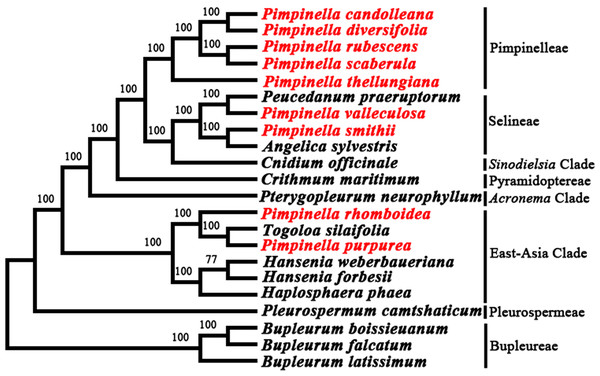
The phylogenetic relationship between nine Pimpinella species and related taxa in the Apioideae was well resolved with high support values (Fig. 5) based on the whole plastome sequence data. It was indicated that five Pimpinella species fell in the tribe Pimpinelleae. P. smithii and P. valleculosa were located in the tribe Selineae. P. rhomboidea and P. purpurea were clustered in the East-Asian clade, which coincided with the results obtained from ITS and cpDNA sequences (rps16 and rpl16 introns) (Wang et al., 2014).
Figure 5: Phylogenetic analysis from the plastomes of nine Pimpinella species and allied taxa.
Discussion
Apiaceae is a family that is famous for its unique fruit characteristics. Pimpinella is a relatively large member of the Apiaceae family and understanding the phylogeny and taxonomy of this genus is important. In this study, the Pimpinella plastomes were determined to be all quadripartite in the genome structure. We determined it had several distinct characteristics. First, the overall sizes of chloroplast genomes varied from 146,432 bp (P. valleculosa) to 165,666 bp (P. purpurea). Second, the numbers of the unique genes encoded by plastomes ranged from 87 (P. purpurea) to 99 (P. smithii and P. valleculosa). Among the closely-related species, the plastomes rarely fluctuated in size and gene content. For example, the plastome sizes of nine Chamaesium species reported (Guo et al., 2020) ranged from 152,703 bp to 155,712 bp, and contained 133 genes uniformly (including 95 unique genes). As another example, the chloroplast genome lengths of five Bupleurum species ranged from 155,621 bp to 156,108 bp, with similar genome structure (Li et al., 2020). In most cases, the variations in the lengths of the plastomes resulted from the contraction and expansion of the IR region (Downie & Jansen, 2015). IR contraction of ∼1.5 kb in Pimpinella relative to tobacco (Plunkett & Downie, 2000) was suggested based on chloroplast-DNA restriction site analysis. Herein, Pimpinella candolleana, P. diversifolia, P. rubescens, P. scaberula, and P. thellungiana with IR sizes of 24,867–25,112 bp possessed ∼0.2–0.4 kb IR contraction relative to tobacco (Nicotiana tabacum) (GenBank accession No. Z00044). Most notably, the large contractions of the IR regions in P. smithii (∼7.2 kb) and P. valleculosa (∼7.6 kb) made their plastome sizes significantly smaller than the newly identified plastomes. Subsequently, in P. smithii and P. valleculosa, rps19, rpl2, rpl23, and ycf2 had only one copy. The huge expansion (∼5.7 kb) of the IR regions in P. purpurea with a length of 31,049 bp made it the largest of the nine newly-obtained Pimpinella plastomes. At the LSC/IRb border, its IR expanded ∼6.1 kb compared with P. diversifolia, and resulted in many genes (infA, rpl2, rpl14, rpl16, rpl22, rpl36, rps3, rps8, and rps19) contained within the IR region. This kind of large IR expansion (>1,000 bp) was not usual, and also happened to Crithmum maritimum (Apiaceae) (Downie & Jansen, 2015). The expansion may originate from sequence recombination (Downie & Jansen, 2015), although this is waiting to be confirmed by subsequent studies.
The preference for codons ending with A/T in the Pimpinella plastomes was confirmed and also observed in other genera in Apiaceae (Guo et al., 2020; Li et al., 2020; Ren et al., 2020; Liu et al., 2022). The conversion type (C-U) of the RNA editing sites found here was similar to that of many other vascular plants (Wakasugi, Tsudzuki & Sugiura, 2001). Short repeats with 30-40 bp occupied most of these editing sites, which was consistent with the Ligusticum species in Apiaceae (Ren et al., 2020). A large number of SSRs in the newly sequenced Pimpinella chloroplast genomes were discovered to be mononucleotides (A/T), similar to many other angiosperms.
Our analyses support the results of previous studies based on ITS and cp DNA marker (rps16 intron and rpl16 intron) sequences (Wang et al., 2014). These studies found that the Chinese Pimpinella congeners did not cluster in one group, but instead mainly gathered in the Pimpinella core group, accompanied by P. smithii and P. valleculosa clustered within Selineae. P. purpurea and P. rhomboidea diverged earlier and clustered within the East Asian clade. The results clearly show Pimpinella as one non-monophyletic group. Meanwhile, our plastome data included a large amount of phylogenetically informative characteristics with greater support for the present topology compared to previous ones (Wang et al., 2014). They also provide a basis for future taxonomic studies of Chinese Pimpinella. In China, Pimpinella thellungiana is the most closely-related to the type species (P. saxifraga L.) (Wang et al., 2014). Four species clustered with P. thellungiana should be moved into the Pimpinella sensu stricto clade. Moreover, perhaps P. smithii should be transferred into the genus Angelica and P. valleculosa into Peucedanum. As for P. purpurea and P. rhomboidea, the present results were aligned with the treatments (Gui L, 2022, unpublished) to combine these two species into Tongoloa. The greatly divergent plastomes of P. purpurea and P. rhomboidea, compared to the other Pimpinella plastomes examined, may be related to potential chloroplast capture events. ‘Capture’ cases (Rieseberg & Soltis, 1991) have been reported in many different taxa. Chloroplast capture may bring incongruent plastid and nuclear DNA phylogeny reconstruction (Liu et al., 2020). It is necessary to be cautious when completing phylogenetic reconstruction using chloroplast DNA data (Rieseberg, Choi & Ham, 1991). Although chloroplast capture is also present in Apiaceae (Yi, Jin & Wen, 2015; Wen et al., 2021), the improper classification of P. purpurea and P. rhomboidea within genus Pimpinella may be a more accurate explanation, considering their congruent phylogenetic relationships between nuclear and chloroplast phylogenies (Wang et al., 2014). These taxonomic changes depend on a broader sampling for molecular analyses and morphological studies.
Previous studies (Guo et al., 2020; Li et al., 2020; Ren et al., 2020; Liu et al., 2022) have shown promising results in using the complete chloroplast genome sequences to infer phylogenies of genera in Apiaceae. Our study indicated that the plastomes proved to be good markers when used to reconstruct the phylogeny with a larger sampling. In addition, the Pimpinella and allied taxa may have a more complicated phylogeny, and future in-depth studies based on the chloroplast genomes are necessary.
Conclusion
This study is the first attempt to comprehensively examine plastome features and infer phylogeny using plastome data for the Pimpinella genus. The circular DNA of nine newly obtained plastomes contained a LSC region, SSC region, and pair of IRs. The plastomes ranged from 146,432 bp (P. valleculosa) to 165,666 bp (P. purpurea) in length. The plastid genomes of P. candolleana, P. diversifolia, P. rubescens, P. scaberula, and P. thellungiana were comprised of 130 genes, including 84 protein-coding genes, 37 tRNA genes, and eight rRNA genes. However, P. smithii and P. valleculosa contained only 127 genes. Moreover, the plastome of P. rhomboidea consisted of 131 genes. Most intriguingly, the plastid genome of P. purpurea consisted of 139 genes, including 93 protein-coding genes, accompanied by 15 protein-coding genes(infA, ndhB, rpl2, rpl14, rpl16, rpl22, rpl23, rpl36, rps3, rps7, rps8, rps12, rps19, ycf1, and ycf2), and seven tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC) duplicated. Phylogenetic analysis revealed that P. candolleana, P. diversifolia, P. rubescens, P. scaberula, and P. thellungiana were clustered in Pimpinelleae; P. smithii and P. valleculosa were clustered with Selineae members; and P. rhomboidea and P. purpurea in the East-Asian clade were relatively distant from their congeners. This study provides new evidence that chloroplast genomes might be useful when reconstructing the phylogeny of Pimpinella with a larger species sampling, which will be helpful for solving taxonomy problems in the genus.