Abstract
Background:
Uropathogenic Escherichia coli (UPEC) and Klebsiella pneumoniae (K. pneumoniae) are major pathogens which cause urinary tract infections (UTI) in pediatric patients. The presence of extended-spectrum β-lactamases (ESBLs) in these pathogens may further exacerbate infections and hamper successful treatment.Objectives:
We undertook a study to investigate the prevalence of ESBL genetic indicators among K. pneumoniae strains isolated from pediatric patients in Tehran, Iran. Moreover, genotyping of blaCTX-M-15-positive isolates was determined through repetitive extragenic palindromic sequence polymerase chain reactions (REP-PCR).Methods:
A total of 76 non-duplicate K. pneumoniae isolates were collected from outpatients admitted with UTIs at the pediatric nephrology wards of two hospitals in Tehran, Iran. The antibacterial susceptibility of K. pneumoniae isolates was determined by the disk diffusion method. The isolates were examined phenotypically and genotypically for ESBL production using the combined-disk method and PCR, respectively. The blaCTX-M-positive isolates were subjected to minimal inhibitory concentration (MIC) testing for ceftazidime and cefotaxime. The clonal relationships of blaCTX-M-15-positive isolates were determined through REP-PCR.Results:
The highest rates of antibiotic resistance were obtained for ampicillin (92.1%), followed by ceftazidime (40.8%), cefotaxime (40.8%), and aztreonam (39.5%). However, only one isolate (1.3%) was resistant to imipenem. Among the ESBL-positive isolates, blaCTX-M(64.5%) was the most prevalent gene, followed by blaSHV (54.8%) and blaTEM (41.9%). Of 20 blaCTX-M-carrying isolates, 14 isolates showed MICs of 256 μg/mL against cefotaxime. The other six isolates had MICs of 512 μg/mL. However, 16 out of 20 blaCTX-M-carrying isolates exhibited MICs of 128 μg/mL against ceftazidime. The other four K. pneumoniae isolates showed MICs of 256 μg/mL. Of 17 blaCTX-M-15-positive K. pneumoniae isolates, 16 distinct REP-PCR patterns (genotypes) were obtained.Conclusions:
The frequency of blaCTX-Mamong K. pneumoniae isolates was at an alarming rate, indicating that more efforts should be undertaken to track and monitor the spread of K. pneumoniae that produce CTX-M β-lactamases.Keywords
Urinary Tract Infections Pediatric Extended-Spectrum β-Lactamases Klebsiella pneumoniae
1. Background
Urinary tract infections (UTIs) are one of the most common infections in children, with an estimated prevalence of approximately 0.7% per person annually and accounting for more than $180 million spent per year for treatment (1-3). Uropathogenic Escherichia coli (UPEC) and Klebsiella pneumoniae (K. pneumoniae) are major nosocomial pathogens that cause UTIs (4). Up until the age of six, 7% and 2% of girls and boys will experience at least one UTI, respectively (2). Inadequate treatment of UTIs among the pediatric population can result in a high level of morbidity, including renal abscess formation, septicemia, renal scarring, hypertension, and even renal failure (3). In order to minimize the complications of a UTI, early diagnosis and empirical antibiotic prescription are often necessary, even before the return of urine culture results (5). Some UTI-causing pathogenic bacteria carry antibiotic-resistant genetic indicators, such as extended-spectrum β-lactamases (ESBLs) or carbapenemase genes, making for truly multi-drug-resistant (MDR) pathogens (5, 6). These MDR strains are resistant to at least three different classes of antibiotics. Thus, an increasing proportion of UTIs caused by MDR pathogens, including K. pneumoniae, has been a noticeable problem in different parts of the world in recent years (7, 8).
The spread of ESBL-producing gram-negative bacteria is a major concern for the development of therapies for infection (9). For instance, the ESBL-producing Klebsiella species and E. coli are now listed as two of the six drug-resistant pathogens against which new therapies are urgently needed (10). The spread of ESBL-producing K. pneumoniae not only results in clinical failures, but also prolonged hospitalization, higher morbidity, and excess health care costs (11). ESBLs are a group of enzymes with the ability to hydrolyze and cause resistance to various types of β-lactam antibiotics, including third-generation cephalosporins and monobactams (12, 13). Most ESBLs can be categorized into three groups: TEM, SHV, and CTX-M types (14). Although most of the ESBLs are derived from mutations in the classic TEM and SHV enzymes, the CTX-M type β-lactamases have become more important (15). CTX-M β-lactamases have become the most prevalent ESBL enzymes in Enterobacteriaceae during the past decade, particularly in certain European, Asian, and South American countries (12, 16). More specifically, recent reports from Iran have shown that the frequency of blaCTX-Mgenes among Klebsiella isolates is at an alarming rate (17, 18).
Because of the difficulty in managing infections caused by ESBL-producing K. pneumoniae, particularly in pediatric patients, it is very important to survey the ESBL type most prevalent nationally in order to periodically ascertain changing resistance patterns. These surveillances can provide useful information regarding their epidemiology, and can help physicians to choose the most appropriate antibiotic for successful antimicrobial therapy.
2. Objectives
Only a few studies on the prevalence of ESBL genes among K. pneumoniae strains isolated from children with UTIs are available at this time. Therefore, we have investigated the prevalence of these genes among K. pneumoniae strains isolated from pediatric patients in Tehran, Iran. In addition, genotyping of blaCTX-M-15-positive isolates was determined through the method of repetitive extragenic palindromic sequence polymerase chain reaction (REP-PCR).
3. Methods
3.1. Bacterial Isolates
In this cross-sectional study conducted over a period of six months from July 2015 to January 2016, a total of 76 non-duplicate K. pneumoniae isolates were collected from outpatients admitted with UTIs at the pediatric nephrology wards of two hospitals. All of the urine samples were cultured on eosin methylene blue (EMB, Merck KGaA, Darmstadt, Germany) and blood agar. These culture plates were incubated overnight at 37 ± 1°C. The Klebsiella isolates were identified using conventional bacteriological methods and biochemical testing (19). The verified K. pneumoniae isolates were transported to a laboratory where they were frozen at -80°C in trypticase soy broth (TSB, Merck) containing 20% (v/v) glycerol for further analysis.
3.2. Antimicrobial Susceptibility Testing
Testing for the antibiotic susceptibility of the isolates was performed on Mueller-Hinton agar (MHA, Merck) via the disk diffusion method in accordance with the Clinical and laboratory standards institute (CLSI) guidelines (20). The following antibiotics were used: amikacin (30 μg), ampicillin (10 μg), aztreonam (30 μg), cefotaxime (30 μg), ceftazidime (30 μg), ciprofloxacin (5 μg), co-trimoxazole (25 μg), gentamicin (10 μg), and imipenem (10 μg). The antibiotics were purchased from Sigma-Aldrich, Germany. To ensure the accuracy of the results obtained through susceptibility testing, Escherichia coli ATCC 25922 was used for quality control (20).
3.3. Phenotypic Detection of ESBL Production
The phenotypic detection of ESBL producing strains was conducted by employing a combined-disk method that used an antibiotic substrate of ceftazidime (30 μg) or cefotaxime (30 μg), and as an inhibitor of ESBL production, 10 µg of clavulanic acid were used. A difference of ≥ 5 mm between the beta-lactam disk and the disk containing the antibiotic associated with clavulanic acid was taken to be phenotypic confirmation of ESBL production (21). K. pneumoniae ATCC 700603 was used as an ESBL-positive control for the phenotypic confirmatory test.
3.4. Molecular Characterization of ESBL Genes
DNA was extracted from ESBL-positive Klebsiella isolates with a DNA extraction kit (AccuPrep® Genomic DNA Extraction Kit, Bioneer, South Korea) according to the manufacturer’s instructions, and was used for subsequent PCR analysis.
Clinical isolates of ESBL-producing Klebsiella were screened for blaSHV, blaTEM, and blaCTX-Mby PCR. K. pneumoniae ATCC 7881 containing blaSHV, blaCTX-Mand blaTEM genes was used as a positive control. In addition, the isolates testing positive for blaCTX-Mwere further analyzed by PCR with blaCTX-M-15-specific primers (12). A clinical isolate of E. coli carrying blaCTX-M-15 (provided by Dr. Mojtaba Memariani, Tarbiat Modares University, Tehran, Iran) was used as a positive control (12).
The primers used to amplify the genes are listed in Table 1. The amplification reactions were carried out in an Eppendorf thermal cycler (Germany) in a final volume of 25 µL containing 2.5 µL of 10X PCR buffer, 0.8 mg/µL MgCl2, 200 µM of deoxynucleotide triphosphates (dNTPs), 0.5 units of Taq polymerase, 10 pmol of each primer, and 5 µL of sample DNA. The PCR conditions for amplification were as follows: four minutes of initial denaturation at 94°C, followed by 35 cycles of denaturation at 93°C for 30 seconds, annealing different temperatures (at 53 - 58°C, Table 1) for 30 seconds, and extension at 72°C for 40 seconds, ending with a final extension period of 72°C for four minutes. To ascertain the expected sizes of the amplicons, the PCR products were analyzed with agarose gel electrophoresis stained with ethidium bromide (Sigma–Aldrich, Steinheim, Germany) and visualized using an ultraviolet (UV) transilluminator (Tanon, Shanghai, China).
PCR Primers Used in the Study for Detection of ESBL Genes in K. pneumoniae Isolates
| Target Genes | Primer Sequence (5’ to 3’) | Amplicon Size, bp | Annealing Temperature, °C | References |
|---|---|---|---|---|
| blaTEM | F: GAG TAT TCA ACA TTT CCG TGT C | 848 | 53 | (22) |
| R: TAA TCA GTG AGG CAC CTA TCT C | ||||
| blaSHV | F: AAG ATC CAC TAT CGC CAG CAG | 231 | 56 | (22) |
| R: ATT CAG TTC CGT TTC CCA GCG G | ||||
| blaCTX-M | F: TTT GCG ATG TGC AGT ACC AGT AA | 544 | 58 | (23) |
| R: CGA TAT CGT TGG TGG TGC CAT A | ||||
| blaCTX-M-15 | F: CAC ACG TGG AAT TTA GGG ACT | 996 | 55 | (24) |
| R: GCC GTC TAA GGC GAT AAA CA |
3.5. Determination of Minimal Inhibitory Concentrations (MICs)
The blaCTX-M-positive K. pneumoniae isolates were subjected to MIC testing for ceftazidime and cefotaxime via an agar dilution method based on CLSI standard procedure (20). Escherichia coli ATCC 25922 was used for quality control. Briefly, antibiotic solutions (Sigma-Aldrich, Germany) were added to molten Mueller-Hinton agar to provide twofold concentrations ranging from 0.5 to 1,024 μg/mL. Bacterial suspensions were applied to agar plates using a sampler inoculator to yield a final inoculum of 104 colony-forming units (CFUs) per spot (25). The results were read after incubation at 37°C for 18 - 24 hours. The assays were performed in duplicate.
3.6. REP-PCR
The clonal relationships of the blaCTX-M-15-positive isolates were determined by REP-PCR using primers REP-1 (5’-IIIGCGCCGICATCAGGC-3’) and REP-2 (5’-ACGTCTTATCAGGCCTAC-3’), as described previously (26). Amplification reactions were carried out in a 50 µL-volume. Cycling conditions were comprised of an initial denaturation (94°C for 10 minutes), followed by 30 cycles of denaturation (94°C for one minute), annealing (40°C for one minute), extension (65°C for 6 minutes), and a final extension at 65°C for eight minutes. The PCR products were separated by electrophoresis at 50 V for three hours on 1.5% (w/v) agarose gels stained with ethidium bromide (Sigma-Aldrich), and visualized using an UV- transilluminator (Tanon).
QLICs software was used to generate an unweighted pair group method with arithmetic mean (UPGMA) dendrogram. Band identification was performed manually. The Dice similarity coefficient (DSC) was used with position tolerance settings of 1.0%.
3.7. Statistical Analysis
Chi-squared or Fisher’s exact tests (SPSS software, version 17) were used for comparison of the categorical data. P values less than 0.05 were considered to be statistically significant.
4. Results
The mean age of the patients was 4.1 ± 3.4 years. These patients were comprised of 49 (64.5%) girls and 27 (35.5%) boys. The highest rates of antibiotic resistance were obtained for ampicillin (92.1%), followed by ceftazidime (40.8%), cefotaxime (40.8%), and aztreonam (39.5%). However, only one isolate (1.3%) was resistant to imipenem (Figure 1).
Antibiotic Susceptibilities of K. pneumoniae Isolates
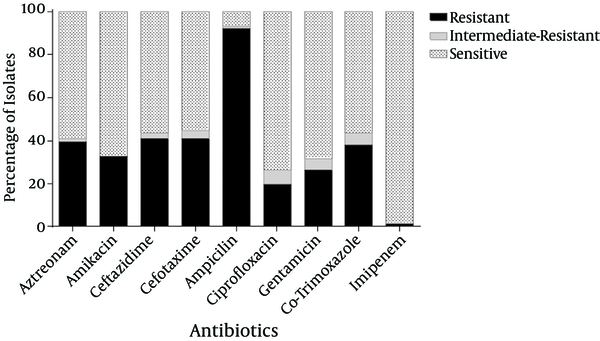
ESBL production was detected in 31 (40.8%) isolates of K. pneumoniae based on their phenotypes using ceftazidime/clavulanic acid and cefotaxime/clavulanic acid. The phenotypically-identified ESBL-producing isolates of K. pneumoniae were subjected to PCR using blaTEM, blaSHV, and blaCTX-M-specific primers. According to the PCR results, among the ESBLs-positive isolates, blaCTX-M(n = 20, 64.5%) was the most prevalent gene, followed by blaSHV (n = 17, 54.8%), and blaTEM (n = 13, 41.9%). Among the ESBL-producing K. pneumoniae, 15 out of 31 (48.4%) and 16 out of 31 (51.6%) carried one and more than one type of ESBL gene, respectively (Table 2). Moreover, out of 20 blaCTX-M-positive isolates, 17 isolates harbored the blaCTX-M-15variant. No significant associations were observed between the genes and the other mentioned categories, including patients’ genders and ages (P > 0.05).
Prevalence of ESBL Genes and Their Combinations Among 31 ESBLs-Positive Isolates
| β-Lactamases Genes | No. (%) |
|---|---|
| One gene | |
| blaCTX-M | 7 (22.6) |
| blaSHV | 5 (16.1) |
| blaTEM | 3 (9.7) |
| Total | 15 (48.4) |
| More than one gene | |
| blaCTX-M+ blaSHV | 6 (19.3) |
| blaCTX-M+ blaTEM | 4 (12.9) |
| blaSHV + blaTEM | 3 (9.7) |
| blaCTX-M+ blaSHV+ blaTEM | 3 (9.7) |
| Total | 16 (51.6) |
Of 20 blaCTX-M-carrying isolates, 14 isolates showed MICs of 256 μg/mL against cefotaxime. The other six isolates had MICs of 512 μg/mL. However, 16 out of 20 blaCTX-M-carrying isolates exhibited MICs of 128 μg/mL against ceftazidime. The other four isolates showed MICs of 256 μg/mL.
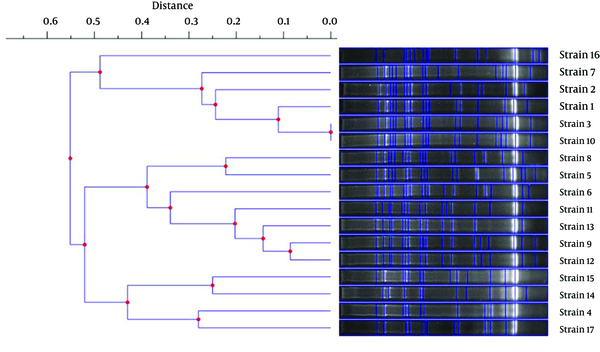
In order to define the clonal patterns of the blaCTX-M-15-carrying isolates, genotyping with REP-PCR was performed. Of 17 blaCTX-M-15-positive K. pneumoniae isolates, 16 distinct REP-PCR patterns (genotypes) were obtained. Only two isolates showed similar patterns (i.e., strain 3 and strain 10), as shown in Figure 2. Furthermore, the levels of similarity between the REP-PCR fingerprints of the isolates range from 45 to 100%.
Dendrogram Constructed With the UPGMA Method Using Genetic Distances Obtained by the REP-PCR Analysis of K. pneumoniae Isolates Carrying blaCTX-M-15
5. Discussion
UTIs are of major clinical importance due to the considerably high morbidity and mortality rates among children affected by them. Knowledge of the etiological agents of UTIs and their antimicrobial resistance patterns in specific geographical locations may aid clinicians when selecting the appropriate empirical antimicrobial therapy (27). Although K. pneumoniae is the second most etiologic agent of community-acquired UTIs after Escherichia coli, the former creates a dilemma for clinicians because of the multi-drug resistance expressed by this pathogen (28). In this study, the prevalence of K. pneumoniae strains resistant to third generation cephalosporins such as ceftazidime and cefotaxime is quite concerning (40.8%). The rate in these findings was higher than those expressed in other reports from Tehran, Iran, and thus an alarming increase in resistance to cephalosporins among K. pneumoniae isolates has been presented (18, 29). However, similar to other reports, imipenem was still the most effective antibiotic against K. pneumoniae isolates (1, 30). Fortunately, resistance to carbapenems remains rare among K. pneumoniae cases in Iran (30, 31).
The emergence and dissemination of multi-drug resistant K. pneumoniae expressing extended-ESBLs can result in clinical failure, prolonged hospitalization, increased morbidity, mortality, and increased health care costs (11). In the present study, the prevalence of ESBL-producing K. pneumoniae was 40.8%. In a study conducted in Turkey, Kizilca et al. showed that the prevalence of ESBL production in K. pneumoniae isolates associated with community acquired-UTIs was 53.2%, which is higher than that of our study (32). The proportion of ESBL production was 41.4% in another study from Greece, which is almost in accordance with the findings of this survey (33). In a recent study from Shahrekord, Iran, the proportion of ESBL producers among K. pneumoniae isolated from both community-acquired and nosocomial UTIs was 58% (34). However, Feizabadi et al. (2010) showed that the prevalence of ESBL production among nosocomial K. pneumoniae isolates was 72.1%, which was much higher than that of our study (31). In another study from Tehran, the rate of ESBL production was 54.9% among K. pneumoniae isolated from children admitted to pediatric hospitals (35). It should be noted that the prevalence of ESBL production in K. pneumoniae varies depending on geographical area, the nature of the institution, the age of population, and patient co-morbidities (36).
Inappropriate use of antibiotics and the transfer of ESBLs via a variety of mobile genetic elements, including transposons, insertion sequences, and integrons, play important roles in dissemination of ESBL-producing bacteria (37). In this study, blaCTX-M(64.5%) was found in the majority of ESBL-producing K. pneumoniae isolates, followed by blaSHV (54.8%) and blaTEM (41.9%). In agreement with the present study, AL-Subol and Youssef found that blaCTX-Mwas the most prevalent among E. coli and K. pneumoniae isolates in Syria, followed by blaSHV and blaTEM (38). The findings of the present study are consistent with an earlier study conducted in Morocco which found that CTX-M enzymes were the most common ESBL types (39). During the past few decades, CTX-M-type ESBLs have undergone rapid and global spreading, and they are now the most prevalent type of ESBL worldwide (21). These higher rates of CTX-M among K. pneumoniae could be attributed to efficient mobile genetic elements, such as integron structures, which may have influenced the rapid and easy dissemination of blaCTX-M(37). According to our results, blaCTX-M-15was the dominant variant among blaCTX-M-positive strains. In a recent study conducted by Najar Peerayeh et al. (2014), blaCTX-M-15 was also the most prevalent ESBL gene (62.5%) among ESBL-producing K. pneumoniae isolated from hospitalized patients in Tehran (18). However, in another study from Tehran (2010), it was found that blaTEM1, blaSHV5, blaSHV11, and blaCTX-M-15were the dominant ESBL genes among nosocomial K. pneumoniae strains (31). CTX-M enzymes confer higher levels of resistance to cefotaxime than to ceftazidime, as we have observed in our study. Our results showed that the proportion of blaCTX-M-carrying isolates with higher MIC values was greater than those of other studies (18, 29). This may suggest the presence of other mechanisms being involved in addition to CTX-M-15. Some studies from Iran showed that blaSHV was the most common ESBL type among K. pneumoniae isolates (31, 40). Comparing our data with previous reports from Iran revealed that there has been a marked change in prevalence of ESBL types. It is also important to note that the co-existence of different ESBL genes within the same isolate have been reported in other countries, as has also been detected in the current study (38, 39).
In this study, genotyping of CTX-M-15-producing K. pneumoniae isolates by REP-PCR showed that they were genetically diverse, indicating that multiple clones within the community have acquired blaCTX-M-15. In a survey performed in Indonesia (41), Severin et al. used REP-PCR for the genotyping of K. pneumoniae strains isolated from an academic hospital over the course of a four-month period. They observed 25 distinct profiles among 69 CTX-M-15-producing K. pneumoniae isolates (41). Using a similar technique, Lim et al. obtained 50 different profiles among 51 non-repeat K. pneumoniae strains isolated from five public hospitals in Malaysia in 2004 (42). In another study from Iran, genotyping of 37 CTX-M-positive K. pneumoniae by REP-PCR revealed 31 different patterns, suggesting that this heterogeneity could be partially attributed to different places and sources of infections (43).
In conclusion, this study provided insight into the current prevalence of ESBL-producing K. pneumoniae isolated from pediatric patients in Tehran, Iran. Our results showed that the frequency of blaCTX-Mamong Klebsiella isolates was at alarming rate, indicating that more efforts should be undertaken to track and monitor the spread of K. pneumoniae that produce CTX-M β-lactamases within both the hospital and community settings. Furthermore, blaCTX-M-15 has emerged as the predominant class of the ESBL gene structure among CTX-M-producing K. pneumoniae isolates within the community in Iran. However, further sequence analyses are necessary for a more comprehensive analysis of ESBL variants. We hope that our findings can be helpful for providing a better understanding of the epidemiology of ESBL genes among uropathogenic isolates of K. pneumoniae in Iran.
Acknowledgements
References
-
1.
Dahle KW, Korgenski EK, Hersh AL, Srivastava R, Gesteland PH. Clinical value of an ambulatory-based antibiogram for uropathogens in children. J Pediatric Infect Dis Soc. 2012;1(4):333-6. [PubMed ID: 23687582]. https://doi.org/10.1093/jpids/pis055.
-
2.
Schmidt B, Copp HL. Work-up of pediatric urinary tract infection. Urol Clin North Am. 2015;42(4):519-26. [PubMed ID: 26475948]. https://doi.org/10.1016/j.ucl.2015.05.011.
-
3.
Gaspari RJ, Dickson E, Karlowsky J, Doern G. Antibiotic resistance trends in paediatric uropathogens. Int J Antimicrob Agents. 2005;26(4):267-71. [PubMed ID: 16154724]. https://doi.org/10.1016/j.ijantimicag.2005.07.009.
-
4.
Yang YS, Ku CH, Lin JC, Shang ST, Chiu CH, Yeh KM, et al. Impact of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae on the outcome of community-onset bacteremic urinary tract infections. J Microbiol Immunol Infect. 2010;43(3):194-9. [PubMed ID: 21291846]. https://doi.org/10.1016/S1684-1182(10)60031-X.
-
5.
Copp HL, Shapiro DJ, Hersh AL. National ambulatory antibiotic prescribing patterns for pediatric urinary tract infection, 1998-2007. Pediatrics. 2011;127(6):1027-33. [PubMed ID: 21555502]. https://doi.org/10.1542/peds.2010-3465.
-
6.
Stillwell T, Green M, Barbadora K, Ferrelli JG, Roberts TL, Weissman SJ, et al. Outbreak of KPC-3 producing carbapenem-resistant klebsiella pneumoniae in a US pediatric hospital. J Pediatric Infect Dis Soc. 2015;4(4):330-8. [PubMed ID: 26582872]. https://doi.org/10.1093/jpids/piu080.
-
7.
Navidinia M, Krimi A, Ahsani R, Fallah F, Adabian S, Malekan MA, et al. Antibiotic susceptibility spectrum in UPEC from urine in children with UTI in Mofid children hospital. J Pure Appl Microbiol. 2012;6(2):751-6.
-
8.
Karimi A, Rahbar M, Fallah F, Navidinia M, Malekan MA. Detection of integron elements and gene groups encoding ESBLs and their prevalence in Escherichia coli and Klebsiella isolated from urine samples by PCR method. Afr J Microbiol Res. 2012;6(8):1806-9.
-
9.
Randrianirina F, Vedy S, Rakotovao D, Ramarokoto CE, Ratsitohaina H, Carod JF, et al. Role of contaminated aspiration tubes in nosocomial outbreak of Klebsiella pneumoniae producing SHV-2 and CTX-M-15 extended-spectrum beta-lactamases. J Hosp Infect. 2009;72(1):23-9. [PubMed ID: 19282056]. https://doi.org/10.1016/j.jhin.2009.02.004.
-
10.
Talbot GH, Bradley J, Edwards JJ, Gilbert D, Scheld M, Bartlett JG, et al. Bad bugs need drugs: an update on the development pipeline from the antimicrobial availability task force of the infectious diseases society of America. Clin Infect Dis. 2006;42(5):657-68. [PubMed ID: 16447111]. https://doi.org/10.1086/499819.
-
11.
Zhou T, Zhang Y, Li M, Yu X, Sun Y, Xu J. An outbreak of infections caused by extensively drug-resistant Klebsiella pneumoniae strains during a short period of time in a Chinese teaching hospital: epidemiology study and molecular characteristics. Diagn Microbiol Infect Dis. 2015;82(3):240-4. [PubMed ID: 25865067]. https://doi.org/10.1016/j.diagmicrobio.2015.03.017.
-
12.
Memariani M, Najar Peerayeh S, Zahraei Salehi T, Shokouhi Mostafavi SK. Occurrence of SHV, TEM and CTX-M beta-lactamase genes among Enteropathogenic Escherichia coli strains isolated From children with diarrhea. Jundishapur J Microbiol. 2015;8(4). ee15620. [PubMed ID: 26034531]. https://doi.org/10.5812/jjm.8(4)2015.15620.
-
13.
Villegas MV, Correa A, Perez F, Miranda MC, Zuluaga T, Quinn JP, et al. Prevalence and characterization of extended-spectrum beta-lactamases in Klebsiella pneumoniae and Escherichia coli isolates from Colombian hospitals. Diagn Microbiol Infect Dis. 2004;49(3):217-22. [PubMed ID: 15246513]. https://doi.org/10.1016/j.diagmicrobio.2004.03.001.
-
14.
Pitout JD, Laupland KB. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis. 2008;8(3):159-66. [PubMed ID: 18291338]. https://doi.org/10.1016/S1473-3099(08)70041-0.
-
15.
Kaur M, Aggarwal A. Occurrence of the CTX-M, SHV and the TEM genes among the extended spectrum beta-lactamase producing isolates of Enterobacteriaceae in a tertiary care hospital of north india. J Clin Diagn Res. 2013;7(4):642-5. [PubMed ID: 23730637]. https://doi.org/10.7860/JCDR/2013/5081.2872.
-
16.
Wang G, Huang T, Surendraiah PK, Wang K, Komal R, Zhuge J, et al. CTX-M beta-lactamase-producing Klebsiella pneumoniae in suburban New York City, New York, USA. Emerg Infect Dis. 2013;19(11):1803-10. [PubMed ID: 24188126]. https://doi.org/10.3201/eid1911.121470.
-
17.
Afzali H, Firoozeh F, Amiri A, Moniri R, Zibaei M. Characterization of CTX-M-type extend-spectrum beta-lactamase producing Klebsiella spp. in Kashan, Iran. Jundishapur J Microbiol. 2015;8(10). ee27967. [PubMed ID: 26587221]. https://doi.org/10.5812/jjm.27967.
-
18.
Peerayeh SN, Rostami E, Siadat SD, Derakhshan S. High rate of aminoglycoside resistance in CTX-M-15 producing Klebsiella pneumoniae isolates in Tehran, Iran. Lab Med. 2014;45(3):231-7. [PubMed ID: 25051075]. https://doi.org/10.1309/LMDQQW246NYAHHAD.
-
19.
Forbes BA, Sahm DF, Weissfeld AS, Trevino EA. Bailey and Scott's diagnostic microbiology, Mosby. St. Louis; 2002.
-
20.
Performance standards for antimicrobial susceptibility testing. Twentieth informational supplement. Document M100-S25. Wayne; 2015.
-
21.
Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. 2005;18(4):657-86. [PubMed ID: 16223952]. https://doi.org/10.1128/CMR.18.4.657-686.2005.
-
22.
Weldhagen GF, Poirel L, Nordmann P. Ambler class A extended-spectrum beta-lactamases in Pseudomonas aeruginosa: novel developments and clinical impact. Antimicrob Agents Chemother. 2003;47(8):2385-92. [PubMed ID: 12878494].
-
23.
Edelstein M, Pimkin M, Palagin I, Edelstein I, Stratchounski L. Prevalence and molecular epidemiology of CTX-M extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Russian hospitals. Antimicrob Agents Chemother. 2003;47(12):3724-32. [PubMed ID: 14638473].
-
24.
Doi Y, Adams-Haduch JM, Endimiani A, Sidjabat HE, Gaddad SM, et al. High prevalence of CTX-M-15-producing Klebsiella pneumoniae among inpatients and outpatients with urinary tract infection in Southern India. J Antimicrob Chemother. 2008;61(6):1393-4. [PubMed ID: 18356153]. https://doi.org/10.1093/jac/dkn109.
-
25.
Wiegand I, Hilpert K, Hancock RE. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3(2):163-75. [PubMed ID: 18274517]. https://doi.org/10.1038/nprot.2007.521.
-
26.
Romero L, Lopez L, Rodriguez-Bano J, Ramon Hernandez J, Martinez-Martinez L, Pascual A. Long-term study of the frequency of Escherichia coli and Klebsiella pneumoniae isolates producing extended-spectrum beta-lactamases. Clin Microbiol Infect. 2005;11(8):625-31. [PubMed ID: 16008614]. https://doi.org/10.1111/j.1469-0691.2005.01194.x.
-
27.
Mirsoleymani SR, Salimi M, Shareghi Brojeni M, Ranjbar M, Mehtarpoor M. Bacterial pathogens and antimicrobial resistance patterns in pediatric urinary tract infections: a four-year surveillance study (2009-2012). Int J Pediatr. 2014;2014:126142. [PubMed ID: 24959183]. https://doi.org/10.1155/2014/126142.
-
28.
El Bouamri MC, Arsalane L, El Kamouni Y, Zouhair S. Antimicrobial susceptibility of urinary Klebsiella pneumoniae and the emergence of carbapenem-resistant strains: A retrospective study from a university hospital in Morocco, North Africa. Afr J Urol. 2015;21(1):36-40.
-
29.
Nasehi L, Shahcheraghi F, Nikbin VS, Nematzadeh S. PER, CTX-M, TEM and SHV beta-lactamases in clinical isolates of Klebsiella pneumoniae isolated from Tehran, Iran. Iran J Basic Med Sci. 2010;13(3):111-8.
-
30.
Ghasemi Y, Archin T, Kargar M, Mohkam M. A simple multiplex PCR for assessing prevalence of extended-spectrum beta-lactamases producing Klebsiella pneumoniae in intensive care units of a referral hospital in Shiraz, Iran. Asian Pac J Trop Med. 2013;6(9):703-8. [PubMed ID: 23827147]. https://doi.org/10.1016/S1995-7645(13)60122-4.
-
31.
Feizabadi MM, Mahamadi-Yeganeh S, Mirsalehian A, Mirafshar SM, Mahboobi M, Nili F, et al. Genetic characterization of ESBL producing strains of Klebsiella pneumoniae from Tehran hospitals. J Infect Dev Ctries. 2010;4(10):609-15. [PubMed ID: 21045352].
-
32.
Kizilca O, Siraneci R, Yilmaz A, Hatipoglu N, Ozturk E, Kiyak A, et al. Risk factors for community-acquired urinary tract infection caused by ESBL-producing bacteria in children. Pediatr Int. 2012;54(6):858-62. [PubMed ID: 22882781]. https://doi.org/10.1111/j.1442-200X.2012.03709.x.
-
33.
Dotis J, Printza N, Marneri A, Gidaris D, Papachristou F. Urinary tract infections caused by extended-spectrum betalactamase-producing bacteria in children: a matched case control study. Turk J Pediatr. 2013;55(6):571-4. [PubMed ID: 24577973].
-
34.
Latifpour M, Gholipour A, Damavandi MS. Prevalence of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae isolates in nosocomial and community-acquired urinary tract infections. Jundishapur J Microbiol. 2016;9(3). ee31179. [PubMed ID: 27226874]. https://doi.org/10.5812/jjm.31179.
-
35.
Derakhshan S, Peerayeh SN, Fallah F, Bakhshi B, Rahbar M, Ashrafi A. Detection of class 1, 2, and 3 integrons among Klebsiella pneumoniae isolated from children in Tehran hospitals. Arch Pediatr Infect Dis. 2014;2(1):164-8.
-
36.
Lukac PJ, Bonomo RA, Logan LK. Extended-spectrum beta-lactamase-producing Enterobacteriaceae in children: old foe, emerging threat. Clin Infect Dis. 2015;60(9):1389-97. [PubMed ID: 25595742]. https://doi.org/10.1093/cid/civ020.
-
37.
Chong Y. Extended-spectrum β-lactamase-producing bacteria: an emerging clinical concern," in science against microbial pathogens: communicating current research and technological advances. Formatex Research Center; 2011.
-
38.
AL-Subol I, Youssef N. Prevalence of CTX-M, TEM and SHV beta-lactamases in clinical isolates of Escherichia coli and Klebsiella pneumoniae isolated from Aleppo University hospitals, Aleppo, Syria. Arch Clin Infect Dis. 2015;10(2).
-
39.
Barguigua A, El Otmani F, Talmi M, Bourjilat F, Haouzane F, Zerouali K, et al. Characterization of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates from the community in Morocco. J Med Microbiol. 2011;60(Pt 9):1344-52. [PubMed ID: 21546559]. https://doi.org/10.1099/jmm.0.032482-0.
-
40.
Ghafourian S, Bin Sekawi Z, Sadeghifard N, Mohebi R, Kumari Neela V, Maleki A, et al. The prevalence of ESBLs producing Klebsiella pneumoniae isolates in some major hospitals, Iran. Open Microbiol J. 2011;5:91-5. [PubMed ID: 21915229]. https://doi.org/10.2174/1874285801105010091.
-
41.
Severin JA, Mertaniasih NM, Kuntaman K, Lestari ES, Purwanta M, Lemmens-Den Toom N, et al. Molecular characterization of extended-spectrum beta-lactamases in clinical Escherichia coli and Klebsiella pneumoniae isolates from Surabaya, Indonesia. J Antimicrob Chemother. 2010;65(3):465-9. [PubMed ID: 20053690]. https://doi.org/10.1093/jac/dkp471.
-
42.
Lim KT, Yeo CC, Yasin RM, Balan G, Thong KL. Characterization of multidrug-resistant and extended-spectrum beta-lactamase-producing Klebsiella pneumoniae strains from Malaysian hospitals. J Med Microbiol. 2009;58(Pt 11):1463-9. [PubMed ID: 19589908]. https://doi.org/10.1099/jmm.0.011114-0.
-
43.
Ardalan N, Ramazanzadeh R. Repetitive element PCR fingerprinting (REP-PCR) in Klebsiella pneumoniae producing CTX-M in Sanandaj. Iran J Public Health. 2014;43(2):88.