Abstract
Background:
Shiga toxin-producing Escherichia coli (STEC) is a food-borne pathogen and infection with this organism causes illnesses such as bloody diarrhea, hemorrhagic colitis and hemolytic-uremic syndrome.Objectives:
Considering the lack of any information about the prevalence rate and the antibiotic resistance pattern of O157:H7 serotype in Tabriz, finding answers to the above mentioned subjects was among the goals of this study.Materials and Methods:
Two hundred E. coli strains from diarrheal or non-diarrheal stools of outpatients and hospitalized cases in Tabriz Imam Reza hospital were isolated between September and December 2014 using MacConkey agar and standard biochemical tests and then cultured on sorbitol MacConkey agar. The sorbitol-negative isolates were confirmed as the O157 serotype using O157 antisera. A multiplex polymerase chain reaction (PCR) method was used for the detection of stx-1, stx-2, eae, and mdh genes and the antibiotic resistance pattern of these isolates was determined using Kirby-Bauer method and clinical and laboratory standards institute (CLSI) standards.Results:
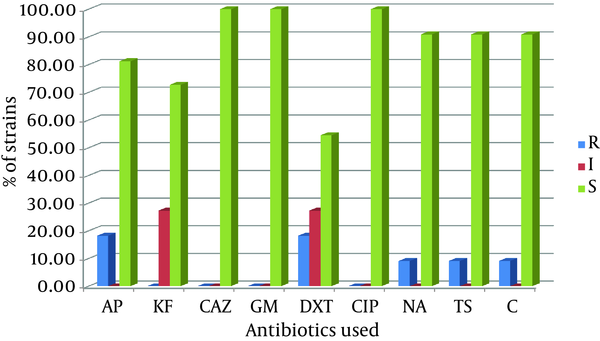
Of the isolates 11 (5.5%) were sorbitol-negative, which were later analyzed by multiplex PCR and the results revealed that 2 (18.18%) isolates contained the stx-1 gene, 10 (90.91%) contained the stx-2 gene, and 5 (45.45%) contained the eae gene. The stx-2 and eae genes were the most commonly encountered virulence factors. All or most of the isolates were susceptible to ceftazidime (100%), gentamicin (100%), ciprofloxacin (100%), nalidixic acid (90.9%), trimetoprim sulfamethoxazole (90.9%), chloramphenicol (90.9%), ampicillin (81.8%), and cephalothin (72.7%). On the contrary, moderate susceptibility of the isolates to doxycycline (54.5%) was observed.Conclusions:
Due to the low frequency of STEC O157 and the high susceptibility rates of the isolates to the tested antibiotics in this study, STEC O157 has not become a major problem in Tabriz yet, but comprehensive microbiological surveillance programs that provide early warning and limit the scale of possible outbreaks would be essential.Keywords
Escherichia coli Microbial Sensitivity Test Multiplex Polymerase Chain Reaction Shiga Toxin
1. Background
Shiga toxin-producing Escherichia coli (STEC) is a food-borne pathogen and several epidemics have been reported worldwide due to this pathogen (1). Person-to-person spread can occur easily due to the very low infectious dose and each nutrient from meat and vegetables to dairy products and water can cause the infection (2). Infection with this organism causes a spectrum of human illnesses, ranging from symptom-free to severe bloody diarrhea, hemorrhagic colitis (HC), and even life-threatening consequences such as hemolytic-uremic syndrome (HUS) (3). More than 100 serotypes capable of producing toxins have been identified. These serotypes pose one or both stx-1, stx-2, or variants of stx-2 genes, which are transferred by a lysogenic phage to the bacteria (4). These toxins primarily attach to the cell surface through a glycolipid receptor called globotriacylceramide (Gb3) and enter the cell by endocytosis; then, during an enzymatic process, remove an adenine form 28s rRNA which inhibits protein synthesis and leads to the cell death (2, 5). In addition to the toxins, another virulence factor associated with STEC is intimin, an outer membrane protein with a molecular weight of 94-97 kDa, which is responsible for adhesion to intestinal epithelial cells and is coded by the eae gene (4, 6).
The identification of the toxin-producing strains is not routinely performed in laboratories, but there are some methods for screening this bacterium. One of these methods is cell culture and observing the cytotoxic effects of the toxins on Vero cells. Unfortunately, this method is time-consuming and has some standardization issues and also requires facilities such as Vero cell cultures, identification of the toxin, and Vero-antitoxins for neutralization (2). Since STEC strains are serologically diverse groups, serological tests are not very helpful. The serotype O157:H7 (also known as enterohemorrhagic E. coli) is the most significant one and more than 100 non-O157 serotypes have been associated with human illnesses (7). It seems that the O157:H7 strain has been emerged from a single cell clone and spread around the world. These strains are genetically homogenous and while most strains of E .coli are capable of fermenting sorbitol, the O157:H7 strains do not have this ability. Therefore, we can isolate this strain by making selective mediums such as sorbitol MacConkey agar (8).
Among the methods of identification, polymerase chain reaction (PCR) is an appropriate method for the detection of toxin. However, according to the genetic diversity of this toxin, it will be difficult to identify the toxin during a single PCR reaction. Since the STEC strains often contain other virulence factors, designing a multiplex PCR method in which some specific genes (eae, stx-1, and stx-2) are simultaneously examined, has a high specificity and sensitivity (2, 7, 9). One of the treatments for infection with this bacterium is antibiotic therapy and the use of antibiotics as additives in animal foods increases bacterial resistance to antimicrobial agents (10).
2. Objectives
Considering the lack of any information about the prevalence rate and the antibiotic resistance pattern of O157:H7 serotype in Tabriz, finding answers to the above mentioned subjects was among the goals of this study.
3. Materials and Methods
3.1. Sample Collection
In this study, the target population was the E. coli strains isolated from diarrheal and non-diarrheal fecal samples of outpatients and hospitalized cases in Tabriz Imam Reza hospital between September and December 2014. The sample size was 200 and a simple random sampling method was employed.
3.2. Isolation and Identification
Inclusion criteria were growth on MacConkey agar medium (HiMedia, India) and complying with standard biochemical tests including triple sugar iron agar (TSI), sulfide indole motility (SIM), citrate agar, and methyl red voges-proskauer (MRVP) (11). The exclusion criteria were inhibition of growth on MacConkey agar and failure to comply with the standard biochemical tests. Patients who had taken antibiotics in the last 48 hours were excluded. Stock cultures were placed in tryptic soy broth (Merck, Germany) glycerol (80:20) medium and maintained at -70°C. Then, the collected samples were inoculated on sorbitol MacConkey agar (Quelab Laboratories Inc., Canada). After 18 hours of incubation at 37°C, sorbitol-negative colonies were identified as O157:H7 strains. All these sorbitol-negative isolates were later examined by O157 antisera (Baharafshan, Iran).
3.3. DNA Extraction and Multiplex Polymerase Chain Reaction
A boiling method with minor modifications was used for DNA extraction by adding 1 - 2 colonies of these sorbitol-negative isolates to 25 µL of tissue buffer (0.5 g sodium dodecyl sulfate (SDS), 0.4 g NaOH, and 200 mL deionized water). The NaOH-SDS solution helped in the lysis of Gram-negative E. coli cells (12). Then, it was boiled using a block heating thermostat (Beco, Germany), at 95°C for 10 minutes and then was centrifuged in a microcentrifuge (Vision, Korea) at 13000 rpm for 1 minute. Finally, 180 µL of deionized water was added to the tube. Concentration and quality of these DNA samples were checked using NanoDrop 1000 spectrophotometer. A ratio of ~ 1.8 absorbance at 260 and 280 nm was generally accepted as pure DNA.
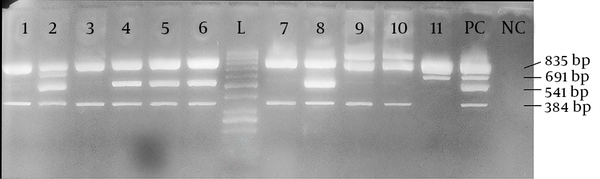
A multiplex PCR with 4 pairs of primers (Takapouzist, Iran) was optimized, as described in STEC center website (13) in a total volume of 25 µL. The final concentrations of the reaction components were as follows: 2 ng of DNA template, 1x PCR buffer, 0.2 mM of each dNTP, 2 mM of MgCl2, 0.2 µM of each primer, and 1 U of Taq DNA polymerase (Jena Bioscience, Germany). Finally, 12.70 µL ddH2O was added to achieve the desired 25 µL volume. The primer sequences as well as each amplicon size are given in Table 1. The mdh primers were related to malate dehydrogenase, a housekeeping gene, which served as internal positive control, confirming the classification of isolates as E. coli. The reaction was carried out by an amplification thermal cycler (Eppendorf, Germany). The thermal profile began by a 10-minute incubation at 94°C and run for 35 cycles with the following parameters: 92°C, one minute; 55°C, one minute; 72°C, 30 seconds. A final stage of 72°C for five minutes was added for completing any partially extended product. The PCR products were later analyzed by electrophoresis on 1.5% agarose gel.
PCR Primers and Conditions Used in This Study
| Primer | Nucleotide Sequence | Target | Amplicon Size, bp |
|---|---|---|---|
| Mdh-F | AGG CGC TTG CAC TAC TGT TA | mdh | 835 |
| Mdh-R | AGC GCG TTC TGT TCA AAT G | mdh | 835 |
| Eae-F | ATT ATG GAA CGG CAG AGG TTA AT | eae | 541 |
| Eae-R | ATC CCC ATC GTC ACC AGA GG | eae | 541 |
| Stx-1-F | CAT TCG CTC TGC AAT AGG TA | Stx-1 family | 691 |
| Stx-1-R | AAC TCG CGA TGC ATG ATG A | Stx-1 family | |
| Stx-2-F | TAT CTG GCG TTA ATG GAG TT | Stx-2 family | 384 |
| Stx-2-R | CCT GTC GCC AST TAT CTG AC |
3.4. Antibiotic Susceptibility Test
The sorbitol-negative strains were examined for resistance against ampicillin (10 µg), cephalothin (30 µg), ceftazidime (30 µg), gentamicin (10 µg), doxycycline (30 µg), ciprofloxacin (5 µg), nalidixic acid (30 µg), cotrimoxazole (25 µg), and chloramphenicol (30 µg) (MAST Group Ltd., UK), using Kirby-Bauer disc diffusion susceptibility test and the characterization of strains as susceptible, reduced susceptibility or resistant was as recommended by the Clinical and Laboratory Standards Institute (CLSI) (14).
4. Results
The isolated strains were tested with sorbitol MacConkey agar and 11 (5.5%) isolates were sorbitol-negative. Among these 11 isolates, 2 (18.2%) were isolated from males, while 9 (81.8%) were isolated from females. The average age of these 11 patients was 31.91 ± 26.31 years; the minimum was one year old and the maximum was 79 years old. Half of the patients were below 35 years of age. Serological test confirmed that all 11 sorbitol-negative isolates were O157 serotypes. By using a multiplex PCR and four pairs of primers, we showed that among these sorbitol-negative isolates, 2 (18.18%) contained the stx-1 gene, 10 (90.91%) the stx-2 gene, and 5 (45.45%) the eae gene. The mdh primers were used as internal positive control for the PCR and all 11 sorbitol-negative isolates were mdh-positive (Figure 1). Among the 200 samples in this study, the overall prevalence of the genes was 2 (1.0%) for stx-1, 10 (5.0%) for stx-2, and 5 (2.5%) for the eae gene. The sorbitol-negative strains were tested for resistance against nine antibiotics. The antibiotic resistance patterns of these isolates are shown in Figure 2.
Agarose Gel Electrophoresis of DNA Fragments Amplified by Multiplex PCR for 11 Isolated Shiga Toxin-Producing Escherichia Coli Strains
The Antibiotic Resistance Pattern of Sorbitol-Negative Strains
5. Discussion
STEC can cause severe human complications and O157:H7 is the most common and well-known serotype worldwide, causing approximately 63000 illnesses, 2100 hospitalizations, and 46 deaths per year in the United States (15). Our study indicated that 5.5% of the E. coli isolates were STEC O157. There are other reports about the prevalence of STEC O157 in other parts of Iran. Salmanzadeh-Ahrabi et al. (16) noted the same low frequency (3.5%) in Tehran. Aslani and Bouzari reported that none of the isolates belonged to STEC O157 in north of Iran (17). Moreover, Mazaheri et al. (18) noted the same results in Tehran, as only 1% of the isolated strains were STEC O157. Finally, Fard et al. (19) reported a frequency of 1.24%. This low prevalence reflects the scarcity of the bacteria in Tabriz and other parts of Iran, suggesting that employing fast and accurate ways to detect these infections in medical laboratories is an essential need.
Between stx-1 and stx-2, stx-2 is the most important virulence factor associated with human diseases and is about 400 folds more toxic to mice than stx-1 (2, 20). Our findings revealed that 10 of 11 (90.91%) STEC O157 strains isolated in this study contained the stx-2 gene. In contrast, only 2 (18.18%) of these strains contained the stx-1 gene. Similar results were noted by Mellor et al. in the United States (73%), Vicente et al. in Brazil (31.5%) and Leotta et al. in both New Zealand (89%) and Argentina (91%) indicated that the prevalence of stx-2 was much higher than that of stx-1 (6, 21, 22). These findings showed that stx-2 played a major role as a virulence factor in STEC O157 infections in Tabriz. Therefore, designing and employing rapid diagnosis tests based on the detection of this toxin can accelerate the speed of infection detection.
Intimin is the key adherence factor for STEC and a strong association has been reported between carriage of eae gene and the ability of STEC strains to cause severe human diseases (23); however, only 5 (45.45%) of these 11 strains contained the eae gene, suggesting that these strains produced additional virulence factors to compensate for the absence of the eae gene. Tahamtan and Namvari reported that 22.23% of the isolates possessed the eae gene (24). Pizarro et al. (11) noted that only 6% of the samples harbored the eae gene, while Bonyadian et al. (25) reported that none of their isolates had the eae gene. As a result, working on some additional adherence factors for STEC strains can be useful. The O157 strains isolated in this study only showed some degrees of resistance against doxycycline, an antibiotic in the tetracycline class. This is in accordance with earlier studies performed in Iran (18, 26). For example, Mazaheri et al. (18) noted that all of the isolated strains were resistant to tetracycline. Jafari et al. (26) also reported a high rate of resistance to tetracycline. In other areas such as China and India, resistance to doxycycline has also been reported (3, 27). This recommends that a tetracycline family antibiotic can be the drug of choice to be used in therapies or even as a prophylactic agent.
Currently, treatment of STEC O157 infection with antibiotics is arguable. In the United States, antibiotic therapy is not recommended for the treatment of O157 STEC infections because of the potential for releasing of shiga toxin and the possibility of HUS (10). However, a chemically synthesized analog of shiga toxin receptor Gb3 that could absorb the toxin is in the clinical trial phase (28). Our study alongside with this clinical trial can be used as a combination therapy and an effective treatment in curing patients with STEC infections. Finding the low frequency of STEC O157 and the high susceptibility rate of isolates to the tested antibiotics in this study, STEC O157 has still not become a major problem in Tabriz. Although, we recommend that comprehensive microbiological surveillance programs that provide early warnings and limit the scale of possible outbreaks would be essential.
Acknowledgements
References
-
1.
Karmali MA. Infection by verocytotoxin-producing Escherichia coli. Clin Microbiol Rev. 1989;2(1):15-38. [PubMed ID: 2644022].
-
2.
Paton JC, Paton AW. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin Microbiol Rev. 1998;11(3):450-79. [PubMed ID: 9665978].
-
3.
Khan A, Das SC, Ramamurthy T, Sikdar A, Khanam J, Yamasaki S, et al. Antibiotic resistance, virulence gene, and molecular profiles of Shiga toxin-producing Escherichia coli isolates from diverse sources in Calcutta, India. J Clin Microbiol. 2002;40(6):2009-15. [PubMed ID: 12037056].
-
4.
Fagan PK, Hornitzky MA, Bettelheim KA, Djordjevic SP. Detection of shiga-like toxin (stx1 and stx2), intimin (eaeA), and enterohemorrhagic Escherichia coli (EHEC) hemolysin (EHEC hlyA) genes in animal feces by multiplex PCR. Appl Environ Microbiol. 1999;65(2):868-72. [PubMed ID: 9925634].
-
5.
Kolling GL, Matthews KR. Export of virulence genes and Shiga toxin by membrane vesicles of Escherichia coli O157:H7. Appl Environ Microbiol. 1999;65(5):1843-8. [PubMed ID: 10223967].
-
6.
Leotta GA, Miliwebsky ES, Chinen I, Espinosa EM, Azzopardi K, Tennant SM, et al. Characterisation of Shiga toxin-producing Escherichia coli O157 strains isolated from humans in Argentina, Australia and New Zealand. BMC Microbiol. 2008;8:46. [PubMed ID: 18366637]. https://doi.org/10.1186/1471-2180-8-46.
-
7.
Paton AW, Paton JC. Direct detection and characterization of Shiga toxigenic Escherichia coli by multiplex PCR for stx1, stx2, eae, ehxA, and saa. J Clin Microbiol. 2002;40(1):271-4. [PubMed ID: 11773130].
-
8.
March SB, Ratnam S. Sorbitol-MacConkey medium for detection of Escherichia coli O157:H7 associated with hemorrhagic colitis. J Clin Microbiol. 1986;23(5):869-72. [PubMed ID: 3519658].
-
9.
Paton AW, Paton JC. Direct detection of Shiga toxigenic Escherichia coli strains belonging to serogroups O111, O157, and O113 by multiplex PCR. J Clin Microbiol. 1999;37(10):3362-5. [PubMed ID: 10488207].
-
10.
Schroeder CM, Zhao C, DebRoy C, Torcolini J, Zhao S, White DG, et al. Antimicrobial resistance of Escherichia coli O157 isolated from humans, cattle, swine, and food. Appl Environ Microbiol. 2002;68(2):576-81. [PubMed ID: 11823193].
-
11.
Pizarro MA, Orozco JH, Degarbo SM, Calderon AE, Nardello AL, Laciar A, et al. Virulence profiles of Shiga Toxin-Producing Escherichia coli and other potentially diarrheagenic E.coli of bovine origin, in Mendoza, Argentina. Braz J Microbiol. 2013;44(4):1173-80. [PubMed ID: 24688508]. https://doi.org/10.1590/S1517-83822014005000010.
-
12.
Wada A, Kono M, Kawauchi S, Takagi Y, Morikawa T, Funakoshi K. Rapid discrimination of Gram-positive and Gram-negative bacteria in liquid samples by using NaOH-sodium dodecyl sulfate solution and flow cytometry. PLoS One. 2012;7(10). eee47093. [PubMed ID: 23077549]. https://doi.org/10.1371/journal.pone.0047093.
-
13.
Michigan State University. STEC Center Website. 2013. Available from: http://www.shigatox.net.
-
14.
Wayne, PA; 2014.Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement.
-
15.
Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, et al. Foodborne illness acquired in the United States--major pathogens. Emerg Infect Dis. 2011;17(1):7-15. [PubMed ID: 21192848]. https://doi.org/10.3201/eid1701.091101p1.
-
16.
Salmanzadeh-Ahrabi S, Habibi E, Jaafari F, Zali MR. Molecular epidemiology of Escherichia coli diarrhoea in children in Tehran. Ann Trop Paediatr. 2005;25(1):35-9. [PubMed ID: 15814047]. https://doi.org/10.1179/146532805X23335.
-
17.
Aslani MM, Bouzari S. An epidemiological study on Verotoxin-producing Escherichia coli (VTEC) infection among population of northern region of Iran (Mazandaran and Golestan provinces). Eur J Epidemiol. 2003;18(4):345-9. [PubMed ID: 12803375].
-
18.
Mazaheri S, Salmanzadeh Ahrabi S, Aslani MM. Shiga toxin-producing Escherichia coli isolated from lettuce samples in tehran, iran. Jundishapur J Microbiol. 2014;7(11). eee12346. [PubMed ID: 25774272]. https://doi.org/10.5812/jjm.12346.
-
19.
Fard AH, Bokaeian M, Qureishi ME. Frequency of Escherichia coli O157:H7 in children with diarrhoea in Zahedan, Islamic Republic of Iran. East Mediterr Health J. 2008;14(5):1022-7. [PubMed ID: 19161073].
-
20.
Kesava Naidu G, RGN GSM, Shivannavar CT. Detection of Shiga toxin genes (stx1 & stx2) and molecular characterization of shiga-toxigenic escherichia coli isolated from divers sources in Gulbarga region, India. Pharmacophore. 2011;2(5):253-65.
-
21.
Mellor GE, Besser TE, Davis MA, Beavis B, Jung W, Smith HV, et al. Multilocus genotype analysis of Escherichia coli O157 isolates from Australia and the United States provides evidence of geographic divergence. Appl Environ Microbiol. 2013;79(16):5050-8. [PubMed ID: 23770913]. https://doi.org/10.1128/AEM.01525-13.
-
22.
Vicentei HIG, do AmaralI LA, Cerqueira AMF. Shigatoxigenic Escherichia coli serogroups O157, O111 and O113 in feces, water and milk samples from dairy farms. Braz J Microbiol. 2005;36(3):217-22. https://doi.org/10.1590/S1517-3822005000300003.
-
23.
Farfan MJ, Torres AG. Molecular mechanisms that mediate colonization of Shiga toxin-producing Escherichia coli strains. Infect Immun. 2012;80(3):903-13. [PubMed ID: 22144484]. https://doi.org/10.1128/IAI.05907-11.
-
24.
Tahamtan Y, Namavari M. Prevalence of O157:H7 and non-O157 E. coli in Iranian domestic sheep. Pak J Biol Sci. 2014;17(1):104-8. [PubMed ID: 24783786].
-
25.
Bonyadian M, Momtaz H, Rahimi E, Habibian R, Yazdani A, Zamani M. Identification & characterization of Shiga toxin-producing Escherichia coli isolates from patients with diarrhoea in Iran. Indian J Med Res. 2010;132:328-31. [PubMed ID: 20847380].
-
26.
Jafari F, Hamidian M, Rezadehbashi M, Doyle M, Salmanzadeh-Ahrabi S, Derakhshan F, et al. Prevalence and antimicrobial resistance of diarrheagenic Escherichia coli and Shigella species associated with acute diarrhea in Tehran, Iran. Can J Infect Dis Med Microbiol. 2009;20(3):e56-62. [PubMed ID: 20808457].
-
27.
Hu GZ, Pan YS, Wu H, Hu H, Xu R, Yuan L, et al. Prevalence of tetracycline resistance genes and identification of tet(M) in clinical isolates of Escherichia coli from sick ducks in China. J Med Microbiol. 2013;62(Pt 6):851-8. [PubMed ID: 23475906]. https://doi.org/10.1099/jmm.0.051896-0.
-
28.
Armstrong GD, Rowe PC, Goodyer P, Orrbine E, Klassen TP, Wells G, et al. A phase I study of chemically synthesized verotoxin (Shiga-like toxin) Pk-trisaccharide receptors attached to chromosorb for preventing hemolytic-uremic syndrome. J Infect Dis. 1995;171(4):1042-5. [PubMed ID: 7706786].