Abstract
Background:
Acinetobacter baumannii is known as a potential pathogen in hospitals and is responsible for the dramatic increase in carbapenem resistance in Iran in the recent years.Objectives:
The current study aimed at determining the genetic association of the isolates by the pulsed-field gel electrophoresis (PFGE) technique, identify international clones, evaluate biofilm formation ability and its relationship with antibiotic resistance.Methods:
In the current study, a total of 48 A. baumannii isolates were collected from 2 hospitals in Tehran, Iran, from 2010 to 2012. Isolates were subjected to antimicrobial susceptibility testing, determination of carbapenemase encoding genes, biofilm formation, and genetic relationships analysis.Results:
The obtained results demonstrated that the rate of resistance to carbapenem, meropenem, imipenem, and doripenem was 76%. The carbapenemase-encoding gene blaOXA-23-like was found in 32 isolates, while blaOXA-40-like (blaOXA-24-like), blaOXA-58-like, blaVIM-type and blaIMP-type were found in 11, 1, 19. and 5 isolates, respectively. When the lineage of the isolates was evaluated by the multiplex polymerase chain reaction (PCR), it was found that 28 isolates belonged to group 1 and 8 isolates to group 2. None of the isolates belonged to group 3. Twelve isolates could not be typed by this method. The study findings interestingly demonstrated that 13 isolates showed no biofilm formation. Data of biofilm formation also demonstrated that 28, 4, and 3 remaining isolates had weak, moderate, and strong biofilm formation, respectively. The pulsed field gel electrophoresis result revealed 11 unique clones.Conclusions:
International clone 1 was the most commonly identified clone in the current study. This clone was mostly associated with blaOXA-23-like gene; therefore, 64% of the isolates in this clone possessed blaOXA-23-like gene.Keywords
Acinetobacter baumannii Electrophoresis Gel Pulsed-Field Biofilm
1. Background
Acinetobacter baumannii emerged as a highly troublesome pathogen for many institutions globally (1). Outbreaks of infections due to this organism are reported in neonatal intensive care units (NICUs) (2), medical and surgical wards, burn units, and are also associated with medical equipment and hands of personnel (3). The rate of antibiotic resistance is high (4). Multidrug-resistant (MDR) A. baumannii is frequently reported as endemic in certain hospital wards and responsible for numerous nosocomial outbreaks worldwide (4, 5). Metallo-beta-lactamases (MBLs) and oxacillinases (OXAs) are the 2 main groups of carbapenemases in A. baumannii. OXA-type beta-lactamases are responsible for the most common type of carbapenem resistance via enzymatic degradation (6). Although MBLs are not the predominant carbapenemases in A. baumannii, VIM, IMP, SIM, and NDM metallo-beta-lactamases contribute to the high-level resistance to carbapenems (7).
Infections can have serious repercussions for patient morbidity and mortality. Patients can acquire the infection from environmental sources or from other patients (8). Acinetobacter spp. frequently causes infections associated with medical devices. Biofilm formation is a well-known pathogenic mechanism in such infections (1). Various typing methods are employed in the epidemiological investigation of outbreaks due to Acinetobacter species. The most widely applied methods focus on variations in the phenotypic properties such as antimicrobial susceptibility, biotypes, serotypes, phage types, or cell envelope protein profiles (9). Many of these traditional typing procedures have insufficient reproducibility, type ability, and discriminatory power.
Molecular techniques such as plasmid typing, ribotyping, and analysis of genomic DNA by pulsed-field gel electrophoresis (PFGE) and by arbitrarily primed polymerase chain reaction (PCR), which directly compare variations in the DNA of bacterial isolates, are applied to epidemiological investigations of Acinetobacter isolates (10, 11).
2. Objectives
The current study aimed at determining the susceptibility rate, PFGE characterization, clonal lineages, and biofilm formation in the clinical isolates of A. baumannii in Tehran hospitals, Iran.
3. Methods
3.1. Sample Collection and Bacterial Identification
A total of 48 clinical samples were collected from wound, trachea, urine, catheter, sputum, and burn from patients admitted to Tehran hospitals from 2010 to 2012. The isolates were identified as A. baumannii according to the analytical profile blaOXA-51-like index 20NE protocol (BioMerieux, France) and genetically confirmed by the presence of gene (12). Then, the isolates were stored at -70°C in nutrient broth plus 30% glycerol.
3.2. Antibiotic Susceptibility Testing
Antibiotic susceptibility was determined by disk diffusion method on Mueller-Hinton agar (Merck, Germany) according to the clinical and laboratory standards institute (CLSI) guidelines for the following antimicrobial agents: imipenem (10 µg), meropenem (10 µg), doripenem (10 µg), piperacillin (100 µg), gentamicin (10 µg), amikacin (30 µg), ciprofloxacin (5 µg), minocycline (30 µg), doxycycline (30 µg), and piperacillin-tazobactam (100/10 µg) (Mast Diagnostics, UK). Escherichia coli ATCC 25922, Escherichia coli ATCC 35218, and also Pseudomonas aeruginosa ATCC 27853 were used as controls (13).
3.3. PCR for Carbapenemase Genes
Genomic DNA was extracted using the boiling method. PCR was employed to detect blaOXA-51-like, the A. baumannii-specific-gene) Table 1. (Isolates were also screened for the other major groups that confer resistance to carbapenems; ie, blaOXA-23-like, blaOXA-40-like (blaOXA-24-like), and blaOXA-58-like by multiplex reaction as well as the blaNDM-type, blaSIM-type, blaIMP-type, and blaVIM-type by single PCR (TAG Copenhagenard, Denmark) (14) (Table 1). The PCR products were separated by 1% agarose gel electrophoresis stained with neutral red (Sigma Aldrich, Germany) and visualized under ultraviolet (UV) light.
. Primers Used for Multiplex Polymerase Chain Reaction
| Prime | Sequence | Size (bp) |
|---|---|---|
| SIM-F | 5’ - GTACAAGGGATTCGGCATCG-3´ | 569 |
| SIM-R | 5’ - GTACAAGGGATTCGGCATCG-3´ | |
| NDMI-F | 5’ - GAGATTGCCGAGCGACTTG-3´ | 497 |
| NDMI-R | 5’ - CGAATGTCTGGCAGCACACTT-3´ | |
| Oxa-51-like-F | 5’ - TAATGCTTTGATCGGCCTTG-3´ | 353 |
| Oxa-51-like-R | 5’ - TGGATTGCACTTCATCTTGG-3´ | |
| Oxa-23-like-F | 5’ - GATCGGATTGGAGAACCAGA-3´ | 501 |
| Oxa-23-like-R | 5’ - ATTTCTGACCGCATTTCCAT-3´ | |
| Oxa-24-like-F | 5’ - GGTTAGTTGGCCCCCTTAAA-3´ | 246 |
| Oxa-24-like-R | 5’ - AGTTGAGCGAAAAGGGGATT-3´ | |
| Oxa-58-like-F | 5’ - AGTATTGGGGCTTGTGCTG-3´ | 599 |
| Oxa-58-like-R | 5’- CCCCTCTGCGCTCTACATAC-3´ | |
| Imp-F | 5’- GAATAGAATGGTTAACTCTC-3´ | 188 |
| Imp-R | 5’- CCAAACCACTAGGTTATC-3´ | |
| Vim-F | 5’ - GTTTGGTCGCATATCGCAAC -3´ | 382 |
| Vim-R | 5’- AATGCGCAGCACCAGGATAG-3´ | |
| Group1 alleles | ||
| Group1ompAF306 | 5’- GATGGCGTAAATCGTGGTA-3´ | 355 |
| Group1and2ompAR660 | 5’ - CAACTTTAGCGATTTCTGG-3´ | |
| Group1csuEF | 5’ - CTTTAGCAAACATGACCTACC-3´ | 702 |
| Group1csuER | 5’ - TACACCCGGGTTAATCGT-3´ | |
| Gp1OXA66F89 | 5’ - GCGCTTCAAAATCTGATGTA-3´ | 559 |
| Gp1OXA66R647 | 5’ - GCGTATATTTTGTTTCCATTC-3´ | |
| Group2 alleles | ||
| Group2omp AF378 | 5’ - GACCTTTCTTATCACAACGA-3´ | 343 |
| GrouPland2ompAR660 | 5’ - CAACTTTAAGCGATTTCTGC-3´ | |
| Group2csuEF | 5’ - GGCGAACATGACCTATTT-3´ | 580 |
| Group2csuER | 5’ - CTTCATGGCTCGTTGGTT-3´ | |
| Gp2OXA69F169 | 5’ - CATCAAGGTCAAACTCAA-3´ | 162 |
| Gp2OXA69R330 | 5’ - TAGCCTTTTTTCCCCATC-3´ |
3.4. Biofilm Formation
Biofilm formation was quantified by the microtiter plate assay method as described previously (15). Briefly, A. baumannii species were grown overnight in trypticase soy broth (TSB) (Merck, Germany) with 0.25% glucose at 37°C. The culture was diluted 1:50 in (TSB). Then, 200 mL of suspension was used to inoculate wells of sterile 96-well polystyrene microtiter plates, followed by incubation at 37°C for 72 hours. After 3 washes with phosphate buffered saline (PBS), any remaining biofilm was stained with crystal violet 1% (w/v) for 25 minutes and wells were washed again with PBS. The dye bound to the adherent cells was re-solubilized with 200 mL of ethanol/acetone (80:20, V/V) and the optical density (OD) was quantified at 570 nm using an enzyme-linked immunosorbent assay (ELISA) reader. Each assay was performed in triplicate. The adherence capabilities of the tested strains were classified into 4 categories; 3 standard deviations (SDs) above the mean OD of the negative control (broth only) was considered as the optical density cutoff (ODc). Isolates were classified as follows: OD ≤ ODc non-adherent bacteria; ODc < OD ≤ 2 × ODc, weakly adherent bacteria; 2 × ODc < OD ≤ 4 × ODc, moderately adherent bacteria; 4 × ODc < OD, strongly adherent bacteria (15).
3.5. Pulsed-Field Gel Electrophoresis
The collected isolates were analyzed by PFGE as described by Durmaz et al. (16). Acinetobacter baumannii ATCC 19606 was included for external reference. Genomic DNA was digested with ApaI (New England Biolabs). The lambda ladder PFGE marker (NEB, US) was used as a molecular size marker. The pulsed-field electrophoresis (Chef Mapper; Bio-Rad Laboratories, Hercules, CA, USA) was performed under the following conditions: temperature 14°C; voltage 6 V/cm; switch angle, 120°C; switch ramp 2.2 - 35 seconds for 19 hours. Gels were stained with ethidium bromide and patterns were visualized under UV light (BIO-RAD, USA). DNA banding patterns were analyzed using Bionumeric 7.0 software (Apllied maths NV, St-Martens-Latem Belgium).
3.6. PCR-Based Sequence Group Typing
Multiplex PCRs to identify groups (international clone) 1 to 3 organisms were performed by amplification of the ompA, csuE, and blaOXA-51-like genes using the primers listed in Table 1 (TAG Copenhagenard, Denmark).
PCR were performed under the following conditions: 94°C for 3 minutes, then, 30 cycles of 94°C for 45 seconds, 57°C for 45 seconds and 72°C for 1 minute, followed by a final extension at 72°C for 5 minutes. Identification of a strain as a member of group 1 or 2 required the amplification of all 3 fragments in the corresponding multiplex PCR, and an absence of any amplification in the other multiplex PCR. Group 3 isolates were defined by the amplification of only the ompA fragment in the group 2 PCR, and the amplification of only the csuE, and blaOXA-51-like fragments in the group 1 PCR (17) (Table 1). The PCR products were separated by 1% agarose gel electrophoresis stained with neutral red (Sigma Aldrich, Germany) and visualized under UV light.
4. Results
4.1. Bacterial Strains
The Isolates were isolated from different clinical samples and wards of Loghman (No. 1) and Milad (No. 2) hospitals, described in Table 2.
Clinical Samples and Different Wards of Hospital
| Clinical Samples | Frequency of Samples (N) | Type of Wards | Frequency of Samples (N) |
|---|---|---|---|
| Wound | 13 | Intensive care unit | 32 |
| Trachea | 24 | Pediatric | 1 |
| Urine | 3 | Medical | 7 |
| Catheter | 2 | Surgery | 5 |
| Sputum | 3 | Ear, nose and throat | 1 |
| Burn | 2 | Emergency | 2 |
| Cerebrospinal fluid | 1 |
4.2. Antibiotic Susceptibility Testing
The disk diffusion susceptibility testing revealed that 100% (n = 48) of the isolates were resistant to piperacillin. The rate of resistance to carbapenem was 76% (meropenem = 77%, imipenem =77%, and doripenem = 75%); among the other tested antibiotics, colistin and polymixin showed the higher rates of antimicrobial activities (isolates were 100% sensitive) (Table 3).
Resistance of Acinetobacter baumannii Isolates to the Tested Antibiotics
| Antibiotic | Resistant, N (%) | Intermediate, N (%) | Susceptible, N (%) |
|---|---|---|---|
| Amikacin | 33 (69) | 0 (0) | 15 (31) |
| Gentamicin | 35 (73) | 3 (6) | 10 (21) |
| Trimethoprim-sulfamethoxazole | 42 (88) | 2 (4) | 4 (8) |
| Ciprofloxacin | 44 (92) | 0 (0) | 4 (8) |
| Ceftazidime | 44 (92) | 0 (0) | 4 (8) |
| Cefepime | 40 (84) | 4 (8) | 4 (8) |
| Cefotaxime | 44 (92) | 3 (6) | 1 (2) |
| Colistin | 0 (0) | 0 (0) | 48 (100) |
| Polymyxin B | 0 (0) | 0 (0) | 48 (100) |
| Piperacillin-tazobactam | 38 (79) | 0 (0) | 10 (21) |
| Piperacillin | 48 (100) | 0 (0) | 0 (0) |
| Minocycline | 4 (8) | 12 (25) | 32 (67) |
| Doxycycline | 27 (56) | 1 (2) | 20 (42) |
| Meropenem | 37 (77) | 4 (8) | 7 (15) |
| Imipenem | 37 (77) | 5 (10) | 6 (13) |
| Doripenem | 36 (75) | 4 (8) | 8 (17) |
Minimum inhibitory concentration (MIC) result for meropenem ranged 0.5 to 256 μg/mL as follows: 10 isolates were sensitive, 3 isolates intermediate, and 35 isolates showed resistance. On the other hand, MIC results for imipenem ranged 0.5 to 64 μg/mL: 11 isolates were sensitive, 4 isolates intermediate, and 33 isolates resistant.
4.3. Prevalence of Carbapenemase-Encoding Genes
All 48 isolates were positive for blaOXA-51-like. The carbapenemase-encoding gene, blaOXA-23-like, was found in 32 isolates (64%). blaOXA-40-like (blaOXA-24-like), blaOXA-58-like, blaVIM-type, and blaIMP-type were found in 11 (22%), 1 (2%), 19 (38%), and 5 (10%) isolates, respectively. None of the isolates had positive PCR results for blaNDM-type and blaSIM-type.
4.4. Biofilm Formation
Among 48 isolates examined in the current study, 13 could not produce biofilm, 28 isolates had weak ability to form biofilm, 4 isolates showed moderate ability, and 3 isolates showed strong ability to form biofilm (Table 4).
Resistance of Acinetobacter baumannii Isolates to carbapenems, Based on the Ability to Form Biofilm
| Antibiotic | Non-Biofilm, Forming (N = 13), N (%) | Weak Biofilm Forming (N =28), N (%) | Moderate Biofilm Forming (N = 4), N (%) | Strong Biofilm Forming (N =3), N (%) | Total, N (%) |
|---|---|---|---|---|---|
| Doripenem | |||||
| Resistant | 8 (22.2) | 23 (63.9) | 2 (5.6) | 3 (8.3) | 36 (100) |
| Intermediate | 1 (25) | 2 (50) | 1 (25) | 0 (0) | 4 (100) |
| Susceptible | 4 (50) | 3 (37.5) | 1 (12.5) | 0 (0) | 8 (100) |
| Imipenem | |||||
| Resistant | 10 (27) | 22 (59.5) | 2 (5.4) | 3 (8.1) | 37 (100) |
| Intermediate | 1 (20) | 3 (60) | 1 (20) | 0 (0) | 5 (100) |
| Susceptible | 2 (33.3) | 3 (50) | 1 (16.7) | 0 (0) | 6 (100) |
| Meropenem | |||||
| Resistant | 9 (24.3) | 23 (62.2) | 2 (5.4) | 3 (8.1) | 37 (100) |
| Intermediate | 2 (50) | 1 (25) | 1 (25) | 0 (25) | 4 (100) |
| Susceptible | 2 (28.6) | 4 (57.1) | 1 (14.3) | 0 (0) | 7 (100) |
4.5. Pulsed-Field Gel Electrophoresis Results
The PFGE results (Figure 1A) revealed 11 clonal complexes with cut off level 74%, and the most prevalent clonal complexes were E, F, and H. The clonal complex E was the most frequent, with 17 clinical isolates. Pulsotype I was sporadic and did not have close genetic similarity with the other groups (Figure 1).
PFGE Clustering Analysis with Cut off Level 74% of 48 A. baumannii Strains, International Clones, and Biofilm Formation
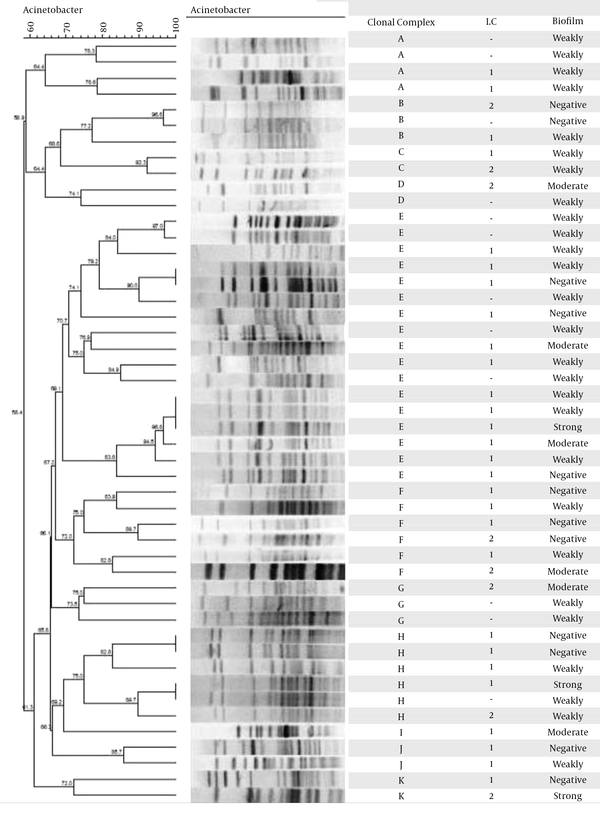
4.6. PCR-Based Sequence Group Typing
All strains belonging to the group 1 of clonal complex yielded all 3 fragments in the group 1 PCR and none of the group 2 PCR; while strains belonging to group 2 gave the expected converse results. Twenty-eight isolates belonged to group 1 and 8 isolates belonged to group 2. None of the isolates belonged to group 3. Twelve isolates could not be typed by this method.
Of the 28 isolates belonging to group 1, eighteen isolates had blaOXA-23-like gene, five isolates had blaOXA-23-like +, VIM+ pattern. The pattern of imipenem resistance among the 28 isolates that belonged to group 1 was as follows: 20 isolates were resistant, 3 isolates had intermediate resistance, and 5 were sensitive; for meropenem: 20 isolates showed resistance, 2 isolates showed intermediate resistance, and 6 isolates were sensitive; for doripenem: 19 isolates were resistant, 2 isolates had intermediate resistance, and 7 isolates were sensitive. Of the 8 isolates that belonged to group 2, according to PCR-based sequence group method, 6 isolates had blaOXA-23-like gene, 3 isolates had blaOXA-23-like +, VIM+ pattern and 1 isolate did not have resistance genes except for blaOXA-51-like; the pattern of resistance for all of the 3 evaluated antibiotics was the same in this group. Seven isolates were resistant to all antibiotics and 1 isolate had intermediate resistance (Tables 5 and 6).
Resistance of Acinetobacter baumannii Isolates to the Tested Antibiotics, Based on International Clones and Beta-Lactamase Genes
| Antibiotic | International Clone 1 (N = 28), N (%) | International Clone 2 (N = 8), N (%) | Other International Clones (N = 12), N (%) | Total, N (%) | Bla-OXA23-like | Bla-OXA24-like | VIM | IMP |
|---|---|---|---|---|---|---|---|---|
| Doripenem | ||||||||
| Resistant | 19 (52.8) | 7 (19.4) | 10 (27.8) | 36 (100) | 27 | 6 | 16 | 4 |
| Intermediate | 2 (50) | 1 (25) | 1 (25) | 4 (100) | 2 | 1 | 0 | 1 |
| Susceptible | 7 (87.5) | 0 (0) | 1 (12.5) | 8 (100) | 3 | 4 | 3 | - |
| Total | 32 | 11 | 19 | 5 | ||||
| Imipenem | ||||||||
| Resistant | 20 (54.1) | 7 (18.9) | 10 (27) | 37 (100) | 26 | 8 | 16 | 5 |
| Intermediate | 3 (60) | 1 (20) | 1 (20) | 5 (100) | 2 | 2 | 2 | - |
| Susceptible | 5 (83.3) | 0 (0) | 1 (16.7) | 6 (100) | 4 | 1 | 1 | - |
| Total | 32 | 11 | 19 | 5 | ||||
| Meropenem | ||||||||
| Resistant | 20 (54.1) | 7 (18.9) | 10 (27) | 37 (100) | 26 | 6 | 16 | 5 |
| Intermediate | 2 (50) | 1 (25) | 1 (25) | 4 (100) | 3 | 2 | 1 | - |
| Susceptible | 6 (85.7) | 0 (0) | 1 (14.3) | 7 (100) | 3 | 3 | 2 | - |
| Total | 32 | 11 | 19 | 5 |
Frequency of Acinetobacter baumannii Isolates in International Clones
| Clonal Complex | International Clone 1 (N = 28) | International Clone 2 (N = 8) | Other International Clones (N = 12) | Total N |
|---|---|---|---|---|
| A | 2 | 0 | 2 | 4 |
| B | 1 | 1 | 1 | 3 |
| C | 1 | 1 | 0 | 2 |
| D | 0 | 1 | 1 | 2 |
| E | 12 | 0 | 5 | 17 |
| F | 4 | 2 | 0 | 6 |
| G | 0 | 1 | 2 | 3 |
| H | 4 | 1 | 1 | 6 |
| I | 1 | 0 | 0 | 1 |
| J | 2 | 0 | 0 | 2 |
| K | 1 | 1 | 0 | 2 |
| 28 | 8 | 12 | 48 |
5. Discussion
Acinetobacter baumannii is one of the main causes of nosocomial infections in the recent years (18). Acinetobacter baumannii is one of the most problematic organisms currently responsible for nosocomial infections, especially in intensive care units (ICUs) (19). Increasing antimicrobial resistance (20) and the ability of A. baumannii to survive on inanimate and dry surfaces are linked to the occurrence of periodic outbreaks observed in various hospitals (21). Acinetobacter baumannii is the most common isolate obtained from skin, blood, sputum, pleural fluid, and urine (22). As many studies show, the organism is most frequently isolated from the respiratory tract than the other body organs among the ICU admitted patients (23, 24).
Acinetobacter baumannii is one of the most frequent causes of nosocomial infections throughout Tehran hospitals, especially in the ICU wards (23). Carbapenem resistance is recently increased by this bacterium; therefore, the current study evaluated the antibiotic sensitivity pattern, carbapenem resistance genes, the ability of biofilm formation, the genetic relationships and the lineages of the species isolated from Tehran hospitals. Most of the samples of the current study were obtained from the ICU wards. The samples were also collected from other wards in high abundance such as medical, surgery, emergency, ear, nose and throat (ENT), and pediatric. Although carbapenems, aminoglycosides, and fluoroquinolones are used to treat A. baumannii infections in different medical settings, resistance to carbapenems is increasing in Iran (25-27).
A worldwide collection of 5127 Acinetobacter spp., collected from 2005 to 2009 from 140 hospitals of 32 countries in North America (17.1%), Europe (22.9%), Latin America (25.2%) and the Asia-Pacific region (34.8%) showed that overall non-susceptibility rate to imipenem and meropenem was 45.9% and 48.2%, respectively. However, the non-susceptibility percentage for imipenem and meropenem, increased from 27.8% and 37.5% in 2005 to 62.4% and 64.4% in 2009, respectively (28).
From 2008 to 2016, thirty-nine studies were performed in Iran on the antibiotic resistance of A. baumannii that more than half of them were performed in Tehran. Nowadays, the rate of carbapenem resistance has increased in Iran. By definition, MDR A. baumannii isolates are resistant to 3 or more agents of different antibiotic classes. In total, 7 studies revealed the presence of MDR isolates. Among the 7 studies, the rates of MDR ranged from 32.7% to 93% in the 584 isolated A. baumannii species sort by time: 2001 to 2007 (1 study, 50%), 2008 to 2009 (1 study, 66%), 2009 to 2010 (1 study, 83%), and 2010 to 2011 (4 studies, 32.7%, 74.9%, 93%, 94.4%) (29).
In a study by Karmostaji et al., in Iran during 2012, on 84 isolates of A. baumannii, 50 (59.52%) species were resistant to imipenem and 74 (88.09%) were resistant to meropenem (30). Also In another study by karmostaji et al., 54.47% of the isolates were resistant to amikacin, 67.47% to imipenem, and 84.55% of the isolates were resistant to meropenem. Among the 123 isolates, 100 (81.3 %) had an acquired blaOXA-23-like carbapenemase, 10 (8.1%) possessed blaOXA-24-like, and 1 (0.81%) blaOXA-58-like carbapenemase (31). The results of the current study showed that the carbapenemase-encoding gene, blaOXA-23-like, was found in 32 (64%) isolates and this finding was consistent with those of other studies reported from Iran. The blaOXA-40-like (blaOXA-24-like), blaOXA-58-like, blaVIM-type, and blaIMP-type genes were found in 11 (22%), 1 (2%), 19 (38%) and 5 (10%) isolates, respectively. However, none of the isolates had positive PCR results for blaNDM-type and blaSIM-type (29). The current study tried to evaluate the results of clonal lineage method with the results obtained by PFGE. In type E, which was the most frequent type in the current study, there were 17 isolates, 13 collected from ICU and 4 from other wards. All 17 isolates were collected in hospital No.1. The origin of 8 isolates was trachea, 5 isolates from wound, 2 isolates from sputum, 1 isolate from urine, and 1 isolate from catheter. Using clonal lineages as a typing method, it was shown that 12 isolates belonged to the group 1 and the method was unable to type 5 other isolates.
Three out of 48 isolates were strong biofilm producers, among which 1 isolate was from hospital No.1 and the other 2 from hospital No.2. Two of these isolates were collected from catheter in ICU and the others were from burn and general wards. These isolates belonged to 3 clonal complexes E, H, and K and by PCR-based sequence group typing method it was found that 2 isolates belonged to group 1 and the other belonged to group 2. All 3 isolates showed high resistance to carbapenem; therefore, MIC of these isolates for imipenem and meropenem were 32 to 128 μg/mL, respectively. All the isolates were MDR. Isolate 3, despite harboring just blaOXA-51-like (carbapenem resistance genes), showed high antibiotic resistance to all 3 antibiotics including meropenem, imipenem, and doripenem. None of the isolates had blaOXA-58-like and blaIMP-type.
All 3 isolates, which were strong biofilm producers, showed resistance to all studied antibiotics. In a similar study conducted by Mahdian et al., 49% of isolates belonged to international clone 1 and 49% to international clone 2; also 74% and 49% of the isolates harbored blaOXA-23-like and blaOXA-24-like, respectively; and 75% and 66% of the isolates were resistant to the imipenem and meropenem, respectively (27). In another study, 58% and 29% of the isolates belonged to international clones 2 and 1, respectively (32). In the study conducted by Kaliterena et al., in Croatia, 51% of the A. baumannii species belonged to class 1 and 27% to class 2 international clone, and the others belonged to other clones (33).
Despite the simplicity and cost-effectiveness of PCR-based sequence group typing method, it could not type all the isolates. However, PFGE as a method can type all isolates.
5.1. Conclusions
International clone 2 was the most commonly detected clonal lineage in the current study. Isolates that belonged to this clone were mostly associated with blaOXA-23-like gene; therefore, 64% of the isolates in this clone possessed blaOXA-23-like gene.
Acknowledgements
References
-
1.
Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21(3):538-82. [PubMed ID: 18625687]. https://doi.org/10.1128/CMR.00058-07.
-
2.
Seifert H, Schulze A, Baginski R, Pulverer G. Comparison of four different methods for epidemiologic typing of Acinetobacter baumannii. J Clin Microbiol. 1994;32(7):1816-9. [PubMed ID: 7929781].
-
3.
Hartstein AI, Morthland VH, Rourke JW, Freeman J, Garber S, Sykes R, et al. Plasmid DNA fingerprinting of Acinetobacter calcoaceticus subspecies anitratus from intubated and mechanically ventilated patients. Infect Control Hosp Epidemiol. 1990;11(10):531-8. [PubMed ID: 2230042].
-
4.
Seifert H, Baginski R, Schulze A, Pulverer G. Antimicrobial susceptibility of Acinetobacter species. Antimicrob Agents Chemother. 1993;37(4):750-3. [PubMed ID: 8494371].
-
5.
Maragakis LL, Perl TM. Acinetobacter baumannii: epidemiology, antimicrobial resistance, and treatment options. Clin Infect Dis. 2008;46(8):1254-63. [PubMed ID: 18444865]. https://doi.org/10.1086/529198.
-
6.
Zarrilli R, Giannouli M, Tomasone F, Triassi M, Tsakris A. Carbapenem resistance in Acinetobacter baumannii: the molecular epidemic features of an emerging problem in health care facilities. J Infect Dev Ctries. 2009;3(5):335-41. [PubMed ID: 19759502].
-
7.
Lin MF, Lan CY. Antimicrobial resistance in Acinetobacter baumannii: From bench to bedside. World J Clin Cases. 2014;2(12):787-814. [PubMed ID: 25516853]. https://doi.org/10.12998/wjcc.v2.i12.787.
-
8.
Villalon P, Valdezate S, Medina-Pascual MJ, Rubio V, Vindel A, Saez-Nieto JA. Clonal diversity of nosocomial epidemic Acinetobacter baumannii strains isolated in Spain. J Clin Microbiol. 2011;49(3):875-82. [PubMed ID: 21177889]. https://doi.org/10.1128/JCM.01026-10.
-
9.
Da Silva G, Dijkshoorn L, van der Reijden T, van Strijen B, Duarte A. Identification of widespread, closely related Acinetobacter baumannii isolates in Portugal as a subgroup of European clone II. Clin Microbiol Infect. 2007;13(2):190-5. [PubMed ID: 17328732]. https://doi.org/10.1111/j.1469-0691.2006.01628.x.
-
10.
Di Popolo A, Giannouli M, Triassi M, Brisse S, Zarrilli R. Molecular epidemiological investigation of multidrug-resistant Acinetobacter baumannii strains in four Mediterranean countries with a multilocus sequence typing scheme. Clin Microbiol Infect. 2011;17(2):197-201. [PubMed ID: 20456455]. https://doi.org/10.1111/j.1469-0691.2010.03254.x.
-
11.
Giannouli M, Cuccurullo S, Crivaro V, Di Popolo A, Bernardo M, Tomasone F, et al. Molecular epidemiology of multidrug-resistant Acinetobacter baumannii in a tertiary care hospital in Naples, Italy, shows the emergence of a novel epidemic clone. J Clin Microbiol. 2010;48(4):1223-30. [PubMed ID: 20181918]. https://doi.org/10.1128/JCM.02263-09.
-
12.
Turton JF, Woodford N, Glover J, Yarde S, Kaufmann ME, Pitt TL. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J Clin Microbiol. 2006;44(8):2974-6. [PubMed ID: 16891520]. https://doi.org/10.1128/JCM.01021-06.
-
13.
Patel J, Cockerill F, Alder J, Bradford P, Eliopoulos G, Hardy D. Performance standards for antimicrobial susceptibility testing; twenty-fourth informational supplement. 34. CLSI standards for antimicrobial susceptibility testing; 2014.
-
14.
Abdalhamid B, Hassan H, Itbaileh A, Shorman M. Characterization of carbapenem-resistant Acinetobacter baumannii clinical isolates in a tertiary care hospital in Saudi Arabia. New Microbiol. 2014;37(1):65-73. [PubMed ID: 24531172].
-
15.
Badmasti F, Siadat SD, Bouzari S, Ajdary S, Shahcheraghi F. Molecular detection of genes related to biofilm formation in multidrug-resistant Acinetobacter baumannii isolated from clinical settings. J Med Microbiol. 2015;64(Pt 5):559-64. [PubMed ID: 25813817]. https://doi.org/10.1099/jmm.0.000058.
-
16.
Durmaz R, Otlu B, Koksal F, Hosoglu S, Ozturk R, Ersoy Y, et al. The optimization of a rapid pulsed-field gel electrophoresis protocol for the typing of Acinetobacter baumannii, Escherichia coli and Klebsiella spp. Jpn J Infect Dis. 2009;62(5):372-7. [PubMed ID: 19762987].
-
17.
Turton JF, Gabriel SN, Valderrey C, Kaufmann ME, Pitt TL. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin Microbiol Infect. 2007;13(8):807-15. [PubMed ID: 17610600]. https://doi.org/10.1111/j.1469-0691.2007.01759.x.
-
18.
Anvarinejad M, Japoni A, Davarpanah MA, Mahmudi H, Mammina C, Vazin A. Phenotypic and Molecular Epidemiology of Acinetobacter calcoaceticus baumannii Complex Strains Spread at Nemazee Hospital of Shiraz, Iran. Jundishapur J Microbiol. 2015;8(6):19180. [PubMed ID: 26322204]. https://doi.org/10.5812/jjm.8(5)2015.19180.
-
19.
Falagas ME, Karveli EA. The changing global epidemiology of Acinetobacter baumannii infections: a development with major public health implications. Clin Microbiol Infect. 2007;13(2):117-9. [PubMed ID: 17328722]. https://doi.org/10.1111/j.1469-0691.2006.01596.x.
-
20.
Van Looveren M, Goossens H, Arpac Steering Group. Antimicrobial resistance of Acinetobacter spp. in Europe. Clin Microbiol Infect. 2004;10(8):684-704. [PubMed ID: 15301671]. https://doi.org/10.1111/j.1469-0691.2004.00942.x.
-
21.
Hamouda A, Evans BA, Towner KJ, Amyes SG. Characterization of epidemiologically unrelated Acinetobacter baumannii isolates from four continents by use of multilocus sequence typing, pulsed-field gel electrophoresis, and sequence-based typing of bla(OXA-51-like) genes. J Clin Microbiol. 2010;48(7):2476-83. [PubMed ID: 20421437]. https://doi.org/10.1128/JCM.02431-09.
-
22.
Shelburne S3, Singh KV, White AC, Byrne L, Carmer A, Austin C, et al. Sequential outbreaks of infections by distinct Acinetobacter baumannii strains in a public teaching hospital in Houston, Texas. J Clin Microbiol. 2008;46(1):198-205. [PubMed ID: 18003801]. https://doi.org/10.1128/JCM.01459-07.
-
23.
Mohajeri P, Farahani A, Feizabadi MM, Ketabi H, Abiri R, Najafi F. Antimicrobial susceptibility profiling and genomic diversity of Acinetobacter baumannii isolates: A study in western Iran. Iran J Microbiol. 2013;5(3):195-202. [PubMed ID: 24475323].
-
24.
Shahcheraghi F, Abbasalipour M, Feizabadi M, Ebrahimipour G, Akbari N. Isolation and genetic characterization of metallo-beta-lactamase and carbapenamase producing strains of Acinetobacter baumannii from patients at Tehran hospitals. Iran J Microbiol. 2011;3(2):68-74. [PubMed ID: 22347585].
-
25.
Najar Peerayeh S, Karmostaji A. Molecular Identification of Resistance Determinants, Integrons and Genetic Relatedness of Extensively Drug Resistant Acinetobacter baumannii Isolated From Hospitals in Tehran, Iran. Jundishapur J Microbiol. 2015;8(7):27021. [PubMed ID: 26421140]. https://doi.org/10.5812/jjm.27021v2.
-
26.
Bahador A, Taheri M, Pourakbari B, Hashemizadeh Z, Rostami H, Mansoori N, et al. Emergence of rifampicin, tigecycline, and colistin-resistant Acinetobacter baumannii in Iran; spreading of MDR strains of novel International Clone variants. Microb Drug Resist. 2013;19(5):397-406. [PubMed ID: 23768166]. https://doi.org/10.1089/mdr.2012.0233.
-
27.
Mahdian S, Sadeghifard N, Pakzad I, Ghanbari F, Soroush S, Azimi L, et al. Acinetobacter baumannii clonal lineages I and II harboring different carbapenem-hydrolyzing-beta-lactamase genes are widespread among hospitalized burn patients in Tehran. J Infect Public Health. 2015;8(6):533-42. [PubMed ID: 26111484]. https://doi.org/10.1016/j.jiph.2015.04.030.
-
28.
Mendes RE, Farrell DJ, Sader HS, Jones RN. Comprehensive assessment of tigecycline activity tested against a worldwide collection of Acinetobacter spp. (2005-2009). Diagn Microbiol Infect Dis. 2010;68(3):307-11. [PubMed ID: 20955916]. https://doi.org/10.1016/j.diagmicrobio.2010.07.003.
-
29.
Moradi J, Hashemi FB, Bahador A. Antibiotic Resistance of Acinetobacter baumannii in Iran: A Systemic Review of the Published Literature. Osong Public Health Res Perspect. 2015;6(2):79-86. [PubMed ID: 25938016]. https://doi.org/10.1016/j.phrp.2014.12.006.
-
30.
Karmostaj A, Najar-Peerayeh S, Salmanian AH. Emergence of tigecycline resistant Acinetobacter baumannii from an intensive care unit (ICU) in Tehran. Jundishapur J Microbiol. 2013;6(3):215-9. https://doi.org/10.5812/jjm.4710.
-
31.
Karmostaji A, Najar-Peerayeh S, Salmanian AH. Distribution of OXA-type class D β-lactamase genes among nosocomial multi drug resistant Acinetobacter baumannii isolated in Tehran hospitals. Jundishapur J Microbiol. 2013;6(5). e8219. https://doi.org/10.5812/jjm.8219.
-
32.
Bahador A, Raoo An R, Farshadzadeh Z, Beitollahi L, Khaledi A, Rahimi S, et al. The Prevalence of IS Aba 1 and IS Aba 4 in Acinetobacter baumannii Species of Different International Clone Lineages Among Patients With Burning in Tehran, Iran. Jundishapur J Microbiol. 2015;8(7):17167. [PubMed ID: 26396712]. https://doi.org/10.5812/jjm.17167v2.
-
33.
Kaliterna V, Kaliterna M, Hrenovic J, Barisic Z, Tonkic M, Goic-Barisic I. Acinetobacter baumannii in Southern Croatia: clonal lineages, biofilm formation, and resistance patterns. Infect Dis (Lond). 2015;47(12):902-7. [PubMed ID: 26365683]. https://doi.org/10.3109/23744235.2015.1078906.