Abstract
Background:
Multidrug resistant (MDR) Acinetobacter baumanii strains have emerged as novel nosocomial pathogens threatening patients’ lives, especially in intensive-care units (ICU). Various types of extended-spectrum β-lactamases (ESBLs) are involved in conferring resistance to β-lactam antibiotics, making their genotypic characterization an essential prerequisite to take proper preventative measures.Objectives:
The aim of this study was to determine the antimicrobial susceptibility and prevalence of blaTEM, blaSHV, blaCTX-M, blaOXA-2, and blaOXA-10 genes among A. baumanii isolates obtained from patients in Tabriz city, North-west Iran.Methods:
The clinical isolates of A. baumanii were collected from patients hospitalized in the Imam Reza hospital of Tabriz. Antimicrobial susceptibility patterns were determined by the disk diffusion method. The frequency of different ESBLS resistance genes were determined by PCR.Results:
Antimicrobial susceptibility testing through the disk diffusion method revealed that the lowest resistance rates were against polymyxin B (16%), colistin (23%), and rifampin (27%); whereas the highest resistance rate was observed against ticarcillin (100%), cefixime (100%), and ceftizoxim (100%). Screening by double disk synergy test showed that 60% of the isolates were ESBL producers. PCR technique on ESBL-positive isolates determined blaSHV gene as the most prevalent (31.6%) and blaOXA-10 as the least prevalent (8.3%) among the studied resistance genes.Conclusions:
The high prevalence of resistance genes supported the essential role of ESBLs in antibiotic resistance of A. baumanii.Keywords
Extended-Spectrum β-Lactamase MDR Genotyping Acinetobacter baumannii
1. Background
Acinetobacter baumannii is a Gram-negative, nonfermentative, obligate aerobic, coccobacilli that have an ubiquitous distribution in nature being recovered from soil or water, human skin, and respiratory tract (1, 2). Acinetobacter baumannii is an important nosocomial pathogen, with a rising prevalence of about 89.2% among hospitalized patients, especially in ICU patients (3). Antimicrobial treatment of these infections may be compromised by the multiple-drug resistance of many strains to β-lactams, aminoglycosides, and fluoroquinolones (4). This bacterium causes a variety of diseases including urinary tract infections, endocarditis, surgical site infections, pneumonia, septicemia, and meningitis (1, 5). Numerous strains of A. baumannii have recently emerged to be resistant against a wide spectrum of antibiotics, particularly β-lactams, which have so far been among drug choices for treatment of A. baumannii infections. Consequently, treatment of these infections has encountered serious problems (2, 6).
Antibiotic resistance genes, including β-Lactamase genes, are the main molecular basis of microbial drug resistance; and plasmids along with the mobile genetic elements such as transposons and integrons are behind the rapid distribution of these multiple resistance genes (7). According to the Amber classification, β-Lactamases are categorized in 4 classes, A-D, based on their amino acid sequence similarities (8). Almost all beta-lactamases including the classic ESBLs, namely TEM, SHV, and CTX-M types, as well as non-classic ESBLs such as VEB and PER types belong to the class A Amber β-lactamases, with the exception of OXA type classic ESBLs like OXA-2 and OXA-10, which categorized in the group D of Amber classification (9, 10). Classic ESBLs have all originated from the parent types in TEM (temoneria), SHV (sulfhydryl variable), and OXA (oxacillinase) families. However, there are newer families of ESBLs discovered during the last 2 - 3 decades, such as VEB (for Vietnamese Extended-spectrum Beta-lactamase) and PER (for Pseudomonas extended resistance) families, among them, VEB-1 and PER-1 being the major types (11). Five different classes of integrons have been identified and INT-1 is the most prevalent type in clinical isolates of gram negative bacilli, including A. baumanii (12, 13).
Determination of antimicrobial resistance patterns and genotypic background of the ESBL production in clinical isolates of MDR bacteria would shed light on their epidemiology and contribute to the wise adoption of proper preventative or therapeutic measures (14). Unfortunately, there is a lack of well documented data on Acinetobacter antimicrobial resistance pattern and gene prevalence in Iran.
2. Objectives
The aim of this study was to determine the antibacterial resistance pattern and molecular prevalence of blaTEM, blaSHV, blaCTX-M, blaOXA-2 and blaOXA-10 genes among the clinical isolates of A. baumanii from patients hospitalized in the Imam Reza hospital of Tabriz, North-west Iran.
3. Methods
3.1. Sampling, Bacterial Isolation and Identification
The study was approved by the ethical committee of Tabriz University of Medical Sciences (code: 76/488). A. baumannii isolates were recovered from various clinical specimens of hospitalized patients in different wards of the Imam Reza hospital of Tabriz as follow: ICU, neurosurgery, internal wards, infectious wards, general surgery unit, and ENT. The clinical specimens were collected from invasive and non-invasive sites including tracheal aspirate, urine, sputum, blood, bronchial washing, catheter, wound, abscess drainage, cerebrospinal fluid (CSF), pleural effusion, and ascitic fluid. The samples were immediately incubated in MacConkey and blood agar (Merck, US) media (37°C for 24 - 48 minutes). The initial identification of isolates was carried out using the standard microbiological and biochemical methods, including Gram-staining, colony morphology, glucose oxidation, citrate utilization, oxidase test, O/F (Oxidation-Fermentation) test, catalase test, and growth ability at 44°C (15, 16).
3.2. Antibiotic Susceptibility Testing
To determine the antibiotic susceptibility pattern of isolates, Kirby Bauer disk diffusion testing was carried out using the clinical laboratory standard institute (CLSI) guidelines (17). Antibiotic disks used were ceftriaxone (30 µg), ceftazidime (30 µg), cefepime (30 µg), tetracycline (30 µg), rifampin (5 µg), gentamicin (10 µg), cephalexin (30 µg), amikacin (30 µg), ciprofloxacin (5 µg), tobramycin(10 µg), levofloxacin (5 µg), ampicillin-sulbactam (10/10 µg), piperacillin (100 µg), piperacillin-tazobactam (100/10 µg), ticarcillin-clavulanic acid (75/10 µg), imipenem (10 µg) meropenem (10 µg), colistin (10 µg), and polymyxin B (300 units) (MAST, UK). The disks were placed on Mueller-Hinton agar (Merck, Germany) plates and inoculated with bacterial suspension equal to 0.5 McFarland at 37°C overnight. The diameter of the zone of growth inhibition was measured using the CLSI guidelines.
3.3. Double-Disk Synergy Test
The double-disk synergy test was used to identify extended-spectrum β-lactamase (ESBL)-producing strains. The antibiotic disks of amoxicillin-clavulanic acid (20 μg/10 μg), cefotaxime (30 μg), ceftazidime (30 μg), and aztreonam (30 μg), obtained from Nissui Pharmaceuticals were used for the test. Amoxicillin-clavulanic acid disk was placed on the core of the plate and the 4 other disks mentioned above were placed in a 10 mm distance from the central disk and 20 mm from each other (18) (Figure 1).
Double Disk Synergy Test on Mueller-Hinton Agar
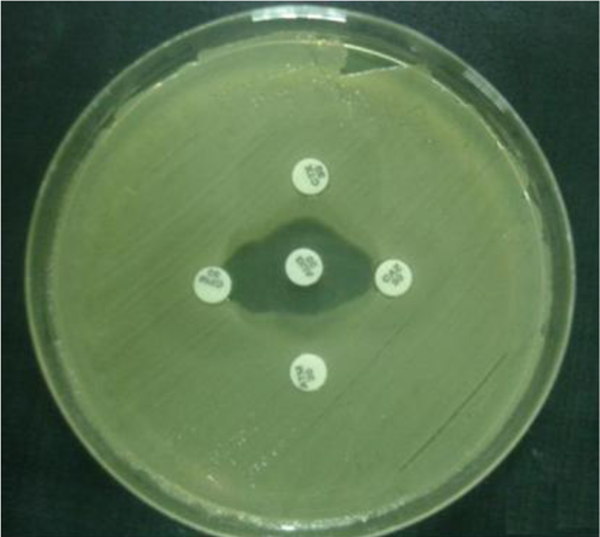
3.4. DNA Extraction and PCR Amplification
DNA was extracted from the isolates using the SDS-Proteinase K and phenol-chloroform method (19). After an overnight culture, fresh colonies was resuspended in TE buffer containing SDS (1%) and proteinase K (10 μg/mL) (Fermentas, Lithuania), and incubated for 3 hours at 40°C, followed by extraction with phenol-chloroform and ethanol precipitation. Finally, the quantity and quality of extracted DNA was checked via spectroscopy and agarose gel electrophoresis, respectively.
The PCR technique was used to detect the presence of ESBL genes (TEM, SHV, CTX-M, OXA2, and OXA10) in the clinical isolates of A. baumannii using specific primers (Table 1). PCR for blaOXA-51-like genes was also carried out to confirm the identity of isolates (20). PCR products were analyzed by agarose gel electrophoresis in TAE buffer. The gel was then stained with ethidium bromide for 20 minutes and visualized by gel documentation system.
The Sequence of Primers Used for PCR Amplification of Resistance Genes
| Primer | Primer Sequences | Annealing Temp, °C | Product Size, bp |
|---|---|---|---|
| TEM-F | 5'-CTTCCTGTTTTTGCTCACCCA-3' | 68 | 717 |
| TEM-R | 5'-TACGATACGGGAGGGCTTAC-3' | ||
| SHV-F | 5'-TCAGCGAAAAACACCTTG-3' | 60 | 471 |
| SHV-R | 5'-TCCCGCAGATAAATCACC-3' | ||
| CTX-M-Fa | 5'- SCSATGTGCAGYACCAGTAA-3' | 60 | 544 |
| CTX-M-R | 5'- CCGCRATATGRTTGGTGGTG-3' | ||
| OXA2-F | 5'- AAGAAACGCTACTCGCCTGC -3' | 60 | 485 |
| OXA2-R | 5'- CCACTCAACCCATCCTACCC -3' | ||
| OXA10-F | 5'-TATCGCGTGTCTTTCGAGTA-3' | 55 | 774 |
| OXA10-F | 5'-TTAGCCACCAATGATGCC-3' | ||
| OXA51-F | 5'-ACAAGCGCTATTTTTATTTCAG-3' | 51 | 641 |
| OXA51-R | 5'-CCCATCCCCAACCACTTTT-3' |
4. Results
4.1. Patients and Bacterial Isolation
A total of 100 A. baumannii isolates were recovered from various clinical specimens including trachea (37%), urine (21%), sputum (9%), blood (7%), catheter (6%), bronchial wash (5%), wound (3%, abscess (3%), and other (6%). The age range of the patients was from 14 to 86 years, where 25% of isolates belonged to patients between the age of 20 - 39 years and 72% were from patients between 40 - 90 years old.
4.2. Bacterial Identification
The results of the biochemical test demonstrated the identity of all the isolates as A. baumannii species. Further PCR analyses showed presence of blaOXA-51-like genes in isolates, which confirmed their identity as A. baumannii.
4.3. Antimicrobial Susceptibility
Results of antimicrobial susceptibility testing showed that the highest antibiotic resistance rate were against ticarcillin (100%), cefixime (100%), and ceftizoxime (100%), followed by aztreonam (97%), cephalexin (97%), cefotaxime (97%), ampicillin (94%), kanamycin (94%), ceftriaxone (94%), ceftazidime (93%), cephalothin (91%), carbenicillin (89%), co-trimoxazole (85%), norfloxacin (84%), ciprofloxacin (80%), chloramphenicol (78%), gentamicin (78%), ofloxacin (71%), tetracycline (65%), tobramycin (63%), meropenem (63%), imipenem (62%), ampicillin-sulbacta (55%) whereas the highest susceptibility rates were noticed for polymyxin B (84%), colistin (77%), and rifampin (73%).
4.4. Genotypic Characterization of ESBL-Positive Isolates
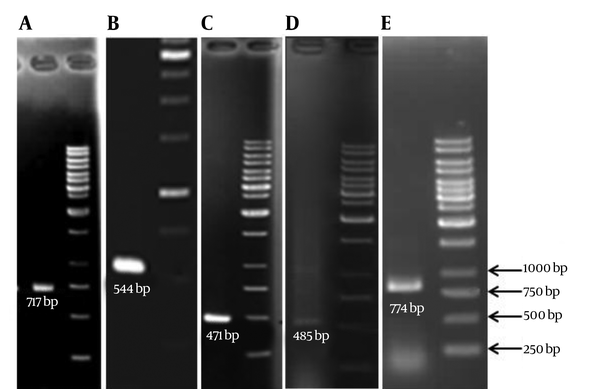
Double-Disk Synergy Test showed that 60% of the isolates were ESBL producers. PCR experiments (Figure 2), for detection of ESBL genotypes of TEM, SHV, CTX-M, OXA-2 and OXA-10 among the ESBL positive isolates revealed that the highest frequency belonged to the SHV (31.6%) whereas the lowest frequency were for OXA10 (8.3%) (Table 2).
PCR Amplification of the A, TEM; B, CTX-M; C, SHV; D, OXA-2; and E, OXA-10 Gene Fragments Demonstrate Sharp Bands Upon Electrophoresis
Prevalence of ESBL Genotypes in the ESBL-Positive Clinical Isolates (N = 60)
| Gene | No. (%) | Gene Coincidence | No. (%) |
|---|---|---|---|
| TEM | 7 (11.6) | TEM + SHV + CTX-M + PER1 + INT-1 | 1 (1.6) |
| SHV | 19 (31.6) | SHV + CTX + INT-1 | 2 (3.3) |
| CTX-M | 8 (13.3) | PER1 + VEB1 + INT-1 | 3 (5) |
| OXA2 | 7 (11.6) | TEM + CTX-M | 3 (5) |
| OXA10 | 5 (8.3) | TEM + SHV | 4 (6.6) |
| OXA2 + OXA10 + INT-1 | 3 (5) | ||
| OXA2 + INT-1 | 7 (11.6) | ||
| OXA10 + INT-1 | 5 (8.3) |
Among 60 ESBL producing isolates, 1 (1.66%) were positive for TEM, SHV, and CTX-M genes, 2 (3.33%) had both CTX-M and SHV, 3 (5%) possessed both CTX-M and TEM genes, and finally 4 isolates (6.66%) contained both SHV and TEM genes together.
5. Discussion
Recent reports have indicated that the antimicrobial resistance rates of Acinetobacter isolates are continuously increasing, consequently posing a growing threat to hospitalized patients (21, 22). Nosocamial strains of Acinetobacter use different antibiotic resistance mechanisms such as reduced access to target caused by alterations in cell wall channels (porins), increased antibiotic expulsion by efflux pumps, enzymatic inactivation by β-lactamases, mutational resistance, and biofilm formation (23). It has been shown that enzymatic degradation by β-lactamases is the most common mechanism in A. baumannii (2).
In this study the results of antimicrobial susceptibility testing showed that all studied strains were resistant against ticarcillin, cefixime, and ceftizoxim (100% resistance); while, 84% of them were susceptible for polymyxin B, 77% for colistin, and 73% for rifampin, indicating the highest susceptibility rates. Similar A. baumanii susceptibility rates had been reported in previous studies in Iran. Shahcheraghi et al. (24) determined resistance rates of 100% against cefixime, 99% against ceftriaxone, and 98% against cefotaxime, along with susceptibility rates of 97% for polymyxin B and 88% for colistin in A. baumanii isolates from Tehran. Mohajeri et al. (25) reported the highest resistance rates against cefotaxime (93%) and ceftriaxone (91%), whilst the highest susceptibility rates were found to colistin (89%) and polymyxin B (86%) in A. baumanii isolates from Kermanshah. These results suggest that polymyxin B and colistin may be helpful in treating A. baumanii-related nosocomial infections in hospital settings.
Findings of this study showed that 60% of the isolates were ESBL-positive, indicating a high frequency of ESBL-associated resistance, whereas reports from India and Turkey indicated a frequency of 28% and 39%, respectively (26, 27). Genotypic screening revealed that 11.6%, 13.3%, and 31.6% of isolates were positive for blaTEM, blaCTX-M, and blaSHV, respectively. By contrast, a study from China reported that the frequency of blaTEM, and blaCTXM positive isolates were 25% and 66%, respectively (28). Furthermore, a study in South America reported that the frequencies of blaTEM, blaCTX-M, and blaSHV genotypes among the studied Acinetobacter spp. were 26.1%, 30.4%, and 8.7%, respectively (29). Nevertheless, our study shows a higher frequency of blaSHV and lower frequencies of blaTEM and blaCTX-M, compared to the mentioned studies.
In our study the prevalence of blaOXA-2 and blaOXA-10 genes were 11.6% and 8.3%, respectively. OXA-type β-Lactamases are characterized by their hydrolysis potency on cloxacillin and oxacillin, which are 50% greater than that for benzylpenicillin (30). Of the 244 known OXA-type β-Lactamases, only 16 of them are known as ESBLs, among which OXA-2 and OXA-10 considered as parent types for the rest (31). Most acquired OXA-type β-lactamases including OXA-2, OXA-10, and their derivatives are associated with class 1 integron or other insertion sequences (32). Integrons play an important role in distributors of acquired drug resistance genes among bacteria and A. baumanii (13). Accordingly, our previous study showed a high prevalence of INT-1 insertion sequence (75%) among the ESBL producing strains (33). Different frequency of INT-1 gene in A. baumanii isolates had been reported in other countries including 27.53% from Spain (8) and 71.4% from Taiwan (13), where the last prevalence rate was similar to our results.
In conclusion, our results revealed a high level of antimicrobial resistance among the studied clinical isolates of A. baumanii, and demonstrated the vital role of the studied ESBL genes in the generation of this antimicrobial resistance. These findings emphasize on the necessity of antimicrobial surveillance in different geographic regions for control of resistance dissemination. Such studies are critical to take proper measures in respect of prevention, patient care or therapy, in order to control further propagation of these severely resistant bacterial strains in community and hospital settings.
Acknowledgements
References
-
1.
Bergogne-Berezin E, Towner KJ. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev. 1996;9(2):148-65. [PubMed ID: 8964033].
-
2.
Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21(3):538-82. [PubMed ID: 18625687]. https://doi.org/10.1128/CMR.00058-07.
-
3.
Boroumand MA, Akhyani H, Sheikhvatan M, Yazdi SH, Saboorian R, Hashemi SH, et al. Evaluation of antimicrobial resistance of Acinetobacter baumannii to Imipenem, Ciporofloxacin and Ceftazidime using E Test. Iran J Public Health. 2009;38(2):130-3.
-
4.
Falagas ME, Bliziotis IA, Siempos ,I. Attributable mortality of Acinetobacter baumannii infections in critically ill patients: a systematic review of matched cohort and case-control studies. Crit Care. 2006;10(2):R48. [PubMed ID: 16563184]. https://doi.org/10.1186/cc4869.
-
5.
Meric M, Kasap M, Gacar G, Budak F, Dundar D, Kolayli F, et al. Emergence and spread of carbapenem-resistant Acinetobacter baumannii in a tertiary care hospital in Turkey. FEMS Microbiol Lett. 2008;282(2):214-8. [PubMed ID: 18371065]. https://doi.org/10.1111/j.1574-6968.2008.01129.x.
-
6.
Shi ZY, Liu PY, Lau Y, Lin Y, Hu BS, Shir JM. Antimicrobial susceptibility of clinical isolates of Acinetobacter baumannii. Diagn Microbiol Infect Dis. 1996;24(2):81-5. [PubMed ID: 9147913].
-
7.
Shaikh S, Fatima J, Shakil S, Rizvi SM, Kamal MA. Antibiotic resistance and extended spectrum beta-lactamases: Types, epidemiology and treatment. Saudi J Biol Sci. 2015;22(1):90-101. [PubMed ID: 25561890]. https://doi.org/10.1016/j.sjbs.2014.08.002.
-
8.
Ribera A, Vila J, Fernandez-Cuenca F, Martinez-Martinez L, Pascual A, Beceiro A, et al. Type 1 integrons in epidemiologically unrelated Acinetobacter baumannii isolates collected at Spanish hospitals. Antimicrob Agents Chemother. 2004;48(1):364-5. [PubMed ID: 14693570].
-
9.
Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. 2005;18(4):657-86. [PubMed ID: 16223952]. https://doi.org/10.1128/CMR.18.4.657-686.2005.
-
10.
Wilke MS, Lovering AL, Strynadka NC. Beta-lactam antibiotic resistance: a current structural perspective. Curr Opin Microbiol. 2005;8(5):525-33. [PubMed ID: 16129657]. https://doi.org/10.1016/j.mib.2005.08.016.
-
11.
Sturenburg E, Mack D. Extended-spectrum beta-lactamases: implications for the clinical microbiology laboratory, therapy, and infection control. J Infect. 2003;47(4):273-95. [PubMed ID: 14556752].
-
12.
Bali EB, Accedil L, Sultan N. Phenotypic and molecular characterization of SHV, TEM, CTX-M and extended-spectrum-lactamase produced by Escherichia coli, Acinobacter baumannii and Klebsiella isolates in a Turkish hospital. Afr J Microbiol Res. 2010;4(8):650-4.
-
13.
Huang LY, Chen TL, Lu PL, Tsai CA, Cho WL, Chang FY, et al. Dissemination of multidrug-resistant, class 1 integron-carrying Acinetobacter baumannii isolates in Taiwan. Clin Microbiol Infect. 2008;14(11):1010-9. [PubMed ID: 19040472]. https://doi.org/10.1111/j.1469-0691.2008.02077.x.
-
14.
Jain A, Mondal R. TEM & SHV genes in extended spectrum beta-lactamase producing Klebsiella species beta their antimicrobial resistance pattern. Indian J Med Res. 2008;128(6):759-64. [PubMed ID: 19246801].
-
15.
Bell SM, Gatus BJ, Pham JN, Rafferty DL. Antibiotic susceptibility testing by the CDS method. A concise laboratory manual. NSW: The antibiotic reference laboratory, South Eastern Area Laboratory Services. Arthur Productions Pty Ltd Sydney; 1999.
-
16.
Hall G. Nonfermenting and miscellaneous Gram negative bacilli. In: Mahon CR, Lehman DC, Manuselis G, editors. Textbook of diagnostic microbiology. Ohio: Saunders-Elsevier; 2007. p. 564-84.
-
17.
Wayne P. Clinical and laboratory standards institute. Performance standards for antimicrobial susceptibility testing. 2007.
-
18.
Nagano N, Nagano Y, Cordevant C, Shibata N, Arakawa Y. Nosocomial transmission of CTX-M-2 beta-lactamase-producing Acinetobacter baumannii in a neurosurgery ward. J Clin Microbiol. 2004;42(9):3978-84. [PubMed ID: 15364979]. https://doi.org/10.1128/JCM.42.9.3978-3984.2004.
-
19.
Yousefi S, Farajnia S, Nahaei MR, Akhi MT, Ghotaslou R, Soroush MH, et al. Detection of metallo-beta-lactamase-encoding genes among clinical isolates of Pseudomonas aeruginosa in northwest of Iran. Diagn Microbiol Infect Dis. 2010;68(3):322-5. [PubMed ID: 20846807]. https://doi.org/10.1016/j.diagmicrobio.2010.06.018.
-
20.
Brown S, Amyes S. OXA (beta)-lactamases in Acinetobacter: the story so far. J Antimicrob Chemother. 2006;57(1):1-3. [PubMed ID: 16332731]. https://doi.org/10.1093/jac/dki425.
-
21.
Livermore DM. The threat from the pink corner. Ann Med. 2003;35(4):226-34. [PubMed ID: 12846264].
-
22.
Tognim MC, Andrade SS, Silbert S, Gales AC, Jones RN, Sader HS. Resistance trends of Acinetobacter spp. in Latin America and characterization of international dissemination of multi-drug resistant strains: five-year report of the SENTRY Antimicrobial Surveillance Program. Int J Infect Dis. 2004;8(5):284-91. [PubMed ID: 15325597]. https://doi.org/10.1016/j.ijid.2003.11.009.
-
23.
Munoz-Price LS, Weinstein RA. Acinetobacter infection. N Engl J Med. 2008;358(12):1271-81. [PubMed ID: 18354105]. https://doi.org/10.1056/NEJMra070741.
-
24.
Shahcheraghi F, Abbasalipour M, Feizabadi M, Ebrahimipour G, Akbari N. Isolation and genetic characterization of metallo-beta-lactamase and carbapenamase producing strains of Acinetobacter baumannii from patients at Tehran hospitals. Iran J Microbiol. 2011;3(2):68-74. [PubMed ID: 22347585].
-
25.
Mohajeri P, Farahani A, Feizabadi MM, Ketabi H, Abiri R, Najafi F. Antimicrobial susceptibility profiling and genomic diversity of Acinetobacter baumannii isolates: A study in western Iran. Iran J Microbiol. 2013;5(3):195-202. [PubMed ID: 24475323].
-
26.
Sinha M, Srinivasa H, Macaden R. Antibiotic resistance profile & extended spectrum beta-lactamase (ESBL) production in Acinetobacter species. Indian J Med Res. 2007;126(1):63-7. [PubMed ID: 17890826].
-
27.
Vahaboglu H, Ozturk R, Aygun G, Coskunkan F, Yaman A, Kaygusuz A. Widespread detection of PER-1-type extended-spectrum beta-lactamases among nosocomial Acinetobacter and Pseudomonas aeruginosa isolates in Turkey: a nationwide multicenter study. Antimicrob Agents Chemother. 1997;41(10):2265-9. [PubMed ID: 9333059].
-
28.
Zhao SY, Jiang DY, Xu PC, Zhang YK, Shi HF, Cao HL, et al. An investigation of drug-resistant Acinetobacter baumannii infections in a comprehensive hospital of East China. Ann Clin Microbiol Antimicrob. 2015;14:7. [PubMed ID: 25643932]. https://doi.org/10.1186/s12941-015-0066-4.
-
29.
Celenza G, Pellegrini C, Caccamo M, Segatore B, Amicosante G, Perilli M. Spread of bla(CTX-M-type) and bla(PER-2) beta-lactamase genes in clinical isolates from Bolivian hospitals. J Antimicrob Chemother. 2006;57(5):975-8. [PubMed ID: 16510850]. https://doi.org/10.1093/jac/dkl055.
-
30.
Bush K, Jacoby GA, Medeiros AA. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39(6):1211-33. [PubMed ID: 7574506].
-
31.
Jacoby G, Bush K. ß-Lactamase classification and amino acid sequences for TEM, SHV and OXA extended-spectrum and inhibitor resistant enzymes. 2014.
-
32.
Poirel L, Naas T, Nordmann P. Diversity, epidemiology, and genetics of class D beta-lactamases. Antimicrob Agents Chemother. 2010;54(1):24-38. [PubMed ID: 19721065]. https://doi.org/10.1128/AAC.01512-08.
-
33.
Farajnia S, Azhari F, Alikhani MY, Hosseini MK, Peymani A, Sohrabi N. Prevalence of PER and VEB Type Extended Spectrum Betalactamases among Multidrug Resistant Acinetobacter baumannii Isolates in North-West of Iran. Iran J Basic Med Sci. 2013;16(6):751-5. [PubMed ID: 23997900].