Abstract
Background:
Amylases play a vital role in biotechnological studies and rank an important position in the world enzyme market (25% to 33%). Bioprocess method of amylase production is more effective than the other sources, since the technique is easy, cost effective, fast, and the enzymes of required properties can be procured.Objectives:
The current study aimed to report the characteristics of novel amylase producing bacterial strains isolated from Taptapani hot spring, Odisha, India.Materials and Methods:
Bacterial strains were isolated by dilution plating method from the water samples collected from Taptapani Hot Spring, Odisha and screened for amylase production through starch hydrolysis. The bacterial isolates were identified morphologically, biochemically, and finally by 16S rDNA profiling.Results:
Based on the morphological, physiological, biochemical characteristics and the molecular characterization, the isolates SS1, SS2, and SS3 were identified as Bacillus barbaricus, Aeromonas veroni, and Stenotrophomonas maltophilia, respectively. The approximate molecular weight of enzymes from SS1, SS2, and SS3 strains were 19 kDa, 56 kDa and 49 kDa, respectively.Conclusions:
The current report isolates, characterizes, and demonstrates the novel heat-adapted amylase-producing bacteria SS1, SS2 and SS3 from Taptapani hot spring, indicating its potentiality and stability under acidic conditions.Keywords
1. Background
The abundance of microorganisms is due to their diversity, but 96% of the microbes cannot be cultured under the laboratory conditions; tracing them out can lead to great achievements in various fields of biotechnology. Regarding the applicability of those microbes to solve environmental problems, it is quite essential to characterize and isolate them (1). Springs are the places where underground water is discharged at specific locations on the earth and they dramatically vary as to the type of water they discharge. Many of the springs are the result of long cracks or joints in sedimentary rock (2). In hot springs the temperature of water lies significantly above the mean of annual air temperature of that region.
Amylases play a vital role in biotechnological studies and rank an important position in the world enzyme market (25% to 33%) (3, 4). Amylases are characterized by their ability to hydrolyze starch to generate glucose, maltose, a mixture of malto-oligosaccharides, and various α-limit dextrin-containing α (1-6) bonds (5). Their wide range of application in nutritional, cosmetic and pharmaceutical processes increases their significance (6). Amylases are classified into endoamylases (α-amylases), exoamylases (β-amylase, glucoamylase), and debranching enzymes (pullulanase, isoamylase) based on their type of action (7). α-amylase (endo 1, 4-α-D-glucanglucohydrolase, EC 3.2.1.1) is an extracellular enzyme that affects starch and degrades it into disaccharide and trisaccharide (8). α-amylase can be derived from various sources such as plants, animals and microorganisms (9).
Bioprocess method of amylase production is more effective than the other sources, since the technique is easy, cost effective, fast, and enzymes of the required properties can be procured. The microbial amylases could be potentially useful in various industrial processes such as sugar, textile, paper, brewing, distilling industries and pharmaceuticals (10). Due to the potential technological significance and the economic benefits of amylases, it has drawn the global attention.
2. Objectives
The current study aimed to report the characteristics of novel amylase producing bacterial strains isolated from Taptapani hot spring, Odisha, India. Production conditions were optimized (temperature, pH, carbon and nitrogen sources) to achieve high enzyme production and better enzyme activity.
3. Materials and Methods
3.1. Isolation Site and Cultivation Conditions
Water samples were collected at the depth of 6 to 10 cm in sterile bottles from Taptapani hot spring, and brought to the Research Laboratory in aseptic conditions. The samples were stored in a refrigerator at 4°C for further processing. Bacteria were isolated by serial dilution and spread plate method. A volume of 0.1 mL of each dilution was transferred to Luria Bertani agar (1% tryptone, 0.5% yeast extract, 0.5% NaCl, 1.5% agar, pH 7-7.5). The samples were spread uniformly using a glass rod, and incubated at 37°C for 24 hours. The bacterial isolates were further subcultured on the respective media in order to obtain pure culture. Pure isolates were refrigerated at 4°C for further studies.
3.2. Screening of Amylase Producing Bacteria
Starch hydrolysis test was used to screen the bacterial isolates showing amylolytic properties. The starch agar plates were streaked by microbial isolates followed by their incubation at 37°C for 24 hours. After incubation, 1% iodine solution (freshly prepared) was flooded on the starch agar plate. The presence of blue color around the growth are as indicated negative result, and a clear zone of hydrolysis surrounding the growth areas indicated a positive result (11).
3.3. Identification of Amylase Producing Bacteria
The colony morphology of the isolates was observed under the microscope with respect to color, shape, size, nature of the colony, and pigmentation. Cultural and physiological characteristics of the amylase producing bacterial isolates were studied to identify the purposes and the obtained results were compared with those of the standard description of Bergey's Manual of Determinative Bacteriology (12).
3.4. Microscopic Observations
Gram staining of the bacterial isolates was performed and observed under a high power magnifying lens in the light microscope. Endospore staining, capsule staining, and motility test were performed to observe the morphology and motility of the cells (1).The shape of the cell and its surface features were explored in more detail by Environmental Scanning Electron Microscopy (FEI QUANTA 200 MARK 2) using the method described by Deflaun et al. (13).
3.5. Antibiotic Assays
The susceptibility to antibiotics was checked on a log phase culture by measuring the Diameter of Zone of Inhibition (DZI) using the disc diffusion technique; antibiotic discs were purchased from Hi-Media Laboratory Pvt. Ltd, Mumbai. India. The selected antibiotic groups, the culture conditions, and the assessment procedure were as Nandy et al. (14).
3.6. Biochemical Characterization
Various biochemical tests like indole, methyl red, Voges Proskauer, simmons citrate, catalase, oxidase, urease, nitrate reduction, gelatin hydrolysis, starch hydrolysis, and SIM (SulphideIndole Motility medium) tests were performed as per the standard methods reported by Deflaun et al. (13) and Nandy et al. (14).
3.7. Growth Profile
The overnight grown culture in LB medium acted as the parent source; 1% inoculation from the parent culture was added to 100 mL of sterile LB medium and incubated at 37°C under 200 rpm shaking. At the regular interval of one hour, 3 mL of culture was aliquoted and the optical density was measured at 660 nm (Systronics UV-Vis spectrophotometer), representing the extent of growth. The effect of various carbon and nitrogen sources on the maximum growth of isolates was studied by adjusting different carbon (sucrose, fructose, dextrose, starch and lactose) and nitrogen sources (meat extract, beef extract, yeast extract and urea, casein) of 1% concentration in the production medium.
3.8. Molecular Characterization
The molecular identification was performed on the basis of sequence analysis of 16S rDNA. Genomic DNA was isolated from the pure culture. The partial sequence of the 16S rDNA gene was amplified by PCR using the universal prokaryotic primers, 5'- ACGGGCGGTGTGTAC -3' and 5'-CAGCCGCGGTAATAC-3', which amplify a 1500-base pair region of the 16S rDNA gene (15). The PCR product was purified and sequenced. Sequencing was done by the Heleni Biomolecule Company, located in Guntur, Andhra Pradesh, India. Partial 16S rDNA gene sequence was required to NCBI-BLAST (http: // www. ncbi.nlm.nih.gov/Blast) and the nearest neighbor of the isolate was determined. The construction of phylogenetic tree was performed by neighbor-joining tree algorithm using bootstrap value of the MEGA4. (http://www.megasoftware.net/) (16).
3.9. Amylase Production
Freshly prepared inoculum was used to inoculate the production medium. To prepare the inoculums, a loopful of bacterial isolate was transferred in 100 mL of the medium containing (g/L) starch 10, peptone 10, yeast extract 20, KH2PO4 0.10, CaCl2.2H2O 0.10, MgSO4.7H2O 0.50 and FeSO4.7H2O 0.02. The flask was loaded on a rotary shaker incubator (Remi instruments Ltd.) at a speed of 200 rpm at 37°C for 24 hours. Amylase production was carried out by submerged fermentation in Erlenmyer Flask. The flask was loaded on a rotary shaker incubator at a speed of 200 rpm at 37°C for 24 hours. After incubation, fermented broth was centrifuged at 8000 rpm for 20 minutes in a cooling centrifuge. After specific time intervals, samples were taken out to determine the enzyme activity and protein concentration.
3.9.1. Amylase Assay
Amylase was determined by spectrophotometric method as described by Sharma (17). One unit is defined as the amount of enzyme required to liberate one µmol of sugar reduction per minute by applying the following formula (18).
IU/mL/min = (Activity of enzyme × 1000)/ (Molecular weight of Maltose × Incubation time)
3.9.2. Estimation of Protein Content
Total amount of protein throughout the experiment was measured according to Lowry et al. (1951) method, using Bovine serum albumin (BSA) as Standard (19).
3.9.3. Partial Purification of Amylase Enzyme
Partial purification of amylase enzyme was achieved by ammonium sulfate precipitation followed by dialysis. The cell free extract was saturated with ammonium sulfate up to 80%. The content was incubated overnight and centrifuged at 10000 rpm for 30 minutes. Supernatant was collected and saturated up to 90% with ammonium sulfate, then the content was centrifuged at 10000 rpm for 30 min and pellet was collected for further analysis. The enzyme mixture was transferred in a dialysis bag and immersed in phosphate buffer at 4°C for 24 hours. The buffer was continuously stirred using a magnetic stirrer throughout the process. Buffer was changed three times during the process in order to obtain proper purification.
3.9.4 Optimization of Temperature and pH for Enzyme Activity
The effect of temperature on enzyme production and activity was studied by adjusting the incubation temperature at 10 to 60°C, and production medium pH at 4.0 to 10.0 (20, 21).
4. Results
4.1. Isolation of Amylase Producing Bacteria
A total 42 different bacterial strains were isolated on the basis of colony morphology. All isolates were primarily screened for amylase production by starch hydrolysis. Among the 42 isolates, six isolates hydrolyzed the starch on starch agar. The isolates SS1, SS2, and SS3 strains showed the maximum clear zone on starch agar plate after pouring 1% iodine solution, indicating maximum amylase activity and hence were selected for further study of amylase production, enzyme activity and optimization.
4.2. Morphological Characterization
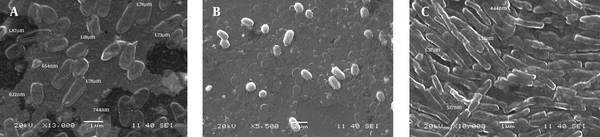
SEM images as shown in Figure 1 (Figure 1a, 1b and 1c) confirm the rod shape of SS3, rod/cocci for SS2 whereas SS1 was irregular in its shape and size. Some more microscopic features of the isolates were identified by the general staining procedures (Table 1). SS1 and SS3 were found Gram positive whereas SS2 was Gram negative. On the other hand SS1 and SS2 showed negative response to acid fast staining but SS3 was found positive for the same test.
Micrograph Showing the Morphological Structure and Dimension of Isolates
| Characteristic | SS1 | SS2 | SS3 |
|---|---|---|---|
| Microscopic characters | |||
| Shape | Irregular | Rod/cocci | Rod |
| Color | Black | White | Yellow |
| Motility | Motile | Motile | Motile |
| Spore formation | Positive | Negative | Negative |
| Gram Staining | Positive | Negative | Positive |
| Acid fast Staining | Negative | Negative | Positive |
| Biochemical characters | |||
| Protease | Positive | Negative | Positive |
| Indole | Negative | Negative | Positive |
| Catalase | Positive | Positive | Positive |
| Lipase | Negative | Negative | Negative |
| Citrate | Negative | Negative | Negative |
| Nitrate Reduction | Negative | Positive | Positive |
| Starch Hydrolysis | Positive | Positive | Positive |
| Hydrogen sulfide | Negative | Negative | Negative |
| Gelatin | Negative | Positive | Positive |
| Voges Proskauer | Positive | Positive | Positive |
| Methyl-red | Positive | Negative | Negative |
| Urease | Negative | Negative | Negative |
| Amylase | Positive | Positive | Positive |
4.3. Biochemical Identification
The strains showed different biochemical profiles which comprise of some positive as well as negative tests (Table 1). All the three strains responded positively to catalase, starch hydrolysis, Voges Proskauer, and amylase tests whereas for lipase citrate, urease and nitrate reductase tests the three isolates showed negative responses. Positivity towards amylase and starch hydrolysis tests confirms the amylase production ability of all the isolates.
4.4. Molecular Identification of Amylase Producing Bacteria
The constructed phylogenetic tree revealed that the isolates SS1, SS2, and SS3 had very close resemblance to the genera Bacillus, Aeromonas and Stenotrophomonas respectively. Thus they were identified as Bacillus barbaricus, Aeromonas veroni, and S. maltophilia, respectively (Figure 2a, 2b and 2c).
Neighbour-Joining Phylogenetic Tree From Analysis of 16S rDNA Gene Sequence of Bacterial Isolates

4.5. Growth Behavior of the Isolates
The three isolates showed distinct growth curves. Their exponential growth phase started after 15 to 20 hours of incubation. The maximum growth for the isolates also varied depending on the use of various carbon and nitrogen sources (Figure 3). Thus growth profiles of the three isolates basically depended upon various environmental conditions.
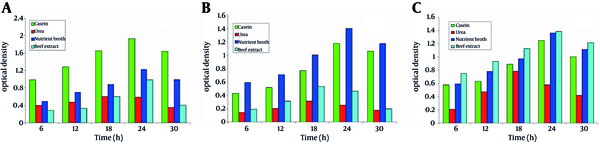
4.6. Effect of Carbon Sources on the Growth of Microbes
The effect of carbon source on growth of the three isolates was characterized using four different sugars at 1 % (w/v) concentration. SS1 showed the maximum growth in the presence of sucrose as carbon source, whereas, the minimum growth was observed in the presence of lactose (Figure 3a). For SS2, xylose had the most significant effect on the growth while fructose had the least (Figure 3b). SS3 showed the maximum growth in the presence of sucrose as carbon source, whereas, the minimum growth was observed in the presence of xylose (Figure 3c)
Effect of Carbon Sources on the Growth of Microbes
4.7. Effect of Nitrogen Sources on the Growth of Microbes
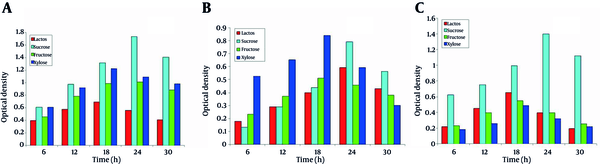
The growth of isolates was studied in the presence of four different nitrogen sources. SS1showed the maximum growth in the presence of casein as nitrogen source; whereas, the minimum growth was observed in the presence of urea (Figure 4a). For SS2, urea showed the least growth (Figure 4b). SS3 showed the maximum growth in the presence of beef extract as nitrogen source, whereas, the minimum growth was observed in the presence of urea (Figure 4c).
Effect of Nitrogen Sources on the Growth of Microbes
4.8. Characterization of Amylase
4.8.1. Electrophoretic Pattern of Partially Purified Amylase
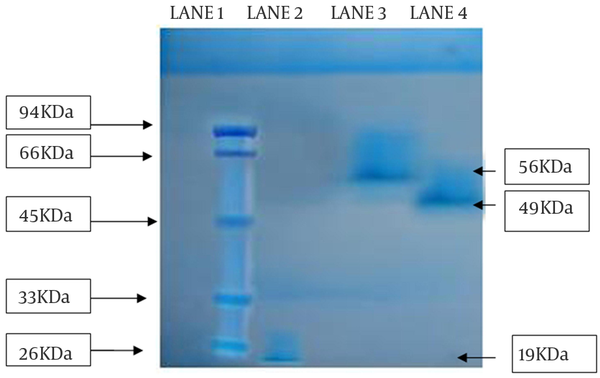
The partial purified amylase was finally subject to SDS-PAGE for molecular weight determination. From the gel, it was observed that the isolated enzyme from all the strains migrated as single bands in the respective lanes, suggesting that the purified proteins were homogeneous. The approximate molecular weight of enzymes from SS1, SS2, and SS3 strains were 19 kDa, 56 kDa and 49 kDa, respectively (Figure 5).
SDS-PAGE Analysis of Partially Purified Amylase
4.8.2. Effect of Temperature on Enzyme Activity
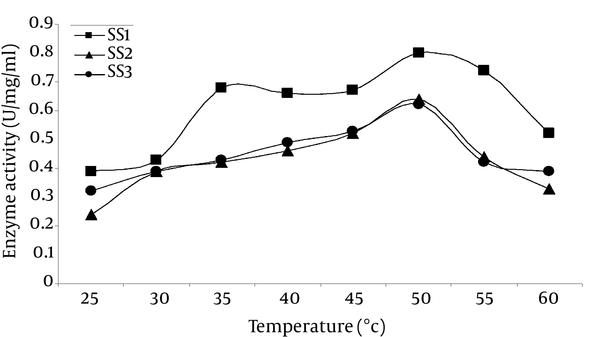
It was observed that the enzyme was optimally active at 50°C (Figure 6), which was very close to that of α-amylase from Bacillus spp. (40°C to 50°C) (22). Strain SS1 showed the highest activity of 0.804 U/mg/mL, while SS2 and SS3 showed 0.693 and 0.597 U/mg/mL, respectively at 50°C. SS1 also showed relatively higher activity of 0.692 U/mg/mL at 35°C compared to the other two strains.
Effect of Temperature on the Activity of Amylase Produced by the Three Bacterial Isolates
4.8.3. Effect of pH on Enzyme Activity
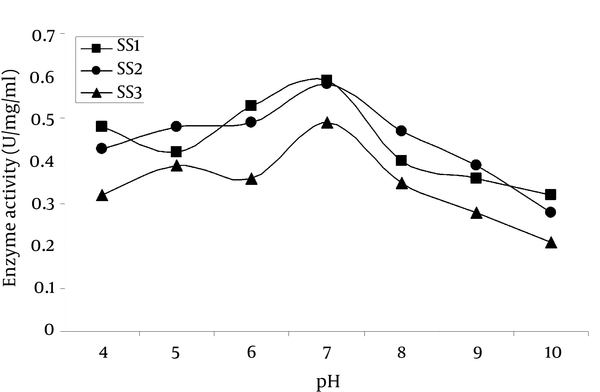
Figure 7 showed that, the amylase exhibited optimum activity at pH 7.0, which was higher than those of α-amylase from Eisenia foetide (pH 5.5) (23) and A. pullulans (pH 4.5) (24). SS1 Strain showed the highest activity of 0.574 U/mg/mL at pH 7.0 while SS2 and SS3 showed 0.540 and 0.481 U/mg/mL, respectively. The enzyme activity profile of SS1 and SS2 strains were almost the same while SS3 showed comparatively less activity in the mentioned pH range.
Effect of pH on the Activity of Amylase Produced by the Three Bacterial Strains
5. Discussion
The thermal active amylases produced by thermotolarent SS1, SS2 and SS3 strains with the most similarity with B. barbaricus, A. veronii, and S. maltophilia were isolated from Taptapani hot spring, Odisha, India. In the previous studies amylase production in several Bacillus species had been examined (25-27) and species of this genus were considered as an ideal host for the industrial production of bulk extracellular amylases. Along with the Bacillus species, the current study introduced two novel bacteria A. veroni and S. maltophilia for extracellular amylase production. In order to use α-amylase in the industry, the enzymes should be stable at different ranges of pH and temperature. The optimal temperature of enzyme activity was 50°C, while the activity decreased at temperatures above 60°C.
This property might limit its use in industrial applications that require high temperatures, but it favors its application in the processes that require complete inactivation of the enzyme with increasing temperature in the process, that is in the baking industry (28). Maximum amylase activity (0.574 U/mg/mL) was obtained using sucrose as carbon source for B. barbaricus and S. maltophilia whereas xylose for A. veroni and casein and beef extract as nitrogen sources for B. barbaricus, S. maltophilia and A. veroni, respectively when the strains were grown at 50°C, and pH 7.0 for 24 hours. The optimal pH for the amylases was 7.0, which was similar to the ones reported for halophilic amylases from various microorganisms (29-31). Amylases with this characteristic may have interesting applications in treatment of saline waters or waste solutions with starch residues and high salt content. As mentioned above, when the SS1, SS2, and SS3 strains were incubated in the presence of antibiotics like oxidase, doxycycline, streptomycin, cephadroxil, and gentamycin, the resistance of the strains in ascending order was SS3 < SS1 < SS2 (Figure 4a, 4b and 4c).
The partially purified amylases showed the molecular weight approximately 56 kDa, 19 kDa, and 49 kDa which were analogous to the result obtained from B. licheniformis, A. hydrophilia, and S. maltophilia α-amylases (32-34). Many researchers reported different molecular mass of α-amylase isolated from different sources (35). The reported isolates, SS1, SS2 and SS3 isolated from Taptapani hot spring were identified as B. barbaricus, A. veroni, and S. maltophilia, respectively. They demonstrated all characteristics of the heat-adapted amylase-producing bacteria. The enzymes produced by them and the ones described here in as Amy SS1, Amy SS2 and Amy SS3 confirm their potentiality and stability even under the acidic conditions. At elevated temperatures, they improve the solubility of starch, decrease the viscosity, limit microbial contaminants and reduce the reaction time (36, 37), thus they have promising industrial applications. These enzymes established resistance in the presence of some antibiotics and may have applications in pharmaceuticals. It is recommended that the future works include the enzyme efficiency increase to work under extremely harsh industrial conditions by genetically engineering the microbes or their products in order to achieve high production potentials and wide industrial applications.
Acknowledgements
References
-
1.
Sarkar AK, Roy S, Pal A, Pakrashi S, Mahra PK, Sahoo S, et al. Microbial biodiversity screening for metal accumulators from mineral ore rich site in Andhra Pradesh, India. OnLine J Biol Sci. 2008;8(2):32. https://doi.org/10.3844/ojbsci.2008.32.40.
-
2.
Sen SK, Mohapatra SK, Satpathy S, Rao GTV. Characterization of hot water spring source isolated clones of bacteria and their industrial applicability. Int J Chem Res. 2010;2(1):1-7.
-
3.
Nguyen QD, Rezessy-Szabó JM, Claeyssens M, Stals I, Hoschke A. Purification and characterisation of amylolytic enzymes from thermophilic fungus; Thermomyces lanuginosus; strain ATCC 34626. Enzyme Microb Technol. 2002;31(3):345-52. https://doi.org/10.1016/S0141-0229(02)00128-X.
-
4.
Ashwini K, Gaurav K, Karthik L, Bhaskara Rao KV. Optimization, production and partial purification of extracellular α-amylase from Bacillus sp. marini. Arch Appl Sci Res. 2011;3(1):33-42.
-
5.
Yang CH, Liu WH. Purification and properties of a maltotriose-producing α-amylase from; Thermobifida fusca. Enzyme Microb Technol. 2004;35(2):254-60. https://doi.org/10.1016/j.enzmictec.2004.05.004.
-
6.
Nigam P, Singh D. Enzyme and microbial systems involved in starch processing. Enzyme Microb Technol. 1995;17(9):770-8. https://doi.org/10.1016/0141-0229(94)00003-A.
-
7.
Goesaert H, Slade L, Levine H, Delcour JA. Amylases and bread firming–an integrated view. J Cereal Sci. 2009;50(3):345-52. https://doi.org/10.1016/j.jcs.2009.04.010.
-
8.
Douglas Crabb W, Mitchinson C. Enzymes involved in the processing of starch to sugars. Trends Biotechnol. 1997;15(9):349-52. https://doi.org/10.1016/S0167-7799(97)01082-2.
-
9.
Pandey A, Nigam P, Soccol CR, Soccol VT, Singh D, Mohan R. Advances in microbial amylases. Biotechnol Appl Biochem. 2000;31 ( Pt 2):135-52. [PubMed ID: 10744959].
-
10.
Oyeleke SB, Oduwole AA. Production of amylase by bacteria isolated from a cassava waste dumpsite in Minna, Niger State, Nigeria. Afr J Microbiol Res. 2009;3(4):143-6.
-
11.
Hamilton LM, Kelly CT, Fogarty WM. Production and properties of the raw starch-digesting α-amylase of; Bacillus; sp. IMD 435. Process Biochem. 1999;35(1):27-31. https://doi.org/10.1016/S0032-9592(99)00028-X.
-
12.
Sharmin S, Hossain MdT, Anwar MN. Isolation and characterization of a protease producing bacteria Bacillus amovivorus and optimization of some factors of culture condition for protease production. J Biol Sci. 2005;5(3):358-62. https://doi.org/10.3923/jbs.2005.358.362.
-
13.
Deflaun MF, Fredrickson JK, Dong H, Pfiffner SM, Onstott TC, Balkwill DL, et al. Isolation and characterization of a Geobacillus thermoleovorans strain from an ultra-deep South African gold mine. Syst Appl Microbiol. 2007;30(2):152-64. [PubMed ID: 16709445]. https://doi.org/10.1016/j.syapm.2006.04.003.
-
14.
Nandy P, Thakur AR, Chaudhuri SR. Characterization of bacterial strains isolated through microbial profiling of urine samples. OnLine J Biol Sci. 2007;7(1):44. https://doi.org/10.3844/ojbsci.2007.44.51.
-
15.
Chaudhuri SR, Pattanayak AK, Thakur AR. Microbial DNA extraction from samples of varied origin. Curr Sci. 2006;91(12):1697.
-
16.
Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596-9. [PubMed ID: 17488738]. https://doi.org/10.1093/molbev/msm092.
-
17.
Sharma K. Manual of Microbiology Tools and Techniques. 1. Delhi India: Ane Books; 2007. p. 141-71.
-
18.
Haq I, Abdullah R, Ashraf H, Athar Shah H. Isolation and screening of fungi for the biosynthesis of alpha amylase. Biotechnol. 2002;2:61-6.
-
19.
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193(1):265-75.
-
20.
Shanmughapriya S, Kiran GS, Selvin J, Thomas TA, Rani C. Optimization, purification, and characterization of extracellular mesophilic alkaline cellulase from sponge-associated Marinobacter sp. MSI032. Appl Biochem Biotechnol. 2010;162(3):625-40. https://doi.org/10.1007/s12010-009-8747-0.
-
21.
Aiyer PVD. Effect of C: N ratio on alpha amylase production by Bacillus licheniformis SPT 27. Afr J Biotechnol. 2004;3(10):519-22.
-
22.
Liu XD, Xu Y. A novel raw starch digesting alpha-amylase from a newly isolated Bacillus sp. YX-1: purification and characterization. Bioresour Technol. 2008;99(10):4315-20. [PubMed ID: 17920264]. https://doi.org/10.1016/j.biortech.2007.08.040.
-
23.
Ueda M, Asano T, Nakazawa M, Miyatake K, Inouye K. Purification and characterization of novel raw-starch-digesting and cold-adapted alpha-amylases from Eisenia foetida. Comp Biochem Physiol B Biochem Mol Biol. 2008;150(1):125-30. [PubMed ID: 18375163]. https://doi.org/10.1016/j.cbpb.2008.02.003.
-
24.
Li H, Chi Z, Wang X, Duan X, Ma L, Gao L. Purification and characterization of extracellular amylase from the marine yeast; Aureobasidium pullulans; N13d and its raw potato starch digestion. Enzyme Microb Technol. 2007;40(5):1006-12. https://doi.org/10.1016/j.enzmictec.2006.07.036.
-
25.
Ghasemi Y, Rasoul-Amini S, Ebrahiminezhad A, Zarrini G, Kazemi A, Mousavi-Khorshidi S, et al. Halotolerant Amylase Production by a Novel Bacterial Strain, Rheinheimera aquimaris. Res J Microbiol. 2010;5(2). https://doi.org/10.3923/jm.2010.144.149.
-
26.
Utong J, Al-Quadan F, Akel H. Effect of various growth conditions on production of extracellular amylase from thermotolerant Bacillus species isolated from hot springs in Jordan. J Boil Sci. 2006;6:621-5.
-
27.
Mishra S, Behera N. Amylase activity of a starch degrading bacteria isolated from soil receiving kitchen wastes. Afr J Biotechnol. 2008;7(18):3326-31.
-
28.
Coronado M, Vargas C, Hofemeister J, Ventosa A, Nieto JJ. Production and biochemical characterization of an alpha-amylase from the moderate halophile Halomonas meridiana. FEMS Microbiol Lett. 2000;183(1):67-71. [PubMed ID: 10650204].
-
29.
Deutch CE. Characterization of a salt-tolerant extracellular a-amylase from Bacillus dipsosauri. Lett Appl Microbiol. 2002;35(1):78-84. [PubMed ID: 12081555].
-
30.
Hutcheon GW, Vasisht N, Bolhuis A. Characterisation of a highly stable alpha-amylase from the halophilic archaeon Haloarcula hispanica. Extremophiles. 2005;9(6):487-95. [PubMed ID: 16075161]. https://doi.org/10.1007/s00792-005-0471-2.
-
31.
Onishi H, Sonoda K. Purification and Some Properties of an Extracellular Amylase from a Moderate Halophile, Micrococcus halobius. Appl Environ Microbiol. 1979;38(4):616-20. [PubMed ID: 16345444].
-
32.
Mahdavi A, Sajedi RH, Rassa M, Jafarian V. Characterization of an α-amylase with broad temperature activity from an acid-neutralizing Bacillus cereus strain. Iran J Biotechnol. 2010;8(2):18-27.
-
33.
Singh L, Ram MS, Agarwal MK, Alam SI. Characterization of Aeromonas hydrophila strains and their evaluation for biodegradation of night soil. World J Microbiol Biotechnol. 2000;16(7):625-30. https://doi.org/10.1023/A:1008968712682.
-
34.
Mohammed K, Arif JM, Ramteke PW. Structural adaptation and biocatalytic activity of microbial α-amylase. Afri J Microbiol Res. 2012;6(2):206-13.
-
35.
Lin LL, Chyau CC, Hsu WH. Production and properties of a raw-starch-degrading amylase from the thermophilic and alkaliphilic Bacillus sp. TS-23. Biotechnol Appl Biochem. 1998;28 ( Pt 1):61-8. [PubMed ID: 9693090].
-
36.
Thippeswamy S, Girigowda K, Mulimani VH. Isolation and identification of alpha-amylase producing Bacillus sp. from dhal industry waste. Indian J Biochem Biophys. 2006;43(5):295-8. [PubMed ID: 17133736].
-
37.
Oyeleke SB, Auta SH, Egwim EC. Production and characterization of amylase produced by Bacillus megaterium isolated from a local yam peel dumpsite in Minna, Niger State. J Microbiol Antimicrob. 2010;2(7):88-92.