Abstract
Background:
The worldwide prevalence of Helicobacter pylori is about 50%. This bacterium needs a number of virulence factors for pathogenesis.Objectives:
This study aimed to determine the prevalence of virulence genes (ureB, cytotoxin-associated gene A [cagA], and vacuolating cytotoxin [vacA]), as well as the antigenic profile in H. pylori strains.Methods:
Eighty-five patients with abdominal pain, including 46 H. pylori-positive and 39 H. pylori-negative cases, were enrolled in this study. The serum levels of interleukin (IL)-17F, tumor necrosis factor α (TNF-α), and interferon γ (IFN-γ) cytokines were measured by multiplex kits and flow cytometry. After molecular identification by the ureC gene, vacA, cagA, and ureB genes were detected by polymerase chain reaction (PCR). Finally, after antigenic extraction, the whole-cell protein was exhibited by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE).Results:
The prevalence of vacA, ureB, and cagA genes were 91.3%, 67.39%, and 50%, respectively. The frequency of genes and cell surface antigens were not significantly different based on the gastritis severity (P > 0.05). IL-17F significantly (P = 0.046) increased in the presence of 19.5 kDa (outer membrane protein [OMP]). Moreover, the OMP antigen significantly enhanced immunoglobulin A (IgA; P = 0.013). In the presence of the 66-kDa (ureB) antigen, the serum level of IFN-γ increased (p = 0.041). Finally, the CagA protein led to increased IgG antibody levels (p = 0.027).Conclusions:
Early detection of H. pylori infection can play a crucial role in managing it. Our results suggest that IL-17F, TNF-α, and IFN-γ cytokines could be diagnostic markers. However, further studies are required to fully investigate this suggestion.Keywords
Helicobacter pylori Cytokines Gastritis Antigenic Profile Virulence Gene
1. Background
The common microorganism of the human stomach is a Gram-negative bacterium, Helicobacter pylori, that colonizes the gastric mucosa of more than half the world population (1, 2). The prevalence of H. pylori is 20% and > 90% in developed and developing countries, respectively (3). This bacterium was detected in a gastric biopsy for the first time in Australia. Urease activity lets the bacterium survive on the gastric epithelium (4). Helicobacter pylori colonization occurs in the gastric mucosa through some virulence genes, such as cytotoxin-associated gene A (cagA) and vacuolating cytotoxin (vacA) (5). The colonization, adhesion, and invasion of H. pylori strains into the gastric epithelial cells is facilitated by different virulence genes (6).
Once H. pylori is located on the gastric lumen, a permanent infection develops, and its long-term presence (if left untreated) leads to several gastro-duodenal diseases, such as gastric ulcer, chronic gastritis, duodenal ulcer, gastric mucosa-associated lymphoid tissue (MALT) lymphoma, and gastric cancer (7). Antibody response to H. pylori has indicated that the level of antibody response provides information beyond the detection of infection. The relationship between high antibody levels was found to be associated with the grade of histological gastritis, gastric mucosal inflammation, mucosal bacterial density, and gastric cancer risk (8). After H. pylori colonization in the stomach, inflammatory responses (production of cytokines) by host immune system cells occur in the gastric mucosa (9).
The virulence of H. pylori varies in different geographical regions. Previous studies have clearly defined that H. pylori virulence factors have a strong effect on pathogenicity and treatment results (10). Some H. pylori virulence factors, including CagA, VacA, H. pyloricag pathogenicity island (cagPAI), and adhesion proteins, are vital in gastric disease pathogenesis (11). Studies have shown that extremely virulent H. pylori strains carry cagPAI, including 31 genes (40 kb region) involved in the host’s inflammatory response and cagA translocation (12). CagA is an extremely studied H. pylori virulence gene. It is placed at the end of cagPAI and encodes an immunodominant protein with a molecular weight of 120 kDa (13).
It has been reported that severe inflammation, gastric cancer, and gastric ulcer are intensely associated with cagA gene expression (14, 15). The presence of cagA in H. pylori strains usually accompanies other virulence factors, including vacA (16). Primary studies on vacA have discovered 2 main polymorphic families of s-region and m-region (17, 18). The m-region and s-region encoded the vacA m1 or m2 and vacA s1 or vacA s2 allele, respectively. According to the literature, vacA m1 strains are more likely to cause gastric epithelial damage than vacA m2 strains (19). In Northeast Asia (such as South Korea and Japan), vacA m1 type strains are more common. In contrast, vacA m2 type strains dominate Southeast Asia (such as Vietnam and Taiwan) (20). The s1 type is further divided into s1a, s1b, and s1c, and the m1 type is divided into m1a and m1b alleles (21).
The majority of prior studies of H. pylori-induced cytokines have focused on cytokine messenger RNA (mRNA) detection or protein quantification in supernatants from in vitro cultures of gastric biopsy specimens, isolated gastric lymphocytes, or gastric epithelial cell lines (22-25). However, in this study, we assessed the levels of cytokines in the blood of patients. In addition, the association between the antigenic profile of bacterial strains and cytokines in the peripheral blood of infected individuals was explored in the current study, which has never been done before. One of the major problems with H. pylori infections is the late diagnosis. We can employ inflammatory cytokines as early detection indicators by measuring their levels in people infected with H. pylori strains having pathogenic antigen patterns.
2. Objectives
This study aimed to evaluate virulence genes, antigen patterns of H. pylori, and levels of some inflammatory cytokines in H. pylori-infected patients.
3. Methods
3.1. Sample Collection
Eighty-five patients with gastritis and dyspepsia were enrolled in this study. Sampling was carried out in Imam Khomeini Hospital, Ahvaz, Iran. Inclusion criteria were patients older than 21 years with dyspepsia and H. pylori infection agreed to participate in the study and signed the consent form. Exclusion criteria were patients who received therapy with nonsteroidal anti-inflammatory drugs (NSAIDs) or antibiotics within 4 weeks of study entrance, patients who reported upper gastrointestinal bleeding, and pregnant females. The rapid urease and histopathological test made the initial diagnosis of H. pylori infection in these patients. The gastritis level was also defined in the samples. After taking the stomach mucus biopsy, 1 part of the biopsy was used for the urease test and culture to approve H. pylori infection. The other part was used in histopathological studies (kept in 10% phosphate-buffered formalin). In addition, 10 mL of blood was taken from the patients, and the serum was instantly extracted using centrifugation at 3,200 rpm (15 min). The extracted serum samples were stored at -70°C. Immunoglobulin G (IgG) and IgA antibodies against H. pylori were measured using ELISA kits (Roche-Germany) (26). Besides the relatively small sample size, the limitation of our study was the lack of previous studies on the topic.
3.2. Helicobacter pylori Isolation and Detection
The biopsy samples were cultured on Brucella agar (Merck, Germany) containing vancomycin (10 µg/mL), trimethoprim (5 µg/mL), amphotericin B (2.5 µg/mL), and sheep blood (5%). After incubation under 10% CO2 and temperature of 37°C conditions for 3 - 5 days, the colony appears, and the usual microbiological tests (such as Gram staining, catalase, urease, and oxidase tests) were performed for H. pylori detection (27-29). Genomic DNA was extracted according to a standard protocol, which was placed in the Pooyagen Azma Company Kit (Tehran, Iran). Then, the purity (A260/A280) and concentration of the extracted DNA were determined using a NanoDrop ND-1000 spectrophotometer.
The quality of the extracted DNA samples was determined using a 2% agarose gel stained with SYBR Green (30). Espinoza et al. advocated using the glmM gene to detect H. pylori in 2011 (31). Indeed, the glmM housekeeping gene is required for bacterial proliferation and cell wall formation, making it unique to H. pylori (32). Therefore, the glmM (ureC) gene was used for the molecular identification of strains; ureC primers are displayed in Table 1. Polymerase chain reaction (PCR) was performed in the final volume of 25 μL containing 12.5 μL master mix 2X (SinaClon, Iran), 1 μL of each primer, 1 μL genomic DNA, and 9.5 μL deionized water. The extracted DNA of Escherichia coli ATCC 25922 was used as a negative control instead of H. pylori genomic DNA. DNA from isolates with known genes was used as a positive control. PCR was accomplished using a thermocycler (BioRad, USA) under initial denaturation conditions of 4 min at 94°C, followed by 30 cycles of denaturation (60 s, 94°C), primer binding (30 s, 51°C), elongation (90 s, 72°C), and final elongation (4 min, 72°C). Ten microliters of the PCR product was subjected to electrophoresis on a 2% agarose gel in 1X TBE buffer at 80 V for 30 min and stained with SYBR Green (30, 33).
The Primer Used in the Study
| Gene | Primer Sequence | Amp. Size (bp) | Reference |
|---|---|---|---|
| ureC-F | 5′- GGATAGACGATGTGATAGG -3′ | 224 bp | (33) |
| ureC-R | 5′- TTGGTTAGGGTGTAAAGC -3′ | ||
| cagA-F | 5′- ATAATGCTAAATTAGACAACTTGAG-3′ | 298 bp | This study |
| cagA -R | 5′- TTAGAATAATCAACAAACATCACGC-3′ | ||
| vacAm1/m2-F | 5′- CAATCTGTCCAATCAAGCGAG-3′ | 567 - 642 bp | (34) |
| vacAm1/m2 -R | 5′- GCGTCAAAATAATTGAAGG-3′ | ||
| ureB-F | 5′- AGTAGCCCGGTAGAACACAACATCCT-3′ | This study | |
| ureB-R | 5′- AGTCCTTTGTCATAAGCCGCTTGG-3′ |
3.3. Virulence Genes Detection and vacA Genotyping
The systems of PCR were the same as mentioned above except for the primers. The amplification condition is illustrated in Table 2. Also, the necessary volume of compounds required in PCR is listed in Table 3. Primers used for cagA, ureB, and genotyping of vacA are presented in Table 1. In the end, the reaction product was evaluated by 2% agarose gel electrophoresis (30, 35).
Amplification Conditions for cagA, ureB, and vacA Genes
| Step | Time |
|---|---|
| Initial denaturation | 94°C, 4 min |
| Cycling (30 cycles) | |
| Denaturation | 94°C, 1 min |
| Annealing (cagA) | 60, 30 s |
| Annealing (ureB) | 60, 30 s |
| Annealing (vacAm1/m2) | 52, 30 s |
| Extension | 72°C, 90 s |
| Final extension | 72°C, 5 min |
The Volume of Compounds Required in PCR
| Compounds | Volume (μL) |
|---|---|
| Distilled water | 7.5 |
| Master mix | 12.5 |
| DNA samples | 3 |
| Round primer mixture | 2 |
| Total | 25 |
3.4. Whole Helicobacter pylori Cell Protein Profile Analysis
As previously described by Huang et al. (36) and Sheykhian et al. (37), whole H. pylori cell protein profiles were analyzed by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE). The bacterial cell was washed by cold phosphate-buffered saline (PBS). After centrifugation, the bacterial pellet was suspended in 10 mL of Tris-HCl buffer (pH 7.8) containing EDTA (1 mM/L) and phenylmethylsulfonylfluoride (PMSF; 1 mM/L). The tube containing bacterial suspension was sonicated for 8 cycles (MSE ultrasonicator). Debris was removed by centrifugation at 1,500g for 15 min. The pellet was resuspended in 10 mL of Tris.HCl (pH 7.8), and then ribonuclease (Rnase) and deoxyribonuclease (DNase; Sigma-Aldrich, USA) were added (0.1 mg/mL). After incubation at 37°C for 2 h, the supernatant was centrifuged at 150,000g and 4°C for 45 min. The H. pylori cell wall pellet was dissolved in 10 mL of sarcosine (2%). After 30 min incubation at room temperature, the outer membrane was pelleted, the supernatant was removed, and the pellet was dissolved in 1 mL of PBS containing 1 mM PMSF (pH 7.8).
3.5. Serum Level of IL-17F, TNF-α, and IFN-γ by Flow Cytometry
The serum level of interleukin (IL)-17F, tumor necrosis factor α (TNF-α), and interferon γ (IFN-γ) were evaluated by flow cytometry using the 13-plex LEGENDplex™ Human Th Cytokine Panel (BioLegend, USA). First, the serum samples were diluted to 1: 6; then, they were tested according to the kit manufacturer’s instructions. Finally, the contents of the well plate were transferred to fluorescence-activated cell sorting (FACS) tubes (Abcam, United Kingdom) to be read by the flow cytometry (BioRad, USA).
3.6. Statistical Analysis
The Mann-Whitney U test and independent sample t-test were used based on the data normality. The Kolmogorov-Smirnov test determined the normality of the data. P values < 0.05 were considered statistically significant.
4. Results
4.1. Study Population
Histopathology, fast urease, and PCR demonstrated that 46 samples were infected with H. pylori (Hp+), while the remaining 39 cases were not infected (Hp-). The participants’ mean age was 43.83 ± 13.41 and 41.28 ± 12.65 years for the Hp+ and HP-, respectively. The Hp+ group included 17 females and 29 males, and the Hp- group included 22 females and 17 males. There was no significant relationship between age and sex in both groups (Table 4). The gastritis severity in the Hp+ group was mild in 23.91% (11/46) and moderate in 76.09% (35/46) of samples. There was also a significant difference between the 2 groups in IgG and IgA levels (Table 4).
Demographic Information of the Helicobacter pylori Positive (Hp+) and H. pylori Negative (Hp-) Groups
4.2. Prevalence of Virulence Genes
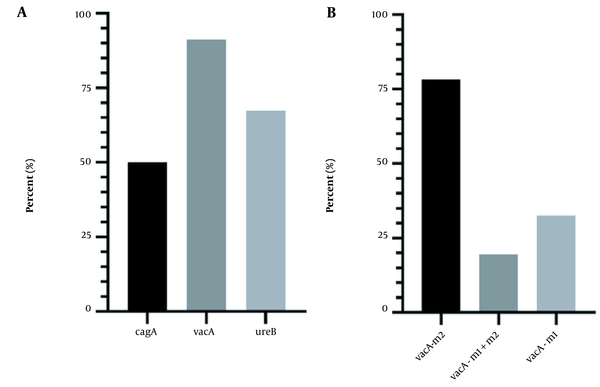
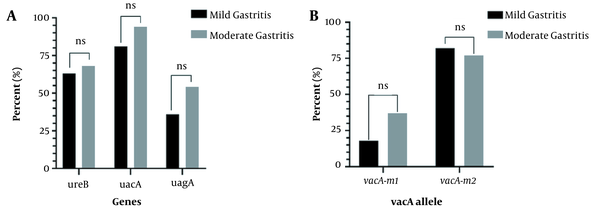
The prevalence rates of virulence genes cagA, vacA, and ureB in the Hp+ group are summarized in Figure 1A. The highest prevalence was related to the vacA gene, with a frequency of 91.3%. VacA gene genotyping was performed (Figure 1B), and the frequency of vacA m1 and vacA m2 was 32.6% and 78.26%, respectively. The abundance of 3 genes and vacA alleles by gastritis severity is also shown in Figure 2. Based on these results, the frequency of genes is not significantly different in patients with mild and moderate gastritis (P > 0.05).
Frequency of studied virulence genes (A) and frequency of vacA gene alleles (B).
Frequency of studied genes (A) and vacA alleles based on the gastritis severity (B) (ns, not significant).
4.3. Antigenic Profile Analysis
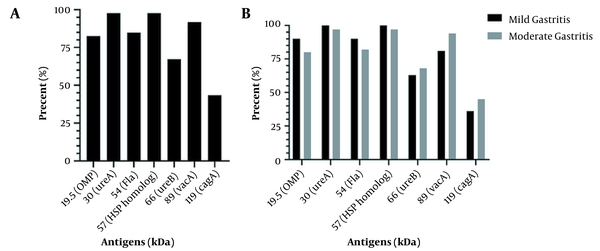
After extraction of whole-cell antigens and analysis by SDS-PAGE (Figure 3), 7 well-known antigens were evaluated in the strains based on the molecular weight (kDa). Figure 4A shows the prevalence of antigen expression in the strains. Also, the relationship between antigens and gastritis severity is shown in Figure 4B. The presence of antigens was not significantly different based on the gastritis severity (P > 0.05).
SDS-PAGE of whole-cell antigens of Helicobacter pylori (L, ladder and 26 - 33, H. pylori strains).
Antigen profile in Helicobacter pylori strains (A) and antigen profile based on gastritis severity (B) (ns, not significant).
4.4. Correlation Between Antibodies, Cytokines’ Levels, and Antigen Profiles
As demonstrated in Table 5, the 3 studied cytokines’ serum levels differ significantly in the Hp+ and Hp- groups (P < 0.05). Also, the effect of H. pylori antigens on the level of antibodies and cytokines’ serum levels are shown in Table 6. According to the results, IL-17F significantly (P = 0.046) increased in the presence of 19.5 kDa [outer membrane protein (OMP)]. Moreover, the OMP antigen enhanced IgA (P = 0.013) significantly. In the presence of the 66-kDa (ureB) antigen, the serum level of IFN-γ increased (P = 0.041). Finally, the CagA protein increased IgG antibody levels (P = 0.027).
IL-17F, TNF-α, and IFN-γ Levels in the Helicobacter pylori Positive (Hp+) and H. pylori Negative (Hp-) Groups
The Antibodies and Cytokines’ Serum Levels in the Presence of Helicobacter pylori Antigens a
| Antigens (kDa) | IL-17F (pg/mL) | TNF-α (pg/mL) | IFN-γ (pg/mL) | IgG (pg/mL) | IgA (pg/mL) |
|---|---|---|---|---|---|
| 19.5 (OMP) | |||||
| Positive | 164.88 | 76.96 | 266.97 | 94 | 104 |
| Negative | 137.28 | 94.5 | 162 | 123 | 68 |
| 30 (UreA) | |||||
| Positive | 141.83 | 79.32 | 253.46 | 100 | 74 |
| Negative | 153 | 111 | 35 | 53 | 69 |
| 54 (Fla) | |||||
| Positive | 145.87 | 82.63 | 266.73 | 102 | 74 |
| Negative | 120.94 | 65.43 | 148.37 | 85 | 76 |
| 57 (HSP homolog) | |||||
| Positive | 141.59 | 80.65 | 251.78 | 100 | 75 |
| Negative | 164 | 51 | 111 | 81 | 59 |
| 66 (UreB) | |||||
| Positive | 211.47 | 123.4 | 520.67 | 100 | 79 |
| Negative | 108.5 | 59 | 117.13 | 99 | 72 |
| 89 (VacA) | |||||
| Positive | 143.85 | 80.18 | 263.34 | 101 | 76 |
| Negative | 123.5 | 78.25 | 95.13 | 78 | 53 |
| 119 (CagA) | |||||
| Positive | 175.86 | 103.39 | 386.33 | 117 | 63 |
| Negative | 116.08 | 62 | 142.86 | 76 | 83 |
5. Discussion
The ureC gene is highly conserved and has been applied to identify H. pylori strains. A previous study reported that the sensitivity and specificity of ureC is more than 90% (38). The present study showed that the prevalence of vacA, ureB, and cagA genes was 91.3%, 67.39%, and 50%, respectively. According to previous studies, cagA has a frequency of 71.4% in Turkey (39), 54% in Sudan (40), 62% in South Africa (41), and 77.27% in India (42). One of the most important virulence factors in H. pylori is an 89-kDa protein, VacA, which can cause cell depletion. In Pandya et al.’s study (42), the prevalence of the vacA gene was 4.54% in India, which is consistent with our results. However, the prevalence of vacA in South Africa (2019) was similar to the present study (90.6%) (41). The vacA polymorphic gene encodes the VacA protein. The m1 genotype of the middle part of the gene is associated with high cytotoxicity. The m2 genotype is found in non-cytotoxic strains (43). Among all vacA positive strains, 78.26% had the m2 allele, and 32.6% had the m1 allele. Another virulence gene in this study was the ureB, which is present in 67.39% of the strains. Urease consists of the main subunits UreA and UreB (44). This enzyme plays a vital role in the colonization of H. pylori. In addition, urea stimulates the production of inflammatory cytokines by mononuclear phagocytes (45).
Surface antigens on the H. pylori cell or secreted from the cell include CagA, VacA, urease subunits (UreA and UreB), heat shock protein (HspA and HspB), subunits of flagellin, catalase, lipopolysaccharide, OMP, and several unknown antigens (46, 47). In this study, known antigens were evaluated in whole isolated strains. In the present study, the CagA antigen was found to be associated with high levels of IgG antibodies. CagA has been identified as a vital virulence factor in H. pylori (48), and CagA antibodies have been observed in patients with gastritis, gastric ulcer, and gastric cancer (49-51). In 2016, Seo et al. showed an association between the CagA antigen and high levels of IgG and IgA antibodies (52). However, in the present study, no association was found between IgA and CagA antigen. In the 2000s, 80% of H. pylori strains carried the cagA gene in Japan and Hong Kong (53, 54), and 94% of 33 Korean children had the cagA gene (55). In Japan, CagA was the most reactive antigen found in all H. pylori-infected serum samples (even from children under 3 years of age) (56). Therefore, the CagA antigen in each region will be important to detect H. pylori infection, and the prevalence of this antigen in strains can be important for diagnosis (56). In Korean studies, the result of a positive serological test for the CagA antibody was considered as H. pylori infection (57). Also, we demonstrated that IgA increased significantly in the presence of OMP (19.5 kDa).
Serum levels of IL-17F, TNF-α, and IFN-γ cytokines were assessed in the study population. There was a significant difference in cytokine levels between patients with and without H. pylori. Thus, the colonization of H. pylori in the stomach can lead to an inflammatory response in the absorption of immune system cells in the gastric mucosa (58). IL-17 mediates the activation of polymorphonuclear neutrophils and leads to gastritis (59). It has been previously reported that there is a significant increase in IL-17 and IFN-γ in the early stages of H. pylori infection (59). IL-17 can stimulate immune cells to release inflammatory mediators, including IL-1, IL-6, and TNF-α (59). All the IL-17F, TNF-α, and IFN-γ cytokines involved in gastritis increased in our studied population. More studies are required to clarify the role of H. pylori virulence factors in the production of cytokines. CagA antigens are more effective in stimulating dendritic cells (DCs) to induce IL-23/IL-17 expression. Also, IL-17 activation by the ERK1/2 MAP kinase pathway is more associated with the CagA antigen (60). We demonstrated that the level of IL-17F in patients with CagA-positive H. pylori was increased (not significantly). Also, the presence of OMP (19.5 kDa) and UreB (66 kDa) antigen caused significant changes in IL-17F and IFN-γ, respectively.
5.1. Conclusions
In conclusion, the antigen profile of H. pylori isolated from Ahwaz, Iran, was shown in the present study. However, for further investigations, we suggest using western blotting and ELISA techniques in addition to SDS-PAGE. As it is clear, early detection of H. pylori infection can play a crucial role in reducing the risks of this bacterium. Therefore, by investigating the levels of inflammatory cytokines and their relationship with bacterial antigen profile, a suitable cytokine can be identified for the rapid diagnosis of H. pylori infection. Our results suggest that IL-17F, TNF-α, and IFN-γ cytokines could be used as a diagnostic marker. However, further investigations are required to approve this suggestion.
References
-
1.
Choi IJ, Kim CG, Lee JY, Kim YI, Kook MC, Park B, et al. Family history of gastric cancer and Helicobacter pylori treatment. N Engl J Med. 2020;382(5):427-36. [PubMed ID: 31995688]. https://doi.org/10.1056/NEJMoa1909666.
-
2.
Tan AH, Lim SY, Mahadeva S, Loke MF, Tan JY, Ang BH, et al. Helicobacter pylori eradication in Parkinson’s disease: a randomized placebo-controlled trial. Mov Disord. 2020;35(12):2250-60. [PubMed ID: 32894625]. https://doi.org/10.1002/mds.28248.
-
3.
Ohno H, Satoh-Takayama N. Stomach microbiota, Helicobacter pylori, and group 2 innate lymphoid cells. Exp Mol Med. 2020;52(9):1377-82. [PubMed ID: 32908209]. [PubMed Central ID: PMC8080604]. https://doi.org/10.1038/s12276-020-00485-8.
-
4.
Zhou Q, Li L, Ai Y, Pan Z, Guo M, Han J. Diagnostic accuracy of the (14)C-urea breath test in Helicobacter pylori infections: a meta-analysis. Wien Klin Wochenschr. 2017;129(1-2):38-45. [PubMed ID: 27848071]. https://doi.org/10.1007/s00508-016-1117-3.
-
5.
Camilo V, Sugiyama T, Touati E. Pathogenesis of Helicobacter pylori infection. Helicobacter. 2017;22(Suppl 1). [PubMed ID: 28891130]. https://doi.org/10.1111/hel.12405.
-
6.
Ranjbar R, Yadollahi Farsani F, Safarpoor Dehkordi F. Antimicrobial resistance and genotyping ofvacA,cagA, andiceAalleles of theHelicobacter pyloristrains isolated from traditional dairy products. J Food Saf. 2018;39(2). e12594. https://doi.org/10.1111/jfs.12594.
-
7.
Ansari S, Yamaoka Y. Helicobacter pylori virulence factor cytotoxin-associated gene A (CagA)-mediated gastric pathogenicity. Int J Mol Sci. 2020;21(19). [PubMed ID: 33050101]. [PubMed Central ID: PMC7582651]. https://doi.org/10.3390/ijms21197430.
-
8.
Butt J, Blot WJ, Shrubsole MJ, Waterboer T, Pawlita M, Epplein M. Differences in antibody levels to H. pylori virulence factors VacA and CagA among African Americans and whites in the Southeast USA. Cancer Causes Control. 2020;31(6):601-6. [PubMed ID: 32222845]. [PubMed Central ID: PMC7286423]. https://doi.org/10.1007/s10552-020-01295-z.
-
9.
Ieni A, Barresi V, Rigoli L, Fedele F, Tuccari G, Caruso RA. Morphological and cellular features of innate immune reaction in Helicobacter pylori gastritis: A brief review. Int J Mol Sci. 2016;17(1). [PubMed ID: 26784180]. [PubMed Central ID: PMC4730350]. https://doi.org/10.3390/ijms17010109.
-
10.
Oktem-Okullu S, Cekic-Kipritci Z, Kilic E, Seymen N, Mansur-Ozen N, Sezerman U, et al. Analysis of correlation between the seven important Helicobacter pylori (H. pylori) virulence factors and drug resistance in patients with gastritis. Gastroenterol Res Pract. 2020;2020:3956838. [PubMed ID: 32908495]. [PubMed Central ID: PMC7475755]. https://doi.org/10.1155/2020/3956838.
-
11.
Sukri A, Hanafiah A, Mohamad Zin N, Kosai NR. Epidemiology and role of Helicobacter pylori virulence factors in gastric cancer carcinogenesis. APMIS. 2020;128(2):150-61. [PubMed ID: 32352605]. https://doi.org/10.1111/apm.13034.
-
12.
Kalali B, Mejias-Luque R, Javaheri A, Gerhard M. H. pylori virulence factors: influence on immune system and pathology. Mediators Inflamm. 2014;2014:426309. [PubMed ID: 24587595]. [PubMed Central ID: PMC3918698]. https://doi.org/10.1155/2014/426309.
-
13.
Backert S, Blaser MJ. The Role of CagA in the gastric biology of Helicobacter pylori. Cancer Res. 2016;76(14):4028-31. [PubMed ID: 27655809]. [PubMed Central ID: PMC5798256]. https://doi.org/10.1158/0008-5472.CAN-16-1680.
-
14.
Saeidi Y, Pournajaf A, Gholami M, Hasannejad-Bibalan M, Yaghoubi S, Khodabandeh M, et al. Determination of Helicobacter pylori virulence-associated genes in duodenal ulcer and gastric biopsies. Med J Islam Repub Iran. 2017;31:95. [PubMed ID: 29951396]. [PubMed Central ID: PMC6014795].
-
15.
Ranjbar R, Farsani FY, Dehkordi FS. Phenotypic analysis of antibiotic resistance and genotypic study of the vacA, cagA, iceA, oipA and babA genotypes of the Helicobacter pylori strains isolated from raw milk. Antimicrob Resist Infect Control. 2018;7:115. [PubMed ID: 30288255]. [PubMed Central ID: PMC6162967]. https://doi.org/10.1186/s13756-018-0409-y.
-
16.
El-Shenawy A, Diab M, Shemis M, El-Ghannam M, Salem D, Abdelnasser M, et al. Detection of Helicobacter pylori vacA , cagA and iceA1 virulence genes associated with gastric diseases in Egyptian patients. Egypt J Med Hum Genet. 2017;18(4):365-71. https://doi.org/10.1016/j.ejmhg.2017.04.003.
-
17.
Salari MH, Shirazi MH, Hadaiti MA, Daryani NA. Frequency of Helicobacter pylori vacA genotypes in Iranian patients with gastric and duodenal ulcer. J Infect Public Health. 2009;2(4):204-8. [PubMed ID: 20701884]. https://doi.org/10.1016/j.jiph.2009.08.004.
-
18.
Atherton JC, Cao P, Peek RJ, Tummuru MK, Blaser MJ, Cover TL. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270(30):17771-7. [PubMed ID: 7629077]. https://doi.org/10.1074/jbc.270.30.17771.
-
19.
Wei GC, Chen J, Liu AY, Zhang M, Liu XJ, Liu D, et al. Prevalence of Helicobacter pylori vacA, cagA and iceA genotypes and correlation with clinical outcome. Exp Ther Med. 2012;4(6):1039-44. [PubMed ID: 23226771]. [PubMed Central ID: PMC3494117]. https://doi.org/10.3892/etm.2012.704.
-
20.
Subsomwong P, Miftahussurur M, Vilaichone RK, Ratanachu-Ek T, Suzuki R, Akada J, et al. Helicobacter pylori virulence genes of minor ethnic groups in North Thailand. Gut Pathog. 2017;9:56. [PubMed ID: 29046726]. [PubMed Central ID: PMC5637267]. https://doi.org/10.1186/s13099-017-0205-x.
-
21.
Mashak Z, Jafariaskari S, Alavi I, Sakhaei Shahreza M, Safarpoor Dehkordi F. Phenotypic and genotypic assessment of antibiotic resistance and genotyping of vacA, cagA, iceA, oipA, cagE, and babA2 alleles of Helicobacter pylori bacteria isolated from raw meat. Infect Drug Resist. 2020;13:257-72. [PubMed ID: 32099418]. [PubMed Central ID: PMC6996226]. https://doi.org/10.2147/IDR.S233612.
-
22.
Basso D, Scrigner M, Toma A, Navaglia F, Di Mario F, Rugge M, et al. Helicobacter pylori infection enhances mucosal interleukin-1 beta, interleukin-6, and the soluble receptor of interleukin-2. Int J Clin Lab Res. 1996;26(3):207-10. [PubMed ID: 8905454]. https://doi.org/10.1007/BF02592984.
-
23.
Yamaoka Y, Kita M, Kodama T, Sawai N, Imanishi J. Helicobacter pylori cagA gene and expression of cytokine messenger RNA in gastric mucosa. Gastroenterology. 1996;110(6):1744-52. [PubMed ID: 8964399]. https://doi.org/10.1053/gast.1996.v110.pm8964399.
-
24.
Milic L, Karamarkovic A, Popadic D, Sijacki A, Grigorov I, Milosevic E, et al. Altered cytokine expression in Helicobacter pylori infected patients with bleeding duodenal ulcer. BMC Res Notes. 2019;12(1):278. [PubMed ID: 31092295]. [PubMed Central ID: PMC6521506]. https://doi.org/10.1186/s13104-019-4310-4.
-
25.
Bagheri V, Memar B, Momtazi AA, Sahebkar A, Gholamin M, Abbaszadegan MR. Cytokine networks and their association with Helicobacter pylori infection in gastric carcinoma. J Cell Physiol. 2018;233(4):2791-803. [PubMed ID: 28121015]. https://doi.org/10.1002/jcp.25822.
-
26.
Lagunes-Servin H, Torres J, Maldonado-Bernal C, Perez-Rodriguez M, Huerta-Yepez S, Madrazo de la Garza A, et al. Toll-like receptors and cytokines are upregulated during Helicobacter pylori infection in children. Helicobacter. 2013;18(6):423-32. [PubMed ID: 23869400]. https://doi.org/10.1111/hel.12067.
-
27.
Hasanzadeh L, Ghaznavi-Rad E, Soufian S, Farjadi V, Abtahi H. Expression and antigenic evaluation of VacA antigenic fragment of Helicobacter Pylori. Iran J Basic Med Sci. 2013;16(7):835-40. [PubMed ID: 23997913]. [PubMed Central ID: PMC3758054].
-
28.
Ghorbani F, Gheisari E, Dehkordi FS. Genotyping of vacA alleles of Helicobacter pylori strains recovered from some Iranian food items. Trop J Pharm Res. 2016;15(8):1631. https://doi.org/10.4314/tjpr.v15i8.5.
-
29.
Yahaghi E, Khamesipour F, Mashayekhi F, Safarpoor Dehkordi F, Sakhaei MH, Masoudimanesh M, et al. Helicobacter pylori in vegetables and salads: genotyping and antimicrobial resistance properties. Biomed Res Int. 2014;2014:757941. [PubMed ID: 25184146]. [PubMed Central ID: PMC4145543]. https://doi.org/10.1155/2014/757941.
-
30.
Safarpoor Dehkordi F, Tavakoli-Far B, Jafariaskari S, Momtaz H, Esmaeilzadeh S, Ranjbar R, et al. Uropathogenic Escherichia coli in the high vaginal swab samples of fertile and infertile women: virulence factors, O-serogroups, and phenotyping and genotyping characterization of antibiotic resistance. New Microbes New Infect. 2020;38:100824. [PubMed ID: 33364031]. [PubMed Central ID: PMC7750135]. https://doi.org/10.1016/j.nmni.2020.100824.
-
31.
Espinoza MG, Vazquez RG, Mendez IM, Vargas CR, Cerezo SG. Detection of the glmM gene in Helicobacter pylori isolates with a novel primer by PCR. J Clin Microbiol. 2011;49(4):1650-2. [PubMed ID: 21289140]. [PubMed Central ID: PMC3122814]. https://doi.org/10.1128/JCM.00461-10.
-
32.
De Reuse H, Labigne A, Mengin-Lecreulx D. The Helicobacter pylori ureC gene codes for a phosphoglucosamine mutase. J Bacteriol. 1997;179(11):3488-93. [PubMed ID: 9171391]. [PubMed Central ID: PMC179139]. https://doi.org/10.1128/jb.179.11.3488-3493.1997.
-
33.
Moradipour A, Khosravi A, Mehrabi M. Analyzing the abundance of the gene glmM in subjects with a positive Helicobacter pylori stool antigen test (HPSA) and correlating glmM abundance with serum levels of cytokines TNF-α and IL-1β. J Ilam Univ Med Sci. 2018;25(5):26-33. https://doi.org/10.29252/sjimu.25.5.26.
-
34.
Safaralizadeh R, Basiri Z, Hosseinpour-Feizi MA, Jabarpour-Boniadi M, Motaghi B, Nemati M. [Correlation of Helicobacter pylori vac A s, m region genotypes with different gastrodoudenal diseases in east Azerbaijan patients]. J Kerman Univ Med Sci. 2015;21(1):21-31. Persian.
-
35.
Safarpoor Dehkordi F, Gandomi H, Basti AA, Misaghi A, Rahimi E. Phenotypic and genotypic characterization of antibiotic resistance of methicillin-resistant Staphylococcus aureus isolated from hospital food. Antimicrob Resist Infect Control. 2017;6:104. [PubMed ID: 29034091]. [PubMed Central ID: PMC5628482]. https://doi.org/10.1186/s13756-017-0257-1.
-
36.
Huang J, Keeling PW, Smyth CJ. Identification of erythrocyte-binding antigens in Helicobacter pylori. J Gen Microbiol. 1992;138(7):1503-13. [PubMed ID: 1512579]. https://doi.org/10.1099/00221287-138-7-1503.
-
37.
Sheykhian A, Zahir MH, Shokri F, Malekzadeh R, Siavashi F, Mustafaie A. Extraction of the outer membrane proteins of H. pylori and evaluation of their presence in stool of the infected individuals. Iran Biomed J. 2004;8(2):83-8.
-
38.
Brooks HJ, Ahmed D, McConnell MA, Barbezat GO. Diagnosis of helicobacter pylori infection by polymerase chain reaction: is it worth it? Diagn Microbiol Infect Dis. 2004;50(1):1-5. [PubMed ID: 15380272]. https://doi.org/10.1016/j.diagmicrobio.2003.11.010.
-
39.
Ozbey G, Aygun C. Prevalence of genotypes in Helicobacter pyloriisolates from patients in eastern Turkey and the association of these genotypes with clinical outcome. Braz J Microbiol. 2012;43(4):1332-9. [PubMed ID: 24031961]. [PubMed Central ID: PMC3769024]. https://doi.org/10.1590/S1517-838220120004000014.
-
40.
Hassan HG, Idris AB, Hassan MA, Altayb HN, Yasin K, Beirage N, et al. Genetic Diversity of the cagA gene of Helicobacter pylori strains from Sudanese Patients with Different Gastroduodenal Diseases. medRxiv. 2019;Preprint. https://doi.org/10.1101/19007435.
-
41.
Idowu A, Mzukwa A, Harrison U, Palamides P, Haas R, Mbao M, et al. Detection of Helicobacter pylori and its virulence genes (cagA, dupA, and vacA) among patients with gastroduodenal diseases in Chris Hani Baragwanath Academic Hospital, South Africa. BMC Gastroenterol. 2019;19(1):73. [PubMed ID: 31088381]. [PubMed Central ID: PMC6518451]. https://doi.org/10.1186/s12876-019-0986-0.
-
42.
Pandya HB, Agravat HH, Patel JS. Prevalence of specific Helicobacter Pylori cagA, vacA, iceA, ureC genotypes and its clinical relevance in the patients with acid-peptic diseases. J Clin Diagn Res. 2017;11(8):DC23-6. [PubMed ID: 28969123]. [PubMed Central ID: PMC5620763]. https://doi.org/10.7860/JCDR/2017/27812.10457.
-
43.
Cover TL, Holland RL, Blanke SR. Helicobacter pylori Vacuolating Toxin. In: Backert S, Yamaoka Y, editors. Helicobacter pylori Research: From Bench to Bedside. Tokyo, Japan: Springer; 2016. p. 113-41. https://doi.org/10.1007/978-4-431-55936-8_5.
-
44.
Valenzuela-Valderrama M, Cerda-Opazo P, Backert S, Gonzalez MF, Carrasco-Veliz N, Jorquera-Cordero C, et al. The Helicobacter pylori urease virulence factor is required for the induction of hypoxia-induced factor-1alpha in gastric cells. Cancers (Basel). 2019;11(6). [PubMed ID: 31185594]. [PubMed Central ID: PMC6627347]. https://doi.org/10.3390/cancers11060799.
-
45.
Donelli LCG. Virulence factors of Helicobacter pylori. Microb Ecol Health Dis. 2009;12(2):259-62. https://doi.org/10.1080/089106000750060512.
-
46.
Herbrink P, van Doorn LJ. Serological methods for diagnosis of Helicobacter pylori infection and monitoring of eradication therapy. Eur J Clin Microbiol Infect Dis. 2000;19(3):164-73. [PubMed ID: 10795588]. https://doi.org/10.1007/s100960050454.
-
47.
Yilmaz O, Sen N, Kupelioglu AA, Simsek I. Detection of H. pylori infection by ELISA and Western blot techniques and evaluation of anti CagA seropositivity in adult Turkish dyspeptic patients. World J Gastroenterol. 2006;12(33):5375-8. [PubMed ID: 16981271]. [PubMed Central ID: PMC4088208]. https://doi.org/10.3748/wjg.v12.i33.5375.
-
48.
Knorr J, Ricci V, Hatakeyama M, Backert S. Classification of Helicobacter pylori virulence factors: Is CagA a toxin or not? Trends Microbiol. 2019;27(9):731-8. [PubMed ID: 31130493]. https://doi.org/10.1016/j.tim.2019.04.010.
-
49.
Soltermann A, Koetzer S, Eigenmann F, Komminoth P. Correlation of Helicobacter pylori virulence genotypes vacA and cagA with histological parameters of gastritis and patient's age. Mod Pathol. 2007;20(8):878-83. [PubMed ID: 17541440]. https://doi.org/10.1038/modpathol.3800832.
-
50.
Hatakeyama M. Helicobacter pylori CagA and gastric cancer: a paradigm for hit-and-run carcinogenesis. Cell Host Microbe. 2014;15(3):306-16. [PubMed ID: 24629337]. https://doi.org/10.1016/j.chom.2014.02.008.
-
51.
Jafarzadeh A, Hassanshahi GH, Nemati M. Serum levels of high-sensitivity C-reactive protein (hs-CRP)in Helicobacter pylori-infected peptic ulcer patients and its association with bacterial CagA virulence factor. Dig Dis Sci. 2009;54(12):2612-6. [PubMed ID: 19160050]. https://doi.org/10.1007/s10620-008-0686-z.
-
52.
Seo JH, Lim CW, Park JS, Yeom JS, Lim JY, Jun JS, et al. Correlations between the CagA antigen and serum levels of anti-Helicobacter pylori IgG and IgA in children. J Korean Med Sci. 2016;31(3):417-22. [PubMed ID: 26955243]. [PubMed Central ID: PMC4779867]. https://doi.org/10.3346/jkms.2016.31.3.417.
-
53.
Yamazaki S, Yamakawa A, Okuda T, Ohtani M, Suto H, Ito Y, et al. Distinct diversity of vacA, cagA, and cagE genes of Helicobacter pylori associated with peptic ulcer in Japan. J Clin Microbiol. 2005;43(8):3906-16. [PubMed ID: 16081930]. [PubMed Central ID: PMC1233989]. https://doi.org/10.1128/JCM.43.8.3906-3916.2005.
-
54.
Wong BC, Yin Y, Berg DE, Xia HH, Zhang JZ, Wang WH, et al. Distribution of distinct vacA, cagA and iceA alleles in Helicobacter pylori in Hong Kong. Helicobacter. 2001;6(4):317-24. [PubMed ID: 11843964]. https://doi.org/10.1046/j.1523-5378.2001.00040.x.
-
55.
Ko JS, Kim KM, Oh YL, Seo JK. cagA, vacA, and iceA genotypes of Helicobacter pylori in Korean children. Pediatr Int. 2008;50(5):628-31. [PubMed ID: 19261108]. https://doi.org/10.1111/j.1442-200X.2008.02641.x.
-
56.
Akada J, Okuda M, Hiramoto N, Kitagawa T, Zhang X, Kamei S, et al. Proteomic characterization of Helicobacter pylori CagA antigen recognized by child serum antibodies and its epitope mapping by peptide array. PLoS One. 2014;9(8). e104611. [PubMed ID: 25141238]. [PubMed Central ID: PMC4139317]. https://doi.org/10.1371/journal.pone.0104611.
-
57.
Jeong HL, Jung Y, Jun J, Yeom JS, Park JS, Seo J, et al. Comparison of four commercial ELISA kits and in-house immunoblotting for diagnosis of Helicobacter pylori infection. Pediatr Gastroenterol Hepatol Nutr. 2012;15(2):85. https://doi.org/10.5223/pghn.2012.15.2.85.
-
58.
Bagheri N, Salimzadeh L, Shirzad H. The role of T helper 1-cell response in Helicobacter pylori-infection. Microb Pathog. 2018;123:1-8. [PubMed ID: 29936093]. https://doi.org/10.1016/j.micpath.2018.06.033.
-
59.
Bagheri N, Azadegan-Dehkordi F, Shirzad H, Rafieian-Kopaei M, Rahimian G, Razavi A. The biological functions of IL-17 in different clinical expressions of Helicobacter pylori-infection. Microb Pathog. 2015;81:33-8. [PubMed ID: 25773771]. https://doi.org/10.1016/j.micpath.2015.03.010.
-
60.
Kabir S. The role of interleukin-17 in the Helicobacter pylori induced infection and immunity. Helicobacter. 2011;16(1):1-8. [PubMed ID: 21241406]. https://doi.org/10.1111/j.1523-5378.2010.00812.x.