Abstract
Keywords
Antiepileptic Drugs Lamotrigine Phenobarbital Tau Protein Molecular Docking Surface Plasmon Resonance
1. Background
Tau is an intrinsically disordered protein that belongs to the family of microtubule-associated proteins (MAPs), which are considered for microtubule stabilization and assembly (1-3). Hyperphosphorylation and aggregation of tau protein cause an increment of solubility of the protein, which can form the neurofibrillary tangles (NFTs) by binding to microtubules (4). Neurofibrillary tangles lead to cell death, and eventually, neurodegenerative diseases are known as tauopathies, such as Alzheimer’s disease (5), Parkinson’s (6), Down’s syndrome (7), dementia pugilistic (8), dementia with argyrophilic grains (9), Pick’s disease (10), etc.
The blood-brain barrier (BBB) prevents the brain uptake of most drugs from the blood, making it difficult to develop new treatments for brain diseases (11). The BBB inhibits the movement of chemicals and medications by generating strong connections between endothelial cells in the central nervous system (CNS) vessels (12). Therapeutic agents and drugs with BBB permeability can be utilized to deal with neurodegenerative disorders. This main issue has been identified as a potential research subject among the researchers.
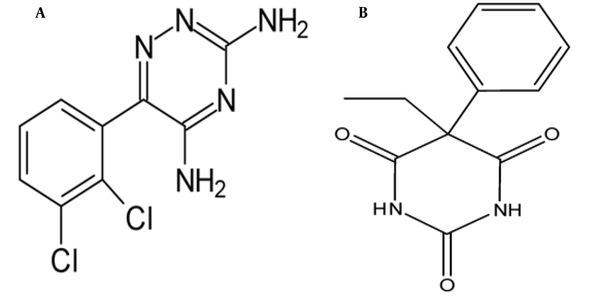
Lamotrigine (LTG) (Figure 1A) and phenobarbital (PHB) (Figure 1B) are known as anticonvulsant and antiepileptic drugs and are commonly used for different types of seizures (13-15). It has been turned out that LTG and PHB can penetrate the BBB, enter the brain, and directly impact the brain (11, 16). Due to the potential of these drugs, investigation of the interaction and their effects on tau protein can provide useful information for a better understanding of the treatment for neurodegenerative diseases.
The chemic structures of A, lamotrigine; and B, phenobarbital
Different binding properties, such as the binding mechanism, the binding constants, and the number of binding sites, have been investigated for LTG/PHB-tau protein interactions.
The molecular modeling method has also been utilized in this study to know the appropriate binding site of LTG/PHB on tau protein.
2. Methods
2.1. Materials
Recombinant human tau-412 (1 mg/mL), LTG (MW 256.09 g/mol), and PHB (MW 232.23 g/mol) were obtained from Sigma-Aldrich (St. Louis, MO, USA). N-hydroxysuccinimide (NHS), dimethylsulfoxide (DMSO), PBS buffer, N-ethyl-N-(3-dimethyl aminopropyl) carbodiimide (EDC), and ethanolamine-HCl were purchased from Merck Company (Darmstadt, Germany). The stock solutions of LTG and PHB were prepared by dissolving the adequate amounts of their powders in PBS buffer and DMSO (1: 1 v/v).
2.2. Apparatus and Procedure
2.2.1. Fluorescence Study
Using a 1-cm quartz cell on a Jasco spectrofluorimeter (FP‐ 750, Kyoto, Japan) in the range of 280 - 400 nm by exciting at 280 nm at three different temperatures (298, 310, and 316 K), fluorescence spectra of the samples were recorded. For this purpose, a fixed concentration of tau protein was titrated with additional concentrations of LTG (4, 19.5, 39, 58.5, and 78 μM) and PHB (4, 8.5, 13, 17, and 21.5 μM).
To minimize the inner-filter effect, Equation 1 was used to correct the fluorescence intensity of tau protein:
Fcorr is the corrected fluorescence intensities, and Fobs correspond to observed fluorescence intensities. Aem and Aex denote the absorbance values of the samples at emission and excitation wavelengths of tau protein, respectively.
2.2.2. Tau Protein Immobilization Procedure
By utilizing the SPR method, LTG and PHB interactions with tau protein were evaluated. In this research, the dual-flow channels SPR device (MP-SPR Navi 210A, BioNavis Ltd., Tampere-region, Finland) and a carboxymethyl dextran (CMD)-modified gold chip were applied. Before inserting the CMD chip into the SPR instrument, the apparatus was washed using a 10-2 M acetate buffer solution (pH 4.5). When the sensogram was achieved to a steady baseline, the chip surface was cleaned by injecting 0.1 M NaOH and 2 M NaCl at a flow rate of 30 μL.min-1. The Carboxymethyl dextran gold chip was activated after 7 min of injection with a flow rate of 30 μL.min-1 of a mixture of NHS (0.05 M) and EDC (0.2 M) solution (1: 1). One mg.mL-1 of the protein has been prepared in PBS buffer solution (pH = 7.0) for the tau protein immobilization on the CMD-sensor chip. Finally, ethanolamine-HCl solution (0.1 M, pH 8.5) inhibited inactive and non-specific groups on the chip surface.
2.2.3. Interactions of Lamotrigine and Phenobarbital with Immobilized Tau Protein
To investigate the interaction of LTG and PHB with tau protein, four different concentrations of LTG and PHB solutions (25, 50, 100, 150 μM) in PBS (pH 7.4) were injected for 3 min at the flow rate of 30 μL.min-1. To calculate the binding kinetic parameters and affinity of tau protein-LTG/PHB complexes, a Trace Drawer™ of SPR Navi™ was employed.
2.2.4. Molecular Docking Studies
The Auto dock 4.2 software was used to conduct additional research on the possible orientations and binding sites of LTG and PHB on tau protein. The 3D structure of tau protein (PDB ID: 5O3L) was obtained from Protein Data Bank (http://www.rcsb.org), and only chain A monomer of tau protein decamer was chosen for theoretical studies. In addition, utilizing the PubChem (https://pubchem.ncbi.nlm.nih.gov) chemical structures of LTG (DIB: 3878) and PHB (DIB: 4763) were obtained and then optimized using Gaussian 05 software package (17-19). Then, the only polar hydrogen atoms were added to the protein structure, the water molecules were removed, and the ligands root were detected. Furthermore, the Lamarckian genetic algorithm (LGA) has been utilized to figure out the best binding site of LTG and PHB on the tau protein. To find the possible binding site of LTG and PHB on the tau protein, firstly, a blind docking with the grid size of 126 × 126 × 126 and 0.431 Å grid spacing was done. Then, the subsequent docking studies were performed using the lowest docked conformation with a grid size of 90 × 90 × 90 and 0.375 Å grid spacing to determine exact binding sites. Finally, the output data analysis was done employing Discovery Studio Client 4.1 and UCSF Chimera software.
3. Results and Discussion
3.1. Tau Protein Fluorescence Quenching
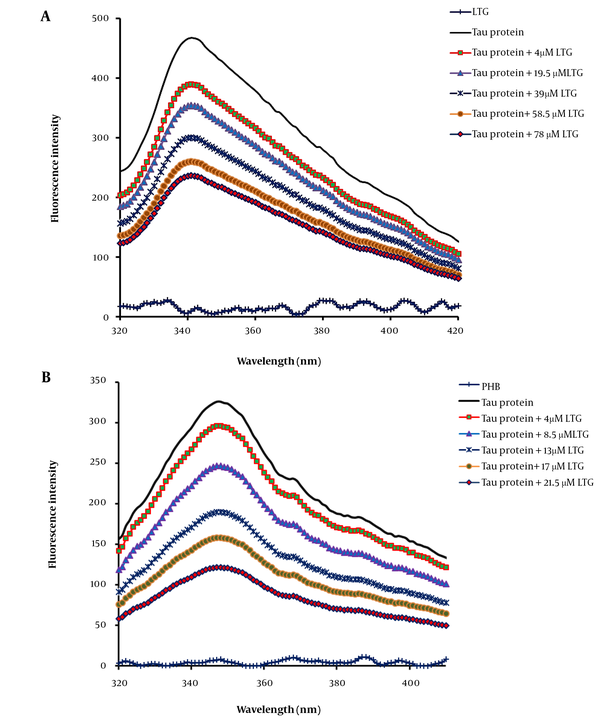
In general, the observed intrinsic fluorescence emission of a protein is related to the existence of three fluorophores, including tyrosine (Tyr), tryptophan (Trp), and phenylalanine (Phe) residues in the protein structure. Tau protein shows a strong intrinsic fluorescence peak mainly originating from the Tyr residues in its structure (20-22). In the present work, tau was titrated with additional concentrations of LTG and PHB, and the fluorescence intensities of the samples were recorded from 280 nm to 400 nm at three different temperatures (Figure 2A and B). The results indicated the significant fluorescence quenching of tau protein upon interaction with LTG and PHB, confirming tau protein-LTG/PHB complex formation (23). Tau protein in the free form showed a maximum fluorescence at 349 nm with an intensity of 523. Upon interaction with ligands, the emission intensity of the protein decreased (quenched). The interaction, complex formation, and structural changes of tau protein in the presence of ligands (LTG and PHB) were validated by reducing the fluorescence intensity of tau protein from 523 to 112.
Tau protein and A, lamotrigine (LTG)/B, phenobarbital (PHB) interactions as determined by intrinsic fluorescence emission intensity in the presence of additional concentrations of LTG (0, 4, 19.5, 39, 58.5, and 78 μM) and PHB (0, 4, 8.5, 13, 17 and 21.5 μM) in phosphate buffer (pH 7.4) at three different temperatures, 298, 310, and 316 K
Fluorescence quenching occurs via three different mechanisms, including dynamic quenching, static quenching, and combined static and dynamic quenching, which can be recognized based on their different responses to temperature. In static quenching type (the formation of the ground-state complex), increasing temperature causes the decrease of quenching rate constants (Ksv); however, the reverse effect can occur in dynamic quenching type (results from collisional encounters) (24, 25). To assess the mechanism of quenching of tau protein following the interaction with LTG and PHB (quenchers), the fluorescence spectra of free tau protein and LTG/PHB-tau protein were recorded at three distinct temperatures and further analyzed employing the Stern-Volmer equation (26) presented as:
where F0 is the fluorescence intensities without quenchers and F is the fluorescence intensities with quenchers. Kq shows the bimolecular quenching rate constant [Q], and KSV correspond to the concentration of LTG/PHB, and the Stern–Volmer dynamic quenching constant, respectively. In addition, τ0 demonstrates the average fluorophore lifetime in the excited state (equal to 10-8 s).
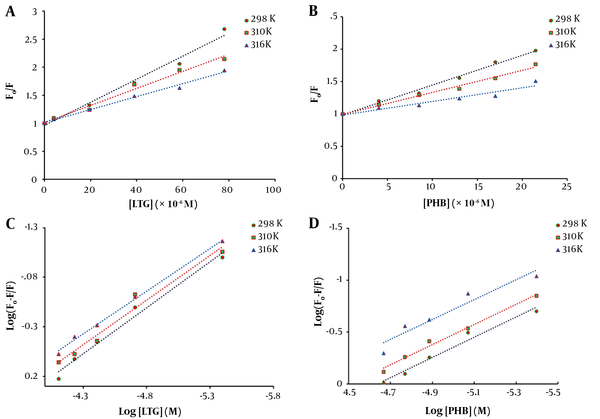
The related curves (F0/F vs. [Q]) were plotted for three different temperatures (Figure 3A and B), and the values of Ksv were calculated using the slopes of the plots and listed in Table 1. Good linearity of the Stern-Volmer plots was shown in the results, confirming the existence of only one type of quenching mechanism (in the case of combined static and dynamic quenching mechanism, the Stern-Volmer plot is non-linear). In addition, decreased Ksv values suggested that LTG and PHB quenched the emission intensity of tau protein via a static quenching mechanism.
Fluorescence quenching Stern-Volmer plot of tau protein with increasing concentrations of A, lamotrigine (LTG) and B, phenobarbital (PHB) at three different temperatures (298, 310, and 316 K) and various concentrations and double logarithmic curves for tau protein in the presence of different concentrations of C, LTG; and D, PHB at three various temperatures (298, 310, and 316 K)
The Stern-Volmer Quenching Constants (KSV) and the Bimolecular Quenching Constants (Kq) of the System of Lamotrigine/Phenobarbital-Tau Protein and Tau Protein at Three Different Temperatures
| Sample and Temperature (K) | Stern-Volmer Quenching Constants (× 104 M-1) | Bimolecular Quenching Constants (× 1012 M-1.s-1) | R2 |
|---|---|---|---|
| Lamotrigine-tau | |||
| 298 | 2.05 | 2.05 | 0.9844 |
| 310 | 1.52 | 1.52 | 0.9841 |
| 316 | 1.15 | 1.15 | 0.9926 |
| Phenobarbital-tau | |||
| 298 | 4.59 | 4.59 | 0.9866 |
| 310 | 3.42 | 3.42 | 0.9881 |
| 316 | 2.12 | 2.12 | 0.9301 |
3.2. Binding Constants and Binding Sites
The binding constant (Kb) is known as the different binding affinities of ligands binding to a protein. The following equation was used to obtain the binding parameters, including Kb and the number of binding sites (n):
The curves of log [(F0-F)/F] against log [Q] were plotted at 298, 310, and 316 K, and the values of Kb and n for LTG/PHB-tau protein complexes were calculated using the intercept and slope of the plots, respectively (Figure 3C and D). The obtained results are presented in Table 2. A decrease in Kb values with increasing temperature indicated the lower stability of LTG/PHB-tau protein complexes at higher temperatures. Also, the binding constants of LTG/PHB-tau protein complexes were in the order of 104, indicating the stronger binding affinity between LTG-tau protein and PHB-tau protein (27, 28). Furthermore, the values of n were very close to 1, showing that LTG and PHB only had one binding site on the tau protein.
Thermodynamic Parameters, Binding Constants (Kb), and the Stoichiometry of Binding (n) for Lamotrigine/Phenobarbital-Tau Protein at Temperatures Obtained from Fluorescence Spectroscopy
| System and Temperature (K) | Binding Constants (× 104 M-1) | Stoichiometry of Binding | ∆H (kJ.mol-1) | ∆S (kJ.K-1.mol-1) | ΔG (kJ.mol-1) | R2 |
|---|---|---|---|---|---|---|
| Lamotrigine-tau | -46.76 | -0.080 | ||||
| 298 | 0.938 | 0.92 | -22.92 | 0.9833 | ||
| 310 | 0.573 | 0.90 | -21.96 | 0.9711 | ||
| 316 | 0.300 | 0.86 | -21.48 | 0.9964 | ||
| Phenobarbital-tau | -36.70 | -0.035 | ||||
| 298 | 3.415 | 0.97 | -26.27 | 0.9735 | ||
| 310 | 2.349 | 0.96 | -25.85 | 0.988 | ||
| 316 | 1.191 | 0.95 | -25.64 | 0.9224 |
3.3. Thermodynamic Parameters Analysis
The type of intermolecular bonds can be determined using thermodynamic parameters (∆H and ∆S). The acting forces between a ligand and a macromolecule mainly include hydrogen, electrostatic bonds, van der Waals forces, and hydrophobic interactions. The van’t Hoff equation (Equation 4) has been employed to calculate the thermodynamic parameters at various temperatures (298 K, 310 K, and 316 K). In addition, ∆G values can be obtained by using Equation 5:
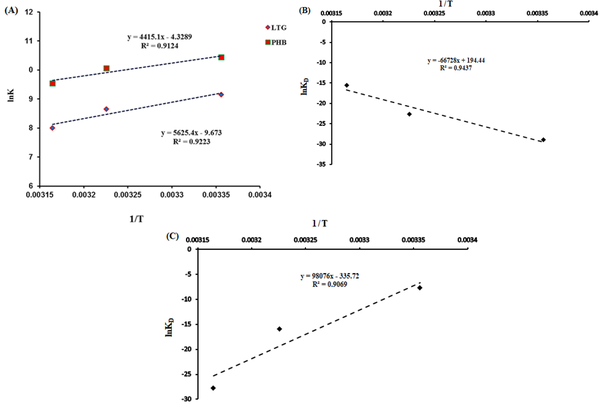
where ∆G, ∆S, and ∆H are the Gibbs free energy change, the entropy change, and enthalpy change, respectively. T and R stand for temperature and universal gas constant (R = 8.314 J.mol-1.K-1), respectively. The ∆H and ∆S values were calculated by using the slope and intercept of lnKD against 1/T (Figure 4A). Table 2 shows the obtained thermodynamic parameters for LTG-tau and PHB-tau complexes (based on fluorescence data analysis). In general, the type of the interaction force between the ligand and protein can be determined based on the sign and amount of thermodynamic parameters: ∆S < 0 and ∆H < 0 indicate that van der Waals forces and hydrogen bonds are the main forces. ∆H > 0 and ∆S > 0 show that the most important forces for complex formation are the hydrophobic interactions, and ∆H < 0 and ∆S > 0 suggest that electrostatic interactions are dominant (29). Based on the fluorescence data, both ∆H and ∆S are negative for LTG-tau and PHB-tau complexes. Hence, the main forces in LTG-tau and PHB-tau interactions are van der Waals forces and hydrogen bonds.
A, van’t Hoff plots for the interactions of phenobarbital (PHB) and lamotrigine (LTG) with tau protein obtained from fluorescence data analysis; B, the plots for PHB to tau protein; and C, LTG to tau protein obtained from surface plasmon resonance analyses
3.4. Surface Plasmon Resonance Parameter Analyses
Optical biosensors, which are SPR-based, are highly used to specify a wide variety of kinetic parameters in macromolecular interactions with ligands. This method has been made unique to a particular analyte by carefully constructing the sensing layer next to the plasmonic or metal layer without the need for any labeling processes, giving helpful information about the binding site of a ligand on a protein structure (30-32).
To assess the affinities of the ligand (LTG/PHB) and tau protein, SPR data analysis may easily get the association (ka) and dissociation (kd) rate constants as kinetic parameters (biomacromolecule). Figure 5 demonstrates the SPR signals related to the interaction of LTG with tau protein at 298, 310, and 316 K at different concentrations. Table 3 displays the protein kinetic results for LTG/PHB-tau. Association rate constant and kd show the number of complex molecules (tau-LTG/PHB) formed per sec and the fraction of decayed complexes in each sec, respectively (33), and the equilibrium constants (KD) value is measured by using the following equation:
Table 3 shows that the KD values for LTG-tau and PHB-tau protein complexes decreased and increased with increasing temperature, respectively. As shown in Table 3, the KD value of LTG was more than that of PHB. So, it can be concluded that LTG has a lower affinity toward tau protein (in comparison with PHB).
Sensorgram of lamotrigine-tau protein at different concentrations (25, 50, 100, and 150 μM) at three different temperatures 298 K, 310 K, 316 K
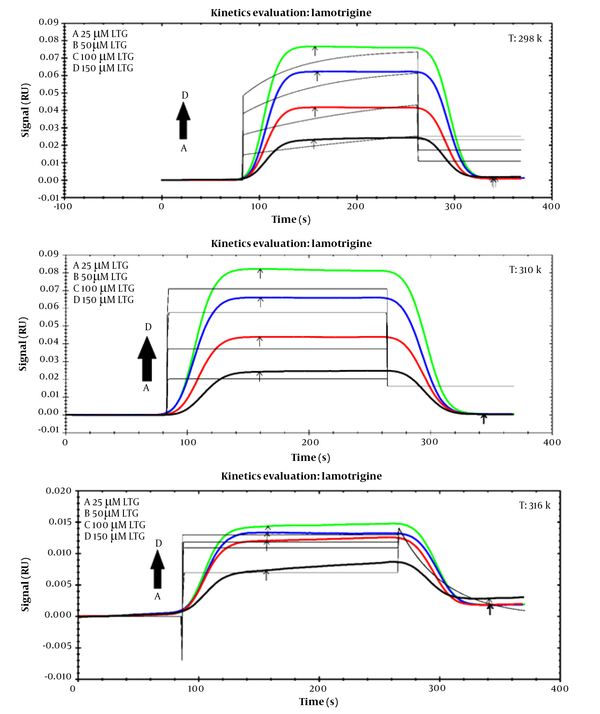
Association Rate Constants (ka), Dissociation Rate Constants (kd), Equilibrium Constants (KD), and Thermodynamic Parameters at Three Temperatures for Lamotrigine-Tau/Phenobarbital-Tau Calculated from Surface Plasmon Resonance Data Analyses
| System and Temperature (K) | Association Rate Constants (M-1.s-1) | Dissociation Rate Constants (s-1) | Equilibrium Constants (M) | ∆H (kJ.mol-1) | ∆S (kJ.K-1.mol-1) | ΔG (kJ.mol-1) |
|---|---|---|---|---|---|---|
| Lamotrigine-tau | 554.77 | 1.616 | ||||
| 298 | 1.20 × 102 | 3.37 × 10-11 | 2.80 × 10-13 | 73.208 | ||
| 310 | 1.44 × 104 | 2.18 × 10-6 | 1.51 × 10-10 | 53.816 | ||
| 316 | 1.41 × 105 | 2.61 × 10-2 | 1.85 × 10-7 | 44.12 | ||
| Phenobarbital-tau | -815.40 | -279.11 | ||||
| 298 | 7.91 × 101 | 3.52 × 10-2 | 4.45 × 10-4 | 82359 | ||
| 310 | 2.92 × 105 | 3.71 × 10-2 | 1.27 × 10-7 | 85708 | ||
| 316 | 3.25 × 106 | 3.06 × 10-6 | 9.42 × 10-13 | 87383 |
On the other hand, thermodynamic parameters were calculated by using the SPR data analysis, and the obtained results indicated that both ∆H and ∆S are positive for the LTG-tau complex, and both ∆H and ∆S are negative for the PHB-tau complex (Figure 4B and C and Table 3). Hence, the main forces for LTG-tau system are hydrophobic interactions, and those of PHB-tau system are van der Waals forces and hydrogen bonds.
3.5. Molecular Docking Study
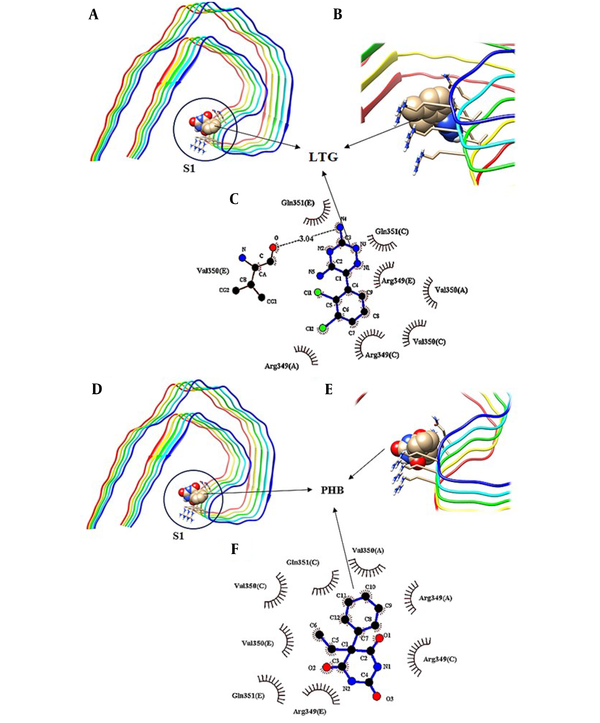
To further investigate the binding sites of LTG/PHB on tau protein, molecular docking investigations were carried out by employing the AutoDock 4.2 program. There are generally three different binding sites on tau protein for ligands, known as S1, S2, and S3 pockets (34). Figure 6 shows the molecular docking results of LTG-tau protein and PHB-tau protein complexes. According to these figures and in agreement with fluorescence data analysis, LTG and PHB can bind to tau protein at a single site. Also, different amino acid residues are involved in complex formation between LTG/PHB and tau protein. As shown in Figure 6A - C, LTG binds to the S1 pocket on tau protein by interacting with Val 350 (E) (via hydrogen bonds) and Val 350 (A), Val 350 (C), Arg 349 (A), Arg 349 (C), Arg 349 (E), Gln 351 (C) and Gln 351 (E) (via hydrophobic interaction). Also, Figure 6D - F show that PHB interacts with tau protein in the S1 pocket and amino acid residues, including Val 350 (A), Val 350 (C), Val 350 (E), Arg 349 (A), Arg 349 (C), Arg 349 (E), Gln 351 (C), and Gln 351 (E) (via hydrophobic interactions) which are important in the formation of the relevant complex. The obtained results in this study showed that LTG and PHB bind to the S1 pocket with a free binding energy of -4.06 kcal.mol-1 and -5 kcal.mol-1, respectively.
Molecular docking model of the binding of lamotrigine (LTG) to tau protein, A, the binding site; B and C, detailed illustration of LTG-tau protein complex, and also D, the binding mode of phenobarbital (PHB) with tau protein; E and F, detailed illustration of PHB-tau protein complex in S1 pockets.
4. Conclusions
In the current study, the interaction of LTG and PHB with tau protein has been studied using fluorescence, SPR, and molecular docking modeling for the first time. Fluorescence data analysis showed that LTG and PHB can quench the fluorescence intensity of tau protein via a static quenching mechanism. Furthermore, the main forces in the LTG/PHB-tau complex formation were hydrogen bonds and van der Waals forces, and the stability of the produced complexes decreased with increasing temperature. Analysis of SPR data at three different temperatures revealed that binding of LTG and PHB to tau protein can cause a decrease and increase in KD values with increasing temperature, respectively. Therefore, the affinity of LTG decreases, and PHB increases with increasing temperature. Molecular docking results (in agreement with experimental results) revealed that there is only one binding site on the S1 pocket of tau protein for both drugs (LTG and PHB). This study provides valuable information about the interactions of LTG/PHB and tau, which could simplify further investigations on the properties and pharmacology of LTG/PHB.
References
-
1.
Baas PW, Qiang L. Tau: It's Not What You Think. Trends Cell Biol. 2019;29(6):452-61. [PubMed ID: 30929793]. [PubMed Central ID: PMC6527491]. https://doi.org/10.1016/j.tcb.2019.02.007.
-
2.
Ahmadi S, Ebralidze II, She Z, Kraatz H. Electrochemical studies of tau protein-iron interactions—Potential implications for Alzheimer’s Disease. Electrochim Acta. 2017;236:384-93. https://doi.org/10.1016/j.electacta.2017.03.175.
-
3.
Brunello CA, Merezhko M, Uronen RL, Huttunen HJ. Mechanisms of secretion and spreading of pathological tau protein. Cell Mol Life Sci. 2020;77(9):1721-44. [PubMed ID: 31667556]. [PubMed Central ID: PMC7190606]. https://doi.org/10.1007/s00018-019-03349-1.
-
4.
Yu L, Wen G, Zhu S, Hu X, Huang C, Yang Y. Abnormal phosphorylation of tau protein and neuroinflammation induced by laparotomy in an animal model of postoperative delirium. Exp Brain Res. 2021;239(3):867-80. [PubMed ID: 33409674]. https://doi.org/10.1007/s00221-020-06007-2.
-
5.
Jouanne M, Rault S, Voisin-Chiret AS. Tau protein aggregation in Alzheimer's disease: An attractive target for the development of novel therapeutic agents. Eur J Med Chem. 2017;139:153-67. [PubMed ID: 28800454]. https://doi.org/10.1016/j.ejmech.2017.07.070.
-
6.
Zhang X, Gao F, Wang D, Li C, Fu Y, He W, et al. Tau Pathology in Parkinson's Disease. Front Neurol. 2018;9:809. [PubMed ID: 30333786]. [PubMed Central ID: PMC6176019]. https://doi.org/10.3389/fneur.2018.00809.
-
7.
Lee NC, Yang SY, Chieh JJ, Huang PT, Chang LM, Chiu YN, et al. Blood Beta-Amyloid and Tau in Down Syndrome: A Comparison with Alzheimer's Disease. Front Aging Neurosci. 2016;8:316. [PubMed ID: 28144219]. [PubMed Central ID: PMC5239773]. https://doi.org/10.3389/fnagi.2016.00316.
-
8.
Chaudhary N, Nagaraj R. Tau fibrillogenesis. Subcell Biochem. 2012;65:75-90. [PubMed ID: 23225000]. https://doi.org/10.1007/978-94-007-5416-4_4.
-
9.
Grinberg LT, Wang X, Wang C, Sohn PD, Theofilas P, Sidhu M, et al. Argyrophilic grain disease differs from other tauopathies by lacking tau acetylation. Acta Neuropathol. 2013;125(4):581-93. [PubMed ID: 23371364]. [PubMed Central ID: PMC3692283]. https://doi.org/10.1007/s00401-013-1080-2.
-
10.
Metrick MA, Ferreira NDC, Saijo E, Kraus A, Newell K, Zanusso G, et al. A single ultrasensitive assay for detection and discrimination of tau aggregates of Alzheimer and Pick diseases. Acta Neuropathol Commun. 2020;8(1):22. [PubMed ID: 32087764]. [PubMed Central ID: PMC7036215]. https://doi.org/10.1186/s40478-020-0887-z.
-
11.
Pardridge WM. Drug transport across the blood-brain barrier. J Cereb Blood Flow Metab. 2012;32(11):1959-72. [PubMed ID: 22929442]. [PubMed Central ID: PMC3494002]. https://doi.org/10.1038/jcbfm.2012.126.
-
12.
Nagpal K, Singh SK, Mishra DN. Drug targeting to brain: a systematic approach to study the factors, parameters and approaches for prediction of permeability of drugs across BBB. Expert Opin Drug Deliv. 2013;10(7):927-55. [PubMed ID: 23330786]. https://doi.org/10.1517/17425247.2013.762354.
-
13.
Poureshghi F, Ghandforoushan P, Safarnejad A, Soltani S. Interaction of an antiepileptic drug, lamotrigine with human serum albumin (HSA): Application of spectroscopic techniques and molecular modeling methods. J Photochem Photobiol B. 2017;166:187-92. [PubMed ID: 27951518]. https://doi.org/10.1016/j.jphotobiol.2016.09.046.
-
14.
de Almeida LCN, de Andrade Marques B, Silva RL, Hamoy AO, de Mello VJ, Borges RDS, et al. New nanocarried phenobarbital formulation: Maintains better control of pentylenetetrazole-Induced seizures. Biotechnol Rep (Amst). 2020;28. e00539. [PubMed ID: 33145190]. [PubMed Central ID: PMC7596104]. https://doi.org/10.1016/j.btre.2020.e00539.
-
15.
Asadi M, Dadfarnia S, Haji Shabani AM, Abbasi B. Simultaneous extraction and quantification of lamotrigine, phenobarbital, and phenytoin in human plasma and urine samples using solidified floating organic drop microextraction and high-performance liquid chromatography. J Sep Sci. 2015;38(14):2510-6. [PubMed ID: 25953277]. https://doi.org/10.1002/jssc.201500237.
-
16.
Romermann K, Helmer R, Loscher W. The antiepileptic drug lamotrigine is a substrate of mouse and human breast cancer resistance protein (ABCG2). Neuropharmacology. 2015;93:7-14. [PubMed ID: 25645391]. https://doi.org/10.1016/j.neuropharm.2015.01.015.
-
17.
Shariati B, Yektadoost E, Behzadi E, Azmoodeh E, Attar F, Sari S, et al. Interaction of silica nanoparticles with tau proteins and PC12 cells: Colloidal stability, thermodynamic, docking, and cellular studies. Int J Biol Macromol. 2018;118(Pt B):1963-73. [PubMed ID: 30009913]. https://doi.org/10.1016/j.ijbiomac.2018.07.041.
-
18.
Pagadala NS, Syed K, Tuszynski J. Software for molecular docking: a review. Biophys Rev. 2017;9(2):91-102. [PubMed ID: 28510083]. [PubMed Central ID: PMC5425816]. https://doi.org/10.1007/s12551-016-0247-1.
-
19.
Nasiri F, Dehghan G, Shaghaghi M, Datmalchi S, Iranshahi M. Probing the interaction between 7-geranyloxycoumarin and bovine serum albumin: Spectroscopic analyzing and molecular docking study. Spectrochim Acta A Mol Biomol Spectrosc. 2021;254:119664. [PubMed ID: 33743310]. https://doi.org/10.1016/j.saa.2021.119664.
-
20.
Rashtbari S, Khataee S, Iranshahi M, Moosavi-Movahedi AA, Hosseinzadeh G, Dehghan G. Experimental investigation and molecular dynamics simulation of the binding of ellagic acid to bovine liver catalase: Activation study and interaction mechanism. Int J Biol Macromol. 2020;143:850-61. [PubMed ID: 31739034]. https://doi.org/10.1016/j.ijbiomac.2019.09.146.
-
21.
Rashtbari S, Dehghan G, Yekta R, Jouyban A. Investigation of the binding mechanism and inhibition of bovine liver catalase by quercetin: Multi-spectroscopic and computational study. Bioimpacts. 2017;7(3):147-53. [PubMed ID: 29159142]. [PubMed Central ID: PMC5684506]. https://doi.org/10.15171/bi.2017.18.
-
22.
Shaghaghi M, Dehghan G, Rashtbari S, Sheibani N, Aghamohammadi A. Multispectral and computational probing of the interactions between sitagliptin and serum albumin. Spectrochim Acta A Mol Biomol Spectrosc. 2019;223:117286. [PubMed ID: 31302563]. https://doi.org/10.1016/j.saa.2019.117286.
-
23.
Zeinabad HA, Zarrabian A, Saboury AA, Alizadeh AM, Falahati M. Erratum: Interaction of single and multi wall carbon nanotubes with the biological systems: tau protein and PC12 cells as targets. Sci Rep. 2016;6:29644. [PubMed ID: 27650105]. [PubMed Central ID: PMC5030518]. https://doi.org/10.1038/srep29644.
-
24.
Rashtbari S, Dehghan G, Yekta R, Jouyban A, Iranshahi M. Effects of Resveratrol on the Structure and Catalytic Function of Bovine Liver catalase (BLC): Spectroscopic and Theoretical Studies. Adv Pharm Bull. 2017;7(3):349-57. [PubMed ID: 29071216]. [PubMed Central ID: PMC5651055]. https://doi.org/10.15171/apb.2017.042.
-
25.
Khataee S, Dehghan G, Rashtbari S, Yekta R, Sheibani N. Synergistic inhibition of catalase activity by food colorants sunset yellow and curcumin: An experimental and MLSD simulation approach. Chem Biol Interact. 2019;311:108746. [PubMed ID: 31301288]. https://doi.org/10.1016/j.cbi.2019.108746.
-
26.
Shaghaghi M, Rashtbari S, Vejdani S, Dehghan G, Jouyban A, Yekta R. Exploring the interactions of a Tb(III)-quercetin complex with serum albumins (HSA and BSA): spectroscopic and molecular docking studies. Luminescence. 2020;35(4):512-24. [PubMed ID: 31883206]. https://doi.org/10.1002/bio.3757.
-
27.
Sood D, Kumar N, Rathee G, Singh A, Tomar V, Chandra R. Mechanistic Interaction Study of Bromo-Noscapine with Bovine Serum Albumin employing Spectroscopic and Chemoinformatics Approaches. Sci Rep. 2018;8(1):16964. [PubMed ID: 30446713]. [PubMed Central ID: PMC6240080]. https://doi.org/10.1038/s41598-018-35384-6.
-
28.
Pawar SK, Jaldappagari S. Interaction of repaglinide with bovine serum albumin: Spectroscopic and molecular docking approaches. J Pharm Anal. 2019;9(4):274-83. [PubMed ID: 31452966]. [PubMed Central ID: PMC6702422]. https://doi.org/10.1016/j.jpha.2019.03.007.
-
29.
Sekowski S, Olchowik-Grabarek E, Wieckowska W, Veiko A, Oldak L, Gorodkiewicz E, et al. Spectroscopic, Zeta-potential and Surface Plasmon Resonance analysis of interaction between potential anti-HIV tannins with different flexibility and human serum albumin. Colloids Surf B Biointerfaces. 2020;194:111175. [PubMed ID: 32544793]. https://doi.org/10.1016/j.colsurfb.2020.111175.
-
30.
Pattnaik P. Surface Plasmon Resonance: Applications in Understanding Receptor–Ligand Interaction. Appl Biochem Biotechnol. 2005;126(2):79-92. https://doi.org/10.1385/abab:126:2:079.
-
31.
Pham M, Hussain A, Bui D, Nguyen TT, You S, Wang Y. Surface plasmon resonance enhanced photocatalysis of Ag nanoparticles-decorated Bi2S3 nanorods for NO degradation. Environ Technol Innov. 2021;23. https://doi.org/10.1016/j.eti.2021.101755.
-
32.
Yekta R, Sadeghi L, Dehghan G. The inefficacy of donepezil on glycated-AChE inhibition: Binding affinity, complex stability and mechanism. Int J Biol Macromol. 2020;160:35-46. [PubMed ID: 32454110]. https://doi.org/10.1016/j.ijbiomac.2020.05.177.
-
33.
Maleki S, Dehghan G, Sadeghi L, Rashtbari S, Iranshahi M, Sheibani N. Surface plasmon resonance, fluorescence, and molecular docking studies of bovine serum albumin interactions with natural coumarin diversin. Spectrochim Acta A Mol Biomol Spectrosc. 2020;230:118063. [PubMed ID: 32000060]. https://doi.org/10.1016/j.saa.2020.118063.
-
34.
Abdelaziz G, Shamsel-Din HA, Sarhan MO, Gizawy MA. "Tau Protein Targeting Via Radioiodinated Azure A For Brain Theranostics: Radiolabeling, Molecular Docking, in vitro And in vivo Biological Evaluation". J Labelled Comp Radiopharm. 2020;63(1):33-42. [PubMed ID: 31785209]. https://doi.org/10.1002/jlcr.3819.
