Published online Nov 14, 2019. doi: 10.5662/wjm.v9.i3.32
Peer-review started: August 5, 2019
First decision: August 20, 2019
Revised: August 26, 2019
Accepted: October 15, 2019
Article in press: October 15, 2019
Published online: November 14, 2019
Histopathologically stained archived tissue slides are stored in hospital archives for years to decades. They are the largest available source of biological materials and are a potentially useful resource that can be used for retrospective epidemiological studies. DNA recovered from the slides can be used for several downstream molecular processes including polymerase chain reaction, single nucleotide polymorphism analysis, and whole genome sequencing. The DNA from these slides can be utilized to compare gene signatures of normal and diseased tissues. However, extraction of high-quality DNA from archived stained hematoxylin and eosin (H&E) slides remains challenging.
To standardize a new protocol for extracting DNA from archived H&E-stained tissue slides for further molecular assays.
A total of 100 archived H&E-stained cancer slides were subjected to a total of five methods of DNA extraction. Methods were varied in the deparaffinization step, tissue rehydration, duration of lysis, and presence or absence of proteinase K. The extracted DNA was quantified using a NanoDrop spectrophometer and the quality was analyzed by agarose gel electrophoresis. Then each sample was subjected to polymerase chain reaction (PCR) to amplify the internal control gene GAPDH, thereby confirming the DNA intactness, which could be further utilized for other downstream applications.
Of the five different methods tested, the third method wherein xylene was used for tissue deparaffinization followed by 72 h of digestion and without proteinase K inactivation yielded the highest amount of DNA with good purity. The yield was significantly higher when compared to other methods. In addition, 90% of the extracted DNA showed amplifiable GAPDH gene.
Here we present a step-by-step, cost-effective, and reproducible protocol for the extraction of PCR-friendly DNA from archived H&E-stained cancer tissue slides that can be used for further downstream molecular applications.
Core tip: In our study, we discussed a step-by-step procedure and results for the extraction of PCR-friendly DNA from archived hematoxylin and eosin-stained tissue slides. Such extracted DNA has the potential to be used for further molecular analyses such as mutation studies, whole genome sequencing, and even the identification of differences in gene signatures between diseased and normal states. Our protocol is simple, cost-effective, and can be performed in a basic molecular biology lab with most common reagents.
- Citation: Ramesh PS, Madegowda V, Kumar S, Narasimha S, S R P, Manoli NN, Devegowda D. DNA extraction from archived hematoxylin and eosin-stained tissue slides for downstream molecular analysis. World J Methodol 2019; 9(3): 32-43
- URL: https://www.wjgnet.com/2222-0682/full/v9/i3/32.htm
- DOI: https://dx.doi.org/10.5662/wjm.v9.i3.32
Molecular diagnostics involves the analysis of DNA, RNA, and proteins or metabolites at the molecular level in order to detect genotypes, mutations, or biochemical changes in the body. The main objective is to test for specific states of health or to see if the disease exists. Analysis of DNA reveals a multitude of cellular processes and hence it is essential to look for alternate sources of DNA. Blood and saliva are among the most common sources of DNA[1], but they are not always available. Extraction of DNA from teeth, bone, and hair follicles has been followed routinely[2,3]. Even with all of these plethora of specimens, it warrants testing for new specimens that can be harnessed to obtain DNA. One such potential resource is archived hematoxylin and eosin (H&E)-stained slides.
Histopathologically stained archived tissue slides are stored in hospital archives for years to decades. They are the largest available source of biological materials [other than formalin fixed paraffin embedded (FFPE) tissue blocks] and are potentially useful resource that can be used for retrospective epidemiological studies. The increasing interest in the genetic basis of diseases has increased the value of available clinical samples, including archived tissue slides. Similar to the DNA recovered from FFPE blocks, DNA from slides can also be used for several downstream molecular processes such as polymerase chain reaction (PCR), real time PCR, single nucleotide polymorphism (SNP), and whole genome sequencing[4]. This can aid researchers in unlocking new genetic information that can answer questions related to the molecular basis of cancer and other diseases and also gaining an understanding of the response to specific therapeutic modalities. H&E-stained slides can also be used to acquire information such as the presence or absence of pathogens that would have been present at the time of sectioning. The high-quality DNA from these slides can be used to compare gene signatures of normal and diseased tissues and also to detect mutations. Many studies have tried to optimize the extraction process, but a standard, reproducible, cost-effective protocol does not exist.
Even though molecular biology tools have seen a major evolutionary upgrade in recent years, extraction of DNA from archived tissue slides is problematic because formalin fixation induces protein-nucleic acid crosslinking. As a result of this cross linkage, it is difficult to separate DNA from histones and to obtain pure nucleic acids. These crosslinks are irreversible and not easily undone[5]. The incomplete reversal of crosslinks causes a negative impact on the quantity and quality of the recovered DNA. The fixation of tissues also leads to fragmentation of the nucleic acids that will affect the yield.
Removal of coverslip and deparaffinization is the biggest hurdle in the entire process. To extract DNA from the H&E-stained slide, first the coverslip must be removed and the tissue must be separated from the paraffin and then the tissue must be subjected to rehydration to allow for effective digestion[6]. Deparaffinization is usually performed using solvents such as xylene to dissolve the paraffin. However, researchers have found that using xylene can cause fragmentation of DNA and thus the loss of valuable specimens. A recent study suggested the use of mineral oil to melt away the paraffin[7]. After removal of the coverslip, it is essential to trim off the excess wax surrounding the tissue. However, some of the specimens are brittle due to age or poor storage conditions. This makes paraffin removal a little more difficult and can cause lower yields[8].
Molecular studies are highly dependent on the quality and quantity of the extracted nucleic acid, and hence it is quintessential to develop an extraction procedure that is suitable for downstream molecular processes. The goal of this study was to conduct a proof-of-principle study, as a new protocol for DNA extraction from H&E cancer slides based on the existing phenol chloroform isoamyl alcohol (PCI) method, which could be further used in molecular analysis.
A successful DNA isolation requires four essential steps: Effective disruption of cells or tissue; denaturation of nucleoprotein complexes; inactivation of nucleases; and maintaining the quality and integrity of the isolated DNA, which will directly affect the results of successful scientific research[9].
The three successfully employed DNA extraction techniques are: A salting out method using NaCl, a column-based method which the majority of commercially available kits use, and the PCI method. The PCI extraction method is by far the most commonly used DNA extraction method in the literature. Although newer procedures have been developed in recent years, the PCI method continues to be used routinely, possibly because it offers an economical advantage over commercially available kits[10]. For this reason, the PCI method was employed in our study but with a few modifications from the traditional one.
A total of 100 histopathologically confirmed H&E-stained slides were handpicked by a pathologist from JSS Medical College Hospital (Mysore, India). The age of the slides ranged from 1 to 2 years old. All slides were prepared from FFPE tissues that had been fixed in 10% buffered neutral formalin processed and embedded manually. The slides were carefully examined by a histopathologist and were confirmed cancer cases.
Xylene (Product No. 35415; Thermo Fisher Scientific, Waltham, MA, United States); Scalpel/razor blade; ethanol (Cat No. 58051; Jebsen & Jessen GmbH & Co., Hamburg, Germany); Tris HCl (Cat No. 34969; Sisco Research Laboratories Pvt. Ltd., New Delhi, India); EDTA dipotassium salt extrapure (Cat No. 62196; Sisco Research Laboratories Pvt. Ltd.); NaCl (Cat No. 15915; Qualigens Fine Chemicals, Mumbai, India); sodium lauryl sulphate (Cat No. 32096; Sisco Research Laboratories Pvt. Ltd.); proteinase K (Cat No. RM2957; HiMedia Laboratories, LLC, Mumbai, India); phenol molecular biology grade (Cat No. 17286; Sisco Research Laboratories Pvt. Ltd.); chloroform molecular biology grade (Cat No. 96764; Sisco Research Laboratories Pvt. Ltd.); isoamyl alcohol extrapure (Cat No. 69931; Sisco Research Laboratories Pvt. Ltd.); sodium acetate anhydrous extrapure (Cat No. 40104 K05; SDFCL, Bengaluru, India); nuclease-free water (Cat No. ML024; HiMedia Laboratories); mineral oil molecular biology grade (Cat No. MB161; HiMedia Laboratories); agarose low EEO (Cat No. MB002; HiMedia Laboratories); Taq polymerase (Cat No. MBT060A; HiMedia Laboratories); deoxynucleotriphosphates (Cat No. MBT078; HiMedia Laboratories); PCR tubes (Cat No. AB0620; Abgene, Portsmouth, NH, United States); microcentrifuge tubes (Cat No. 509-GRD-Q; QSP by Thermo Fisher Scientific).
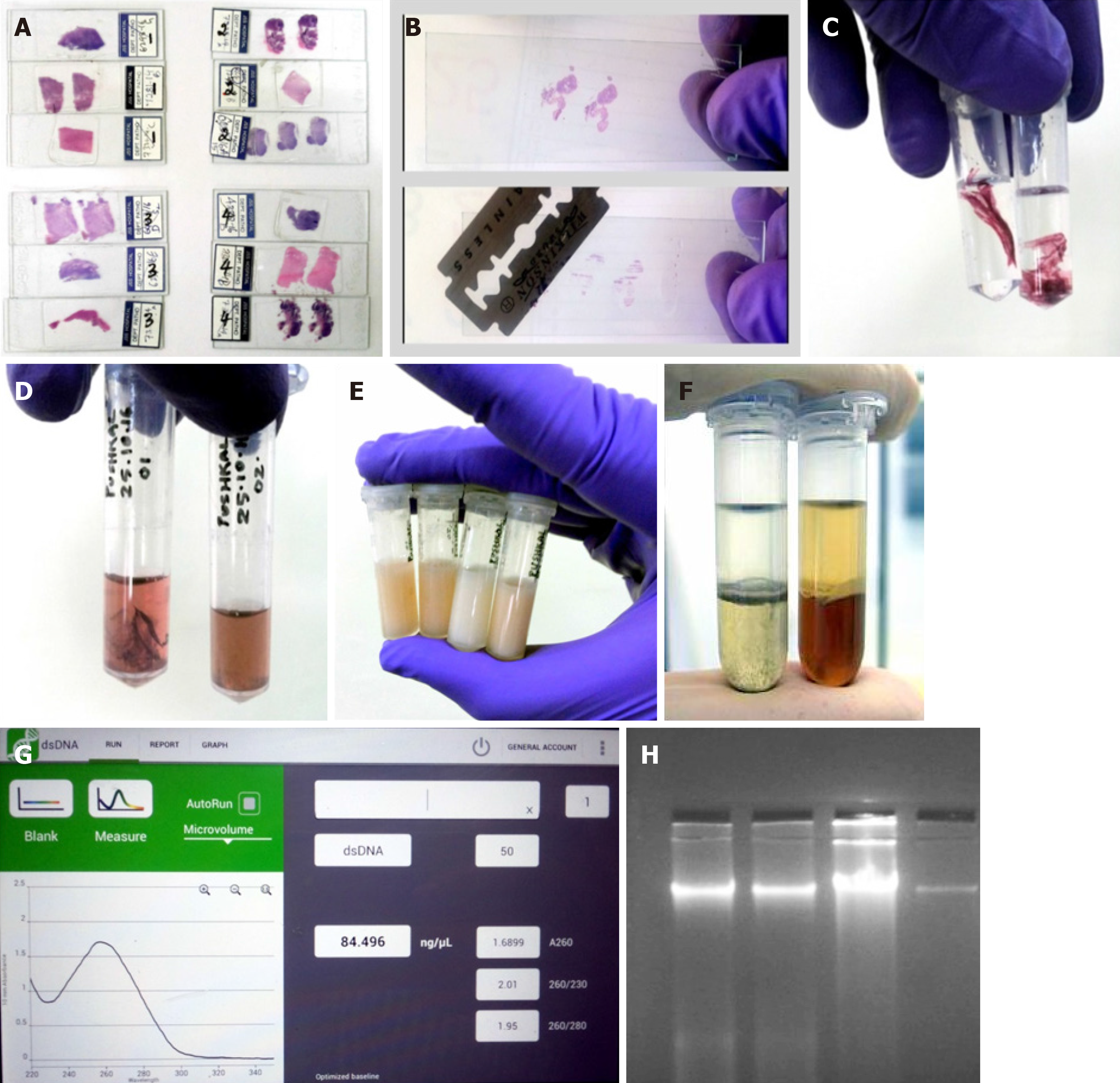
Removal of coverslip is one of the biggest hurdles in the entire process. Generally for any further analysis on the slides, the coverslip has to be removed by immersing the slide in xylene for 4-6 d or until the coverslip falls off spontaneously. Recently, a study by Zhou et al[11] showed that coverslips could be rapidly removed by freezing in liquid nitrogen. However, not all laboratories have access to liquid nitrogen and it possesses a threat to the handler. So we came up with a less harmful idea, namely of using a -80°C freezer and a sterile razor blade. Slides used for the study were photographed and marked using a slide marker. The slides were placed inside a -80°C freezer for 2 h, after which a sterile sharp razor blade was slid under the edges of the coverslip to remove it (Note: Before placing the slides inside the freezer, we made sure the DPX merging the borders of the coverslip was carefully cut out). Sometimes, the coverslip tended to adhere more strongly to the slide; in such cases, the slide was immersed in xylene for 20 min and the coverslip fell off automatically or a spatula could be used to gently push it off. In another method, we also used the liquid nitrogen procedure described by Zhou et al[11], but the slides froze too much making it prone to breakage. Also, we experienced little hardship in using liquid nitrogen as we had a large tank that was not easily accessible to this procedure.
Once the coverslip was removed, the next step was to scrape out the tissue from the slide. It was important to remove the paraffin surrounding the tissue by cutting it out using a sharp sterile blade. Once the tissue was clear of the any residual paraffin, the tissue sections were scraped off by gently, pushing the blade against the slide and moving it from end to end. The scraped tissue was next transferred into a capped tube for further processing (Note: The size of the scraped tissues was not determined as the size varied greatly with each sample. All slides were photographed before processing for future reference).
The H&E-stained slides were prepared from FFPE tissue blocks. So when the sections were taken for the slides, the paraffin residues surrounding the tissue were also fixed to the slide. This paraffin can create problem with the DNA quality; hence it was essential to remove any residual paraffin or even the fixative used. After the tissues from the slides were scraped off, they were transferred into a capped eppendorf tube (2 mL) containing 800 µL 100% pre-warmed xylene (Thermo Fisher Scientific). The tubes were vortexed briefly for 20 s and centrifuged at maximum speed (13500 g) for 5 min. This step ensured that the residual fixative accumulated as supernatant that could be pipetted (Note: To confirm the complete removal of any residual material, the pellet was gently touched with a micropipette tip. If the tissue was free of residue, it would feel soft; otherwise, the xylene wash step was repeated again).
Treatment with solvents such as xylene eventually dehydrates the tissues. So the next step we followed was rehydration of the tissues using a series of different concentrations of ethanol. Ethanol wash in decreasing concentration ensures that the tissue is rehydrated slowly and also removes the stains added during tissue fixation on the slide. A study by Morikawa et al[12] showed that H&E staining does not interfere with DNA testing, so we did not perform any further steps to remove the stain.
The xylene-treated tissues were washed with 800 µL molecular grade ethanol of 100%, 75%, and 50% sequentially. In each step after addition of ethanol, the tubes were vortexed briefly and centrifuged for about 5 min at maximum speed (13500 g). At the end of centrifugation, the supernatant was completely removed. This step was employed as per Pikor et al[13] (Note: After each wash, the supernatant had a slight color. The color intensity decreased with each wash indicating removal of the stain). After the last ethanol wash, the pellet was air-dried for about 5 min. This step is essential as any ethanol residue can interfere with the composition of the lysis buffer.
Since our study evaluated new ways for DNA extraction, we looked into mineral oil as a potential candidate to carry out the deparaffinization. Even though the yield was good by this method, the purity was not up to the mark. Briefly, after coverslip removal, 250-500 µL molecular grade mineral oil was placed on the tissue surface on the slide. Then the slide was placed on a thermoblock heater at 90°C for 20 min. After the prescribed duration of incubation, mineral oil was removed and the deparaffinized tissues were rehydrated in different grades of ethanol.
To examine whether this step of ethanol rehydration was necessary, we tested some slides without any ethanol wash and directly proceeded to lysis after xylene treatment. But this modification did not prove fruitful as a very low yield of DNA was obtained that was not at all amplifiable by PCR.
Tissue pellets obtained from the previous step, i.e. with or without ethanol rehydration, was digested with 500 µL lysis buffer (pH 7.8) and 5 µL proteinase K (200 µg/mL) incubated at 56°C in a shaker incubator for 6 h. The composition of the lysis buffer was 0.5 mol/L Tris HCl, 10 mmol/L EDTA, 100 mmol/L NaCl and 2% SDS. The tubes were agitated continuously at 25 g to ensure proper digestion. The following effects were examined by modifying the protocol as per our needs: effect of incubation time and effect of inactivating proteinase K.
To examine the effects of incubation time, the tubes with tissue pellet, lysis buffer, and proteinase K were incubated for different durations of time. The digestion was carried out for 6h, 12 h, overnight (24 h), 48 h, and 72 h. Tissues of similar sizes were used and the tubes were agitated all along and the temperature was maintained at 56°C (optimum temperature for proteinase K).
The function of proteinase K is to digest proteins and any harmful enzymes such as nucleases. But proteinase K itself being an enzyme may or may not hamper the quality and quantity of the extracted DNA. There is no evidence of proteinase K harming the quality or the quantity of the DNA. So in order to test this, we inactivated proteinase K after the complete incubation period in some of our samples. After the respective incubation period, the tubes were kept at 90°C in a block heater for about 20 min.
We employed the most common, cost-effective method of PCI for DNA purification. Using buffer saturated PCI mixture (24:24:1) and high-speed centrifugation (18500 g for 5 min) a biphasic phase was generated. DNA and RNA remained in the upper aqueous phase whereas proteins, lipids, and polysaccharides were sequestered in the interphase and organic phase. The aqueous phase was collected and subjected to further purification using only the chloroform-isoamyl alcohol mixture. This ensured removal of any phenol residues, which would affect the DNA integrity. After the high-speed centrifugation, the aqueous layer was collected again and was subjected to precipitation.
The aqueous layer obtained from the second phase separation was further subjected to precipitation. Ethanol precipitation is the most commonly used technique for concentrating and de-salting nucleic acid preparations in aqueous solution[14]. The basis of this technique is adding a salt and ethanol to the aqueous solution forces the precipitation of nucleic acids out of the solution. After precipitation, the DNA can be separated from the rest of the solution by centrifugation. The role of the salt is to neutralize the charges on the sugar phosphate backbone. A commonly used salt is sodium acetate. So in this protocol we used a 1/10 volume of sodium acetate-ethanol mixture to precipitate the DNA. After the addition of sodium acetate and ethanol mixture, the tubes were placed in a -20°C freezer for 30 min as the reduced temperature will aid in faster precipitation. The precipitated DNA was recovered by centrifugation at 13500 g for 10 min. The ethanol in the supernatant was discarded and the pellet was washed with 70% ethanol. Washing with ethanol made sure that none of the salt contaminants thrived. Again the tubes were subjected to ice-cold centrifuging to pellet the DNA. Ethanol was removed without disturbing the pellet and the tubes were air-dried. The removal of ethanol completely was essential as any leftovers could inhibit further PCR application. Once the tubes were completely free of ethanol, the pellets were re-suspended in nuclease-free water. The amount of nuclease free water added was directly proportional to the size of the pellet and ranged from 30 to 80 µL.
During several different stages of a molecular biology experiment, it is important to obtain a quick and accurate reading of DNA concentration and yield. The quantity and quality of genomic DNA isolated from these methods were compared using a NanoDrop spectrophotometer (DeNovix Inc., Wilmington, DE, United States). The purity of the extracted DNA was assessed as the ratio of absorbance at 260 nm and 280 nm (A260/280). The workflow of extraction of DNA from archived H&E-stained cancer tissue slides is summarized below (Figure 1).
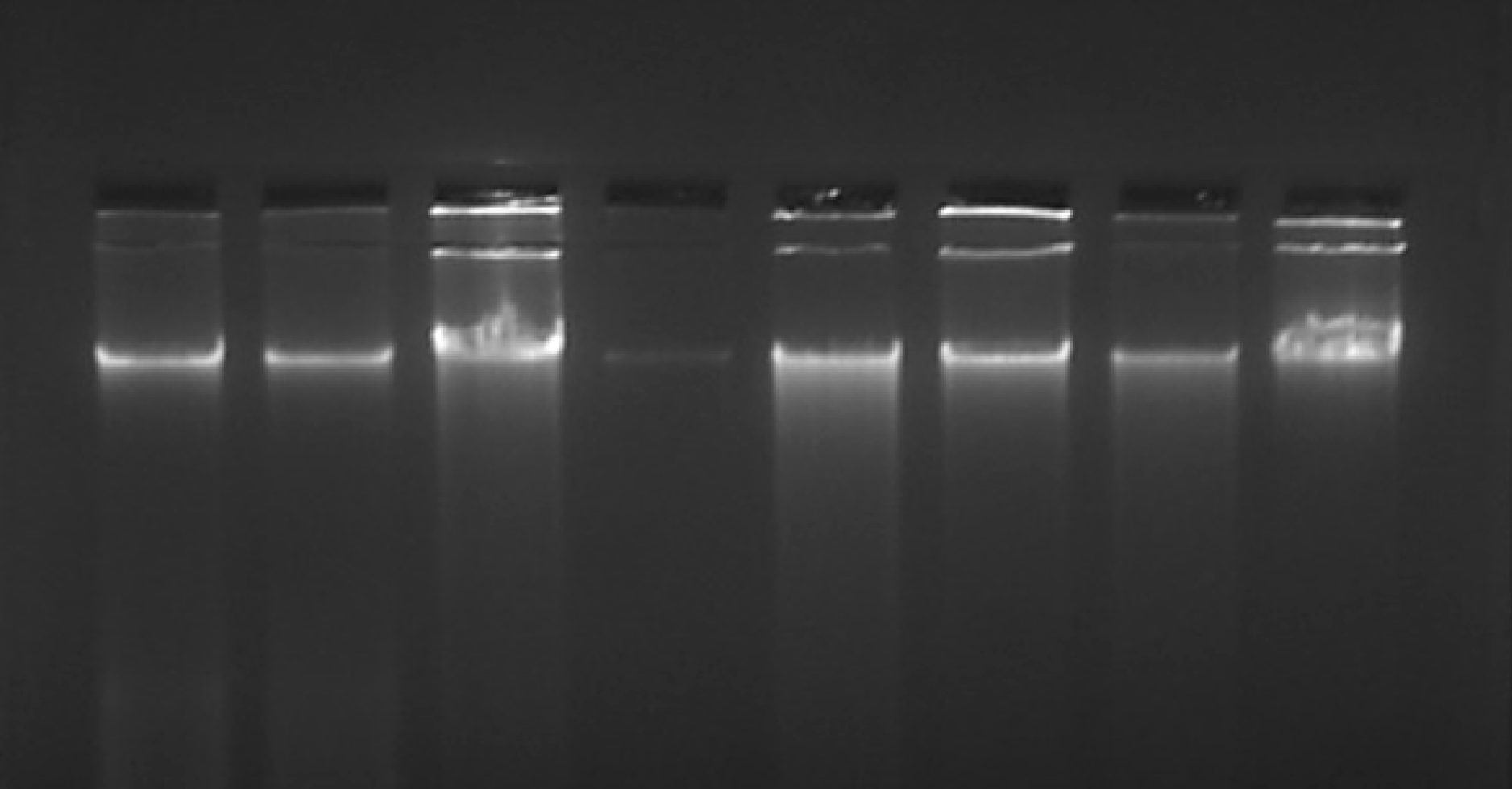
The quality of the extracted DNA was assessed by running gel electrophoresis on a 1% agarose stained with ethidium bromide. Observation of a non-streaky band confirmed the presence of intact DNA.
It was essential to make sure that the extracted nucleic acid was intact before processing for downstream molecular assays. One of the goals of this study was to standardize a protocol to extract PCR-friendly DNA from the archived H&E-stained slides. So we subjected the recovered DNA for the amplification of housekeeping gene such as GAPDH. All of the DNA samples extracted from the slides were subjected to PCR, and only the ones confirmed to be intact were considered for the results.
The PCR was set up using commercially synthesized oligonucleotides for the GAPDH gene. A brief master mix was prepared containing 2.5 U Taq polymerase (Himedia), 2 µmol/L dNTPs, 10× buffer with 25 mmol/L MgCl2 (working = 1× buffer), and 0.4 µmol/L of each of forward and reverse primers. DNA concentration ranging from 50 to 100 ng/reaction was added and the total reaction was made up to 30 µL using PCR-grade water. Amplification was performed in the automated Thermal cycler (Mastercycler gradient, eppendorf) at 95°C for 5 min, followed by 32 cycles at 94°C for 1 min, 58°C for 45 s, 72°C for 1 min and a final extension at 72°C for 2 min. DNA from a known cell line was used as a positive control and to check the adequacy of the program. PCR-grade water was used as a negative control. The primer sequences used were as follows: Forward 5’- GAA ATC CCA TCA CCA TCT TCC AGG-3’; reverse 5’- GAGCCCCAGCCTTCTCCATG-3’. The amplified PCR products were run on a 2% agarose gel stained with ethidium bromide and the gel image was captured on a gel documentation system.
All data were put together in GraphPad Prism 5.0 and the Student’s t-test was performed to assess significance among various methods of DNA extraction.
Our overall strategy was to establish a new protocol to extract DNA from the archived H&E-stained slides. We compared the DNA quantity and purity (using NanoDrop in ng/μL) and quality (using agarose gel electrophoresis) that resulted from different methods of extraction from the slides having H&E-stained tissues. The results of the present study revealed that DNA from H&E slide tissues varied depending on the extraction method but were not degraded (Figure 2). Since our initial standardization showed that only longer duration of digestion improved the DNA yield, we used only a 24 and 72 h digestion protocol and then continued with the PCI extraction method throughout our experiment. Also, we observed that inactivation of proteinase K after digestion did not have much effect on the yield; hence we continued with the PCI extraction method without proteinase K inactivation.
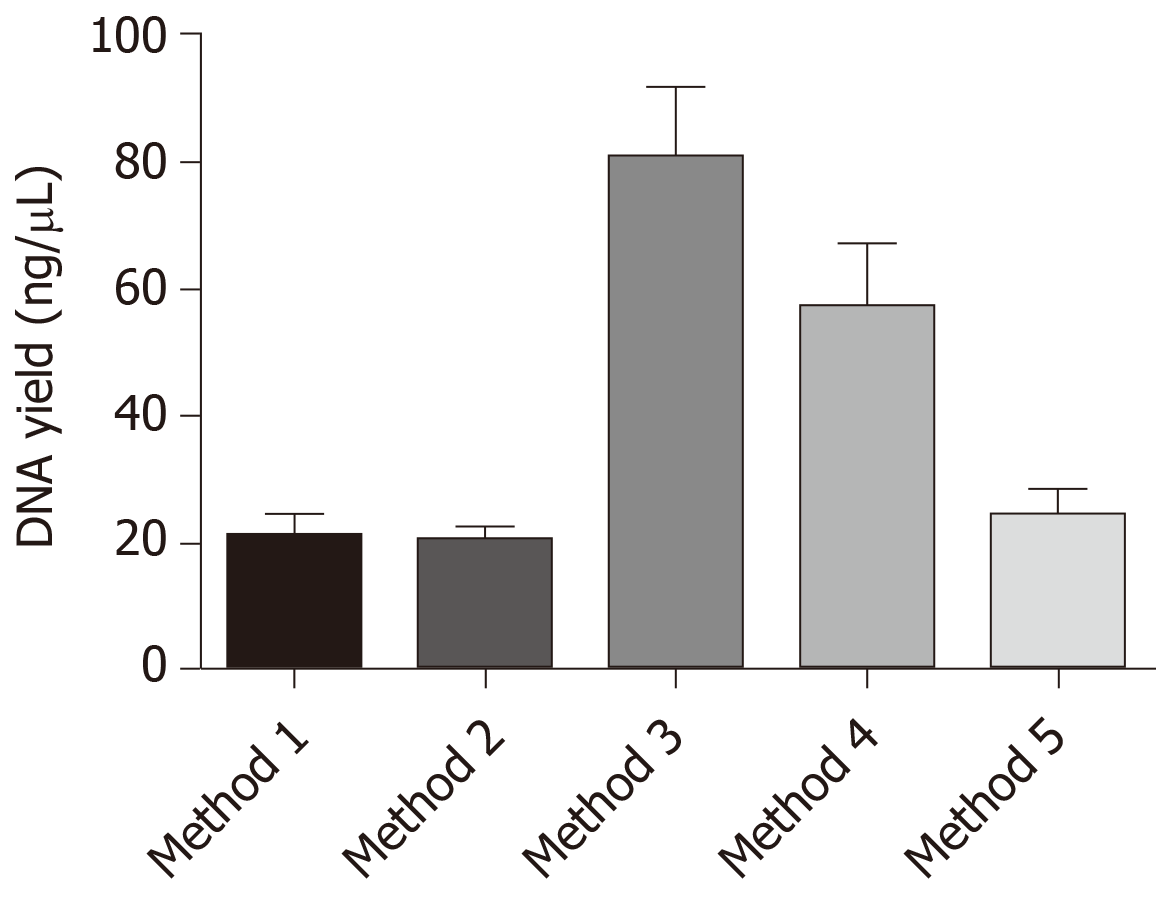
Our data indicated that method 3 with xylene deparaffinization and 72 h digestion showed the highest yield of DNA and was also the best in terms of purity. The samples that were deparaffinized using pre-warmed xylene (about 50°C) produced greater DNA yields than concentrations of nucleic acids produced from the samples that were deparaffinized with mineral oil (molecular grade). The samples deparaffinized with mineral oil also yielded good amount of amplifiable DNA but the purity of the recovered DNA was not up to the mark. It also appears that digestion with proteinase K for duration longer than overnight improved the efficiency of DNA extraction from H&E-stained cancer tissues, and when the tissues were large, overnight digestion seemed to provide successful results (Table 1 and Figure 3).
| Method | Sample size, n | Tissue deparaffinization | Digestion period | Proteinase K inactivation | DNA yield range in ng/µL | DNA purity range, A260/280 |
| 1 | 20 | Xylene | 24 h | No | 7.65-45.23 | 1.42-2.01 |
| 2 | 20 | Xylene | 24 h | Yes | 7.53-33.7 | 1.34-1.94 |
| 3 | 20 | Xylene | 72 h | No | 23.68-208.31 | 1.68-1.89 |
| 4 | 20 | Xylene | 72 h | Yes | 13-149.41 | 1.54-1.93 |
| 5 | 20 | Mineral oil | 72 h | No | 11.74-86.74 | 1.41-1.74 |
The highest mean of DNA recovered and the best purity was observed in methods 3 and 4. So we performed the unpaired t-test to analyze if there was any significance between the results obtained. When we compared the results between the groups, groups 3 and 4 showed significance among all groups (Table 2).
| Sl No. | Unpaired t-test | P value | R value | Significance |
| 1 | Method 1 vs Method 2 | 0.908 | 0.0003 | NS |
| 2 | Method 1 vs Method 3 | < 0.0001 | 0.401 | c |
| 3 | Method 1 vs Method 4 | 0.0017 | 0.23 | b |
| 4 | Method 1 vs Method 5 | 0.524 | 0.01 | NS |
| 5 | Method 2 vs Method 3 | < 0.0001 | 0.418 | c |
| 6 | Method 2 vs Method 4 | 0.0011 | 0.248 | b |
| 7 | Method 2 vs Method 5 | 0.393 | 0.019 | NS |
| 8 | Method 3 vs Method 4 | 0.13 | 0.059 | NS |
| 9 | Method 3 vs Method 5 | < 0.0001 | 0.366 | c |
| 10 | Method 4 vs Method 5 | 0.004 | 0.192 | b |
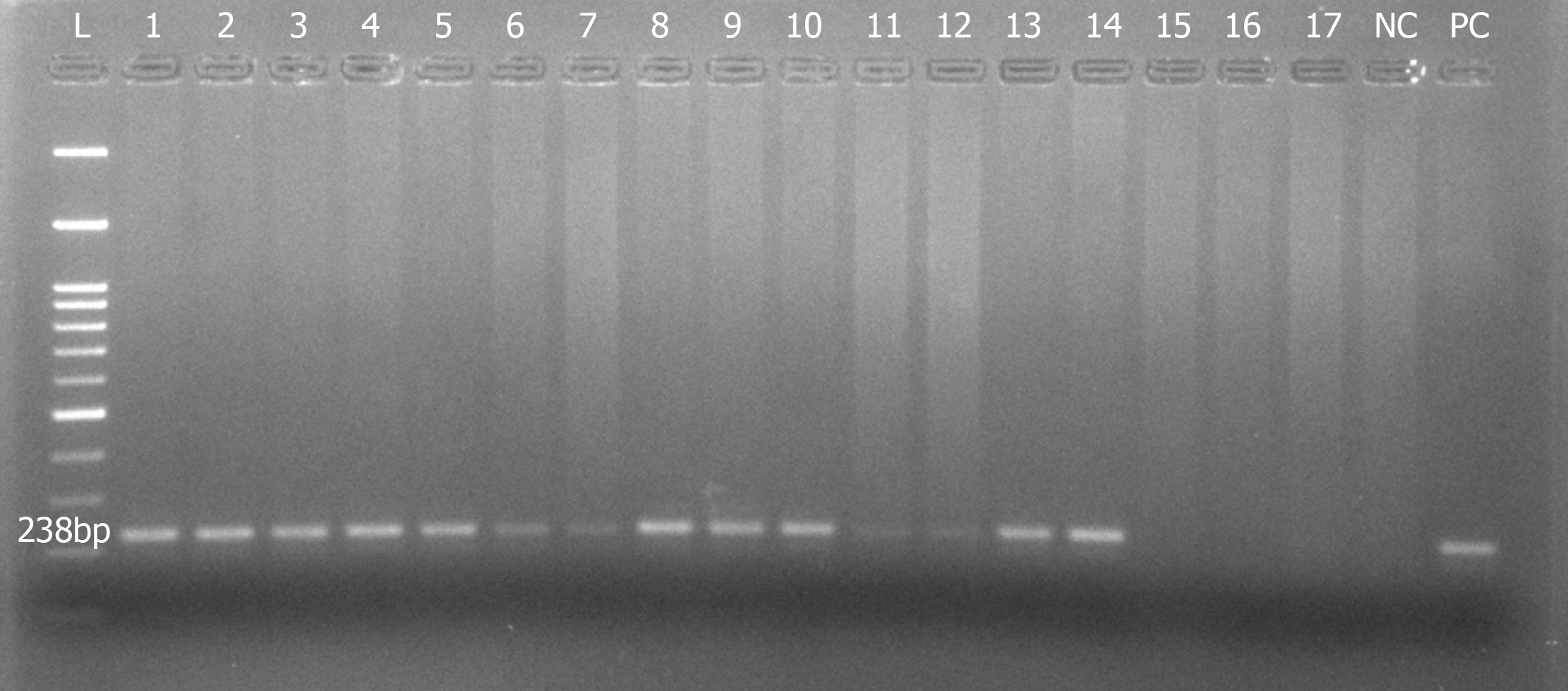
DNA recovered from all groups was subjected to PCR to amplify the GAPDH gene, which was considered to be most stably expressed in almost all tissues and cells. More than 80% of our samples clearly demonstrated amplification of a 238 bp product of GAPDH gene, justifying the intactness of the DNA extracted (Figure 4).
This study was performed to standardize a cost-effective, reproducible protocol for DNA extraction from archived H&E-stained cancer slides and to show that they can be used for downstream molecular analysis. The results demonstrated the efficient amplification of DNA recovered from the archived H&E-stained cancer tissues. The noteworthy point in our protocol is that the samples that were deparaffinized with pre-warmed xylene, digested for up to 72 h, and extracted with the phenol-chloroform method produced the highest DNA yield (Table 1). Although, there are many studies evaluating the potential use of H&E slides for DNA extraction, none of them have assessed if they can be used for downstream molecular processes. Notably, no previous study has standardized a protocol with simplified coverslip removal and deparaffinization. Our data support the idea that archived cancer slides can be harnessed as a potential source for DNA extraction and routine molecular analysis.
Previous studies have evaluated potential extraction of DNA from archived FFPE tissue samples and shown that nucleic acids can be recovered and can be used for PCR and other downstream applications. In the recent past, many commercial kits have also been made available for the extraction of DNA/RNA/protein from the FFPE samples[15-18]. Historically, FFPE specimens have not been considered an ideal source for molecular analysis as nucleic acids may be heavily modified by crosslinking with proteins. However, the discovery of protease digestion releases fragmented nucleic acids, making them suitable for downstream molecular analysis[19]. However, lack of standard protocol and high cost has always limited the use of FFPE specimens. H&E-stained slides, which are made of FFPE blocks, can also serve the same purpose and there are only a handful of studies that have actually looked into it.
As per the goal of our study, we evaluated five different methods of DNA extraction by modifying the coverslip removal, deparaffinization, and digestion steps. High-quality DNA was obtained with a mean yield of 20 ng/µL and more. A study by Sengüven et al[10] analyzed various methods of DNA extraction from archived FFPE specimens, and reported that proper deparaffinization and digestion leads to higher DNA yields. Another study by Snow et al[4] demonstrated the recovery of good yields of DNA from H&E-stained slides using an Qiagen column-based method and Pinpoint slide DNA isolation system with slight modifications. They also used the DNA for microsatellite instability allelic discrimination and variant detection using Sanger sequencing. Both methods described above used commercial kits and again the higher cost got in the way of frequent usage.
Xylene is the most conventional deparaffinization agent used worldwide, but xylene treatment takes time and requires the specimen to be dipped for at least 3-4 d[20]. Several investigators have successfully extracted high-quality DNA using xylene for many years. We optimized the deparaffinization step using pre-heated xylene and obtained significantly improved DNA yields. Shi et al[21] suggested that pre-heating FFPE specimens at higher temperatures in 0.1 mol/L NaOH solution highly increased the efficiency of DNA extraction. In order to avoid toxic chemicals such as xylene, Lin et al[22] demonstrated deparaffinization using mineral oil and then combining with DNA isolation protocol from a commercially available kit. They obtained high-quality DNA, which they tested in a genotyping experiment with 14 microsatellite markers. Contradictorily, in our study when we used mineral oil for deparaffinization, we obtained good DNA yields but the DNA was mostly impure. In addition, some of the samples could not be amplified even with quantitative PCR. That raised a few questions regarding the usage of mineral oil for deparaffinization and to what extent it can undo cross-linkage.
In conclusion, virtually every tissue removed from the body is fixed, paraffin-embedded, and then stored for years to decades. Given the established importance of DNA in molecular biology and its central role in determining fundamental operation of cellular processes, it is essential to look for alternate sources of DNA. Histopathologically stained archived tissue slides are stored in hospital archives for years to decades. They are the largest available source of biological materials and are a potentially useful resource, which can be used for retrospective epidemiological studies. The DNA extracted from fixed tissues can be used for diagnostic applications, such as the detection of mutations and viruses, when the need arises instead of requiring the collection of fresh tissue in anticipation of the need. One mundane but clinically important example is the identification of mislabeled specimens. However, the use of such samples for DNA analysis is limited due to chemical modification by formaldehyde and fragmentation of DNA during tissue processing and storage. Here, we discussed the results from our study for the extraction of PCR-friendly DNA from archived histopathologically stained tissue slides. Hopefully, this simple, cost-effective, and non-laborious protocol can facilitate the molecular analysis of a large number of archived specimens in retrospective studies. Also, similar kind of methodology can be applied for tissues fixed with other fixatives and stained with dyes other than H&E, which may open up the field for future investigations.
Histopathologically stained archived tissue slides are stored in hospital archives for years to decades. They are the largest available source of biological materials and are a potentially useful resource that can be used for retrospective epidemiological studies. DNA recovered from the slides can be used for several downstream molecular processes including polymerase chain reaction, single nucleotide polymorphism analysis, and whole genome sequencing. The slides can also be used to acquire information such as the presence or absence of pathogens that would have been present at the time of sectioning. The DNA from these slides can be utilized to compare gene signatures of normal and diseased tissues.
Generally, the extraction of high-quality DNA from archived stained hematoxylin and eosin (H&E) slides is challenging. Barring commercially available expensive kits, there is a drought of reproducible methods to extract nucleic acids from histopathologically stained tissue slides. The key problem to be addressed here was coming up with new methods for DNA extraction from archived tissue slides that can be easily implemented in molecular biology labs with low resource settings worldwide.
The objective of the study was to standardize a protocol for DNA extraction from archived H&E tissue slides that can be further used for downstream molecular analysis from basic PCR to whole genome sequencing. Also, our objective was to come up with a method that is not only reproducible but also cost-effective.
A total of 100 archived H&E-stained cancer slides were subjected to a total of five methods of DNA extraction. Methods were varied in the deparaffinization step, tissue rehydration, duration of lysis, and presence or absence of proteinase K. The extracted DNA was quantified using a NanoDrop spectrometer and the quality was analyzed by agarose gel electrophoresis. Each sample was subjected to PCR to amplify the internal control gene GAPDH, thereby confirming the DNA intactness that could be further utilized for other downstream applications. Statistical analysis was performed to assess the different methods in terms of yield and purity of the DNA obtained.
Of the five different methods tested, the third method wherein xylene was used for tissue deparaffinization followed by 72 h of digestion and without proteinase K inactivation yielded the highest amount of DNA with good purity. The yield was significantly higher compared to other methods. Also, 90% of the extracted DNA showed amplifiable GAPDH gene indicating the intactness of the DNA, which in turn suggested that this DNA could be used for further molecular analysis.
Our study explored the possible new methods for the extraction of PCR-friendly DNA from archived H&E-stained tissue slides for downstream molecular analysis. We tried and tested alternative methodologies for the removal of coverslip, deparaffinization of the tissues, rehydration and digestion by using simple facilities and common reagents in a basic molecular biology laboratory. We addressed the difficulties in removing the coverslip and deparaffinization of the tissues. Our data indicated that method 3 with xylene deparaffinization and 72 h digestion showed the highest yield of DNA and was also the best in terms of purity. The samples that were deparaffinized using pre-warmed xylene (about 50°C) produced greater DNA yields than concentrations of nucleic acids produced from the samples that were deparaffinized with mineral oil.
In our study, we explored new possibilities of extracting DNA from archived specimens that can be used for molecular analysis. Similar to FFPE tissue blocks, quality H&E tissue slides can be critical in clinical studies and research. Since H&E slides are relatively inexpensive and easy to store, more work can be done with them. Thus recovered DNA can be utilized in the field of oncology for discriminating the mutational profile between the tumor and adjacent normal tissue or even in the field of hematology or immunology to understand the disease state, cause, and possible medication. Based on the preliminary evidence from our study, future research can focus on how to best utilize the discussed methods.
The authors would like to thank the Department of Pathology, JSS Medical College Hospital for providing the archived specimens and the Center of Excellence in Molecular Biology & Regenerative Medicine Laboratory, Department of Biochemistry, JSS Medical College for providing the facilities to conduct the research. Pushkal Sinduvadi Ramesh would like to acknowledge the junior research fellowship from the Council of Scientific and Industrial Research, Government of India.
Manuscript source: Unsolicited manuscript
Specialty type: Medical laboratory technology
Country of origin: India
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ciotti M, Exbrayat JM S-Editor: Ma RY L-Editor: Filipodia E-Editor: Qi LL
| 1. | Abraham JE, Maranian MJ, Spiteri I, Russell R, Ingle S, Luccarini C, Earl HM, Pharoah PP, Dunning AM, Caldas C. Saliva samples are a viable alternative to blood samples as a source of DNA for high throughput genotyping. BMC Med Genomics. 2012;5:19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 93] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 2. | Rohland N, Hofreiter M. Ancient DNA extraction from bones and teeth. Nat Protoc. 2007;2:1756-1762. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 385] [Cited by in F6Publishing: 314] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 3. | Wilson MR, Polanskey D, Butler J, DiZinno JA, Replogle J, Budowle B. Extraction, PCR amplification and sequencing of mitochondrial DNA from human hair shafts. Biotechniques. 1995;18:662-669. [PubMed] [Cited in This Article: ] |
| 4. | Snow AN, Stence AA, Pruessner JA, Bossler AD, Ma D. A simple and cost-effective method of DNA extraction from small formalin-fixed paraffin-embedded tissue for molecular oncologic testing. BMC Clin Pathol. 2014;14:30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 5. | Darst RP, Pardo CE, Ai L, Brown KD, Kladde MP. Bisulfite sequencing of DNA. Curr Protoc Mol Biol. 2010;Chapter 7:Unit 7.9.1-Unit 7.917. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 58] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 6. | Millsaps JL. DNA Extraction from Archived Slides: Analysis and Use in Current Forensic Identification. Available from: trace.tennessee.edu/cgi/viewcontent.cgi?article=3507&context=utk_gradthes. [Cited in This Article: ] |
| 7. | Srinivasan M, Sedmak D, Jewell S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol. 2002;161:1961-1971. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 909] [Cited by in F6Publishing: 860] [Article Influence: 39.1] [Reference Citation Analysis (0)] |
| 8. | Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13387] [Cited by in F6Publishing: 14165] [Article Influence: 393.5] [Reference Citation Analysis (0)] |
| 9. | Tan SC, Yiap BC. DNA, RNA, and protein extraction: the past and the present. J Biomed Biotechnol. 2009;2009:574398. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 402] [Cited by in F6Publishing: 313] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 10. | Sengüven B, Baris E, Oygur T, Berktas M. Comparison of methods for the extraction of DNA from formalin-fixed, paraffin-embedded archival tissues. Int J Med Sci. 2014;11:494-499. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 72] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 11. | Zhou W, Geiersbach K, Chadwick B. Rapid removal of cytology slide coverslips for DNA and RNA isolation. J Am Soc Cytopathol. 2017;6:24-27. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 12. | Morikawa T, Shima K, Kuchiba A, Yamauchi M, Tanaka N, Imamura Y, Liao X, Qian ZR, Brahmandam M, Longtine JA, Lindeman NI. No evidence for interference of H&E staining in DNA testing: usefulness of DNA extraction from H&E-stained archival tissue sections. Am J Clin Pathol. 2012;138:122-9. [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 13. | Pikor LA, Enfield KS, Cameron H, Lam WL. DNA extraction from paraffin embedded material for genetic and epigenetic analyses. J Vis Exp. 2011;pii: 2763. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 60] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 14. | Ofverstedt LG, Hammarström K, Balgobin N, Hjertén S, Pettersson U, Chattopadhyaya J. Rapid and quantitative recovery of DNA fragments from gels by displacement electrophoresis (isotachophoresis). Biochim Biophys Acta. 1984;782:120-126. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 75] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 15. | Schweiger MR, Kerick M, Timmermann B, Albrecht MW, Borodina T, Parkhomchuk D, Zatloukal K, Lehrach H. Genome-wide massively parallel sequencing of formaldehyde fixed-paraffin embedded (FFPE) tumor tissues for copy-number- and mutation-analysis. PLoS One. 2009;4:e5548. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 134] [Cited by in F6Publishing: 141] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 16. | Huijsmans CJ, Damen J, van der Linden JC, Savelkoul PH, Hermans MH. Comparative analysis of four methods to extract DNA from paraffin-embedded tissues: effect on downstream molecular applications. BMC Res Notes. 2010;3:239. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 75] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 17. | Dedhia P, Tarale S, Dhongde G, Khadapkar R, Das B. Evaluation of DNA extraction methods and real time PCR optimization on formalin-fixed paraffin-embedded tissues. Asian Pac J Cancer Prev. 2007;8:55-59. [PubMed] [Cited in This Article: ] |
| 18. | Turashvili G, Yang W, McKinney S, Kalloger S, Gale N, Ng Y, Chow K, Bell L, Lorette J, Carrier M, Luk M, Aparicio S, Huntsman D, Yip S. Nucleic acid quantity and quality from paraffin blocks: defining optimal fixation, processing and DNA/RNA extraction techniques. Exp Mol Pathol. 2012;92:33-43. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 67] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 19. | Gilbert MT, Haselkorn T, Bunce M, Sanchez JJ, Lucas SB, Jewell LD, Van Marck E, Worobey M. The isolation of nucleic acids from fixed, paraffin-embedded tissues-which methods are useful when? PLoS One. 2007;2:e537. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 270] [Cited by in F6Publishing: 269] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 20. | Zhang G, Yu CZ, Su SH, Kalra KL, Zhou D, inventors; Biogenex Laboratories, assignee. Deparaffinization compositions and methods for their use. United States patent US 6632598. 2003;Oct 14. [Cited in This Article: ] |
| 21. | Shi SR, Datar R, Liu C, Wu L, Zhang Z, Cote RJ, Taylor CR. DNA extraction from archival formalin-fixed, paraffin-embedded tissues: heat-induced retrieval in alkaline solution. Histochem Cell Biol. 2004;122:211-218. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 121] [Cited by in F6Publishing: 106] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 22. | Lin J, Kennedy SH, Svarovsky T, Rogers J, Kemnitz JW, Xu A, Zondervan KT. High-quality genomic DNA extraction from formalin-fixed and paraffin-embedded samples deparaffinized using mineral oil. Anal Biochem. 2009;395:265-267. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 74] [Article Influence: 4.9] [Reference Citation Analysis (0)] |