Published online Mar 18, 2021. doi: 10.5500/wjt.v11.i3.16
Peer-review started: December 4, 2020
First decision: December 27, 2020
Revised: January 6, 2021
Accepted: February 12, 2021
Article in press: February 12, 2021
Published online: March 18, 2021
Aim of this frontier review has been to highlight the role of microbiota in healthy subjects and in patients affected by renal diseases with particular reference to renal transplantation. The microbiota has a relevant role in conditioning the healthy status and the diseases. In particular gut microbiota is essential in the metabolism of food and has a relevant role for its relationship with the immune system. The indigenous microbiota in patients with chronic renal failure is completely different than that of the healthy subjects and pathobionts appear. This abnormality in microbiota composition is called dysbiosis and may cause a rapid deterioration of the renal function both for activating the immune system and producing large quantity of uremic toxins. Similarly, after renal trans-plantation the microbiota changes with the appearance of pathobionts, principally in the first period because of the assumption of immunosuppressive drugs and antibiotics. These changes may deeply interfere with the graft outcome causing acute rejection, renal infections, diarrhea, and renal interstitial fibrosis. In addition, change in the microbiota may modify the metabolism of immuno-suppressive drugs causing in some patients the need of modifying the immunosuppressant dosing. The restoration of the indigenous microbiota after transplantation is important, either to avoiding the complications that impair the normal renal graft, and because recent studies have documented the role of an indigenous microbiota in inducing tolerance towards the graft. The use of prebiotics, probiotics, smart bacteria and diet modification may restore the indigenous microbiota, but these studies are just at their beginning and more data are needed to draw definitive conclusions.
Core Tip: Recent studies on the microbiota have documented that a microbiota modification, related to the assumption of immunosuppressive drugs and of antibiotics, as happens in the first period after transplantation may modify the outcomes of the graft. Indeed, dysbiosis may cause acute rejections and reduce the possibility of a tolerance status. In addition, dysbiosis if often the cause of infections and renal fibrosis. Dysbiosis may also cause diarrhea that is a frequent and severe complication in the transplanted patient. Modification of dysbiosis is possible with an appropriate treatment, but studies on this topic are just at their beginning.
- Citation: Salvadori M, Tsalouchos A. Microbiota, renal disease and renal transplantation. World J Transplant 2021; 11(3): 16-36
- URL: https://www.wjgnet.com/2220-3230/full/v11/i3/16.htm
- DOI: https://dx.doi.org/10.5500/wjt.v11.i3.16
The microbiota is defined as the micro-organisms that live in the human body without damaging it in healthy conditions. The most important and the best studied is the microbiota of the digestive system. In particular, the urinary microbiota has also been studied in studies concerning renal diseases and renal transplants.
In recent years the function of the microbiota, particularly the gut microbiota has been extensively examined and the relationship between the microbiota and diseases has been elucidated with particular reference to organs such as the kidney. In this frontier review, the definition of the microbiota and its variety will be provided, along with descriptions of its functions and relationship with the immune system. In addition, the relationship between an abnormal microbiota or pathobionts and renal diseases and renal transplantation has been documented in several studies[1-5]. The relationship between the microbiota and its alterations in patients with kidney disease will be elucidated with particular references to the relationship between the microbiota and renal transplantation.
The words microbiota and microbioma are often mutually used, but they have a different meaning.
The term microbiota refers to all the microorganisms inhabiting some specific niches as gut, skin, lungs and other organs and encompasses bacteria, viruses, fungi and archea. In this review the term microbiota refers principally to bacteria even if in general it strictly refers also to other microorganisms. In a recent study the estimated total number of bacteria for a 70 kg man is approximately 3.8 × 1013 and is approximately of the same order of the number of human cells[6]. The gut microbiota is the most important community because of its quantity and its relationship with kidney disease. The gut microbiota is already present within the first few years of life, and its composition should remain stable in adults, where the dominant bacteria are Bacteroides, Firmicutes and Actinobacteria[7-9]. In the healthy subject the resident microbiota is also called indigenous microbiota. When the indigenous microbiota, due to genetic or environmental factors, cause inflammatory disorders or other diseases, is generally called pathobionts and this condition is called dysbiosis. Pathobionts should be distinguished from acquired infectious agents also called pathogens[10]. Due to the relevance of microbiota both in healthy status and diseases, several national and international scholars performed studies of gut microbiota, such as the Canadian Microbioma Initiative, The Human Meta Genome Consortium Japan, the My New Gut Project of the European Union and the International Human Microbioma Consortium[11-13]. The composition of the gut microbiota under standard conditions is shown in Table 1.
| Intestine sections | Function | Normal flora |
| Stomach | Acid production, pepsin, amylase, CFU < 103/mL | Lactobacillus; Streptococcus; Helycobacter pylori |
| Small intestine: duodenum, jejunum | Pancreatic enzymes, bicarbonate ions, bile salts, CFU: 103-104/mL | Lactobacilli; Enterococci; Streptococci; Actinobacteria |
| Small intestine: ileum | CFU: 103-109/mL | Enterococcus; Bacteroidetes; Lactobacillus; Clostridium; Corynebacteria |
| Large intestine: caecum, colon | Mucus and bicarbonate, CFU:1010-1012/mL | Bacteroidetes; Clostridium; Eubacterium; Ruminococcus; Streptococcus; Enterococcus; Lactobacillus; Fusobacteria |
As mentioned above, the term microbioma has a different meaning than the microbiota and refers to all the microbiota genes and is approximately 150 times larger than the human genome[14,15]. In healthy subjects the gut microbioma is stable and exerts important functions throughout the body as shown in Table 2.
| Protective function | Metabolic function | Structural function |
| Nutrient competition; Barrier fortification; Innate and adaptive immunity activation; Antimicrobial compounds secretion | Vitamin and amino acid biosynthesis; Bile acid biotransformation; Dietary fiber fermentation; Short chain fatty acids production | Mucus layer properties; Crypt and villi development; Villi microvascularization; Tight junction regulation |
Dietary fibers produce energy when metabolized, but not all dietary fibers are metabolized by digestive enzymes[16]. The gut microbiota of the large intestine contains enzymes that are able to metabolize these fibers and recover additional energy[17,18].
Undigested proteins are degraded into peptides, amino acids and other metabolites in the large intestine. Some of these metabolites are dangerous to the body and could cause diseases as colorectal cancers and kidney dysfunction[19]. The MEROPS database documented that the composition of the large intestine microbiota may contains different proteases responsible for inducing the production of different meta-bolites[20,21]. The gut microbiota also exerts important actions on lipids, bile salts and polyphenols.
The structural integrity of the intestinal epithelium is essential to avoid a dangerous increase in permeability. The maintenance of structural integrity is essential for the microbiota. In normal conditions, cytokines produced in the gut may back diffuse in small quantities passing through the gut barrier. The barrier function of the tight junction in dysbiosis condition, may be weakened by several endotoxins of some pathogens as Escherichia coli (E. coli), Clostridium difficile and Clostridium perfrigens. In this condition of dysbiosis, the diffusion of citokines such as interleukin 4, interleukin 1 beta, tubular necrosis factor alpha and interferon gamma is increased[22-26].
The gastrointestinal tract represents a bidirectional barrier between the gut microbiota and the gut immune system[27]. The barrier is composed of three layers: the mucus layer, the antimicrobial peptides (AMPs) and the IgA system.
Mucin glycoproteins secreted by goblet cells form a layer over the epithelia to restrict bacterial adhesion. This layer prevents the adherence of commensal microbiota to gut epithelial cells, limiting the bacterial adhesion[28]. A second layer is represented by AMPs secreted by epithelial cells. AMPs include α and β defensins secreted by the epithelium and mediated by cytosolic nucleotide-binding oligomerization domain-containing protein 2[29,30]. C-type lectins activate Toll-like receptors to limit bacterial penetration through the gut barrier[31].
The third layer is composed of the IgA system. Dendritic cells (DCs) located beneath the epithelial dome of Peyer’s patches take up bacteria, migrate to mesenteric lymph nodes and induce B cells to differentiate into IgA plasma cells that secrete IgA[32,33].
The indigenous microbiota, pathobionts and pathogens promote in the gut the generation of several Th cells among which Th1, Th2, Th17 and Treg. At mucosal sites this may also be due to the production of microbiota metabolites. In particular, the microbiota stimulate epithelial cells to the generation and accumulation of Treg by increase of TGFβ, stimulate macrophages to induce Th17 cells by increase of interleukin 1 beta, and through DNA methylation can induce proliferation of colonic Treg cells. Other actions on immune cells are due to microbiota metabolites as butyrate. Butyrate down regulates IL-10 production from neutrophils and generates an anti-inflammatory activity. Butyrate, down regulating IL-6 from macrophages, induces increased levels of histone acetylation. On the other hand, butyrate, by inhibition of histone deacetylase, inhibits the activation of NF-kB inducing a Th1 cell response[34,35]. The balance of Treg cells and the effector T cells in the intestinal mucosa is related to the ratio between the indigenous microbiota and the pathobionts. In particular the subset of Th1 and Th2 cells activation is characterized by the expression of proinflammatory cytokines including IFNγ, IL4, IL5 and IL13, and IL23[22]. Th 17 cells are characterized by the synthesis of IL-17, which stimulates cells to express the proinflammatory cytokines as IL-6, IL-8, and Il-22[36,37].
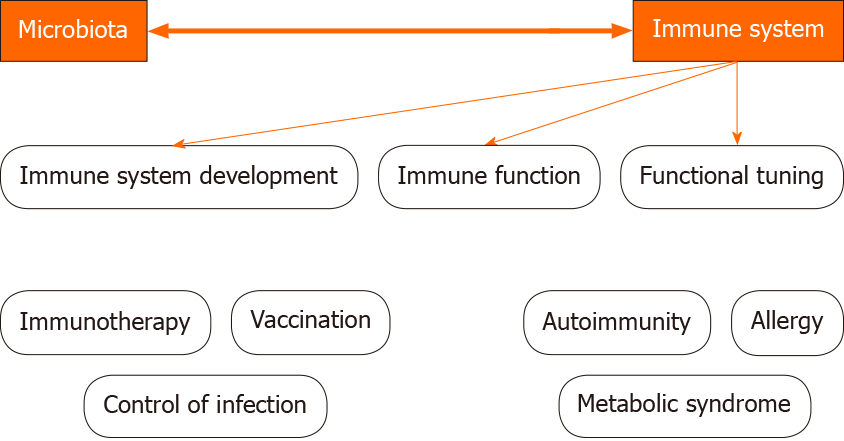
The indigenous microbiota plays a fundamental role in the induction, education and function of the immune system (Figure 1).
The microbiota composition may be modified by several conditions, among which the use of antibiotics, immunosuppressants or diet alterations. In such conditions pathobionts appear and modify the immune system and promote the development of inflammatory diseases[38].
Microbiota-derived Toll-like receptors and NOD ligands and metabolites [such as short-chain fatty acids (SCFAs) and aryl hydrocarbon receptors] may act on local gut cells but also penetrate beyond the mucosa to tune immune cells in peripheral tissues[39].
SCFAs promote DC precursor activation and release into the bloodstream. Microbiota- derived NOD1 Ligands induce mesenchymal cells to produce hemato-poietic growth factors as IL7, stem cell factor (SCF), thrombopoietin, recombinant human flt3-Ligand, IL6[40-42].
In addition, microbiota-derived riboflavin metabolites promote the development of mucosal- associated invariant T cells[43], and commensal bacterial-induced cytokines IL1β and IL23 promote IL17A production from gamma delta T cells[44,45].
Finally, commensal bacterial colonization promotes effector and regulatory T cell responses.
Clostridia colonization promotes retinoic acid receptor-related orphan nuclear receptor gamma (RORγt)[46], and Foxp3+ Treg cell accumulation, which in turn limits colonic Th2 and Th17 cell responses.
Foxp3+ Tregs cells localize in Peyer’s patches and promote B class switching and the production of IgA, which fosters a different microbiota and ensures commensal bacteria compartmentalization from the intestinal epithelium[47].
Under healthy conditions, a balance between antigenic stimuli exists due to the microbiota and the immune response.
However, an aggressive immune response due to the appearance of pathobionts or pathogens in some subjects may cause inflammatory diseases, and a weak response may cause the overgrowth and diffusion of the pathobionts themselves.
Commensal bacteria induce CD4+ cells to differentiate into 4 main subtypes: Th1, Th2, Th17 and Treg. The indigenous microbiota contributes to normalizing the ratio of these subtypes.
Additionally, IgA production contributes to controlling excessive microbiota growth and limiting the growth of pathobionts.
In healthy conditions, segmental filamentous bacteria induce the growth and differentiation of Th17 and Th1 cells[48]. In animal studies has been documented that this is impaired in animals treated with antibiotics while is normal in germ free conditions. Still in the animals, in healthy conditions, Clostridia promote the accumulation of Tregs and production of IL10, which exerts anti-inflammatory effects[49].
Bacterioides fragilis also contributes to maintaining a correct equilibrium between the microbiota and immune system by producing of polysaccharide A and inducing the production of IL10 and Tregs[50].
When the microbiota loses its richness and its correct composition, pathobionts appear and dysbiosis occurs. This change may lead to diseases and kidneys and kidney grafts are among the main targets.
Communication between the gut and kidney occurs either by the activation of the immune system and by microbiota-derived metabolites.
Several studies have documented that the activation of Th17 cells in the gut by the microbiota leads to activation of Th17 cells in the kidney[51]. Chemokine ligand 20/C-C[52] recruits Th17 cells to the kidney.
In animals, the addition of antibiotics reduces Th17 levels and renal damage[53]. The crucial role of Th17 cells in inducing tissue injury is also evidenced by the high levels of Th17 cells in humans with auto-immune kidney diseases and in glomeru-lonephritis[54].
This phenomenon is bidirectional because acute kidney injury (AKI) determines intestinal dysbiosis and T helper Th17 cells, neutrophils and M1 macrophages mediate intestinal inflammation, as well as leaky gut with bacterial translocation. On the other hand, dysbiotic microbiota may exert an adverse effect on kidney injury and the depletion of the pathobionts may mitigate kidney injury[55].
Microbiota-derived metabolites may affect kidney and other organ functions. Indeed, the microbiota may interact with a large number of vital functions in the health body via several metabolites. The targets are host metabolism and immunity as well as cardiovascular and brain functions. Additionally, the microbiota metabolism utilizes enzymes not encoded by the human genome and generates biological products relevant to the host’s health as bile acids, choline, vitamins and SCFAs[56].
SCFAS are among the most relevant metabolites produced by microbiota[57].
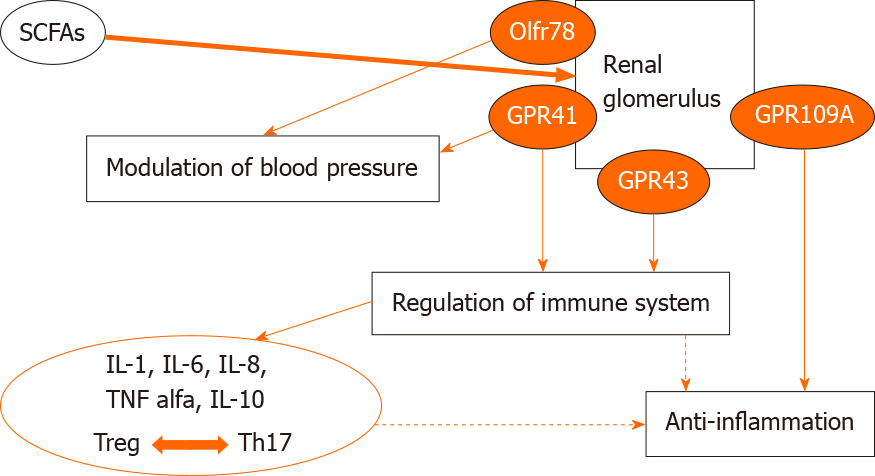
SCFAs activate G protein-coupled receptors (GPR) including GPR41, GPR43 and GPR109A.
The binding of SCFAs to their receptors exerts beneficial effects on the kidney. Indeed, this signaling pathway regulates energy homeostasis[58], stimulates glucagon-like peptide 1 secretion[59], and inhibits the progression of atherosclerosis in mice[60]. The binding of SCFAs to another receptor, Olfr78 exerts beneficial effects on blood pressure[61]. These and other data support a beneficial effect of SCFAs on kidney injury (Figure 2).
In addition, SCFAs also regulate cytokine expression in T cells and the generation of Tregs through histone deacetylase inhibition.
Overall, SCFAs exert a beneficial effect on AKI by reducing the production of cytokines and chemokines such as IL1β, IL6, TNFα and monocyte chemoattractant protein 1[62].
In addition, SCFAs have also extraintestinal actions controlling appetite regulation, glucose and lipid metabolism. This is due to the fact that the above mentioned receptors have also been found in cells as adipocytes, neurons and immune and vascular cells[63].
Equol, produced by certain microbiota subtypes has several beneficial effects, including antiapoptosis, antioxidation, and anti-atherosclerosis, the production of nitric oxide in endothelial cells, antiproliferation and/or migration, and promotion of vascular smooth cells relaxation[64].
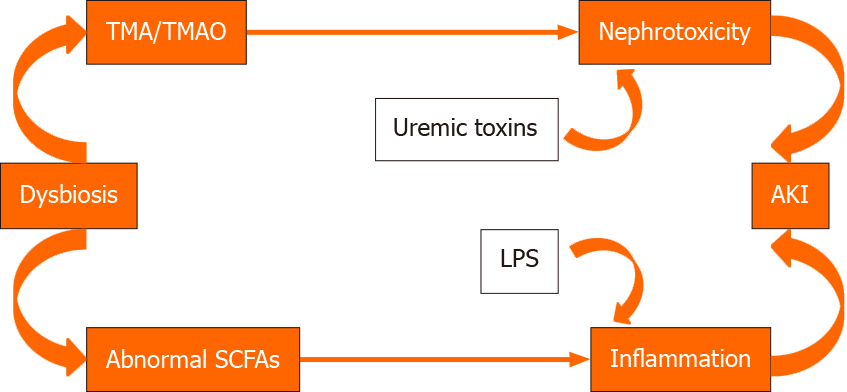
On the contrary, negative effects on vascularization are exerted by metabolites as indoxylsulfate and trimethylamine N oxide (TMAO).
Indoxylsulfate produced by pathobionts as E. coli has deleterious effect on the vascular system. Indoxylsulfate induces apoptosis, senescence, prothrombotic events, proliferation and/or migration and modulation of vascular tone in vascular smooth muscle cells. Similar negative vascular effects are exerted by TMAO.
TMAO is a product of gut bacterial metabolism of choline. Differently from SFCAs it promotes renal interstitial fibrosis[65].
The different effects of these metabolites are shown in Figure 3.
The gut microbiota may also produce uremic toxins that, in the case of dysbiosis, may be produced in high quantities and may damage the kidney[66].
The quorum sensing signals (QS) may be produced either by pathobionts or by indigenous microbiota. Indeed, QS may be divided into two types. Those produced by GRAM- bacteria such as Pseudomonas aeruginosa have negative immune-related processes such as IkK phosphorilation, and activation of mitogen activated protein kinase (MAPK) pathways. These induce NF-kB signaling and chemotaxis. As a result they increase inflammatory genes expression. Differently, the QS signals induced by Bacillus subtilis, have beneficial effects through the induction of p38 MAPK on protein kinase B[57].
Dysbiosis may facilitate AKI either by modifying the SCFAs composition or generating higher quantites of TMAO and uremic toxins. This modification may facilitate the transition from AKI to chronic renal disease (CKD). Indeed, a cross-talk between the intestinal microbiota and the kidney has been observed. During experimental AKI, gut pathobionts may modify immune cells and other pathophysiological mediators to alter the course of AKI. AKI may in turn modify the gut bacterial composition[67,68]. This topic has been extensively studied by Vaziri et al[68] who observed substantial differences in the gut microbiota composition between patients with end stage renal disease and control patients.
This result has been confirmed by Cigarran Guldris et al[69], who substantially found dysbiosis in patients affected by end stage renal disease, due to the presence of pathobionts. Pathobionts modify protein absorption, reduce the utilization of alimentary fibers and are frequently associated with the use of antibiotics[70,71].
In summary, in the healthy subject the indigenous microbiota provides benefits to our health. Indigenous microbiota affects the host by production of metabolites and gut neuropeptides. By sending the informations about the state of inner organs to the brain, they control many important functions as mood, immune response, digestion and heart rate. By this way a bidimensional communication between the gut, its microbioma and the nervous and neuroendocrine systems is established[72].
Different factors, including immunosuppressant and antibiotic therapy, lifestyle and diet, may alter the microbiota and lead to generation of pathobionts and dysbiosis. Dysbiosis disrupts the gut epithelial barrier, induces a loss of barrier integrity and leads to pathogen overgrowth. The leaky gut and increased permeability facilitate the translocation of bacteria and their components into the inner environment. In this dysbiosis situation, the proinflammatory response triggers the elimination of pathogens by intestinal epithelial cells (IL-1, IL-6 and IL-18 secretion), DCs and macrophages that induce the development of the effector CDE4+ cells, Th1 and Th17. Innate immune responses lead to a systemic and allograft inflammation. Moreover, dysbiosis decreases the number of regulatory T cells and increases the number of effector T cells that activate innate immunity. On the other hand, in the colon and liver, dysbiotic gut-derived uremic toxins are further metabolized to TMAO. The accumulation of p-cresyl sulfate in the kidney generates reactive oxygen species that lead to the production of inflammatory cytokines and profibrotic factors. In addition, indoxylsulfate induces inflammation and nephrotoxicity[73-77].
Renal transplant patients, in addition to receiving relevant immunosuppressive therapy in the first period after transplantation, receive several antibiotic treatments as a prophylactic measure to avoid infections.
All these drugs extensively modify the human microbiota, principally at the gut and urinary tract levels. Historically, since the initiation of renal transplantation, when very high doses of cyclosporine A were used, gingival overgrowth was observed as an important side effect. This change was related to modifications of the oral microbiota and generation of pathobionts[78].
In a pilot study, Lee et al[79], performed polymerase chain reaction in samples from 26 kidney transplant recipients and documented a change in the microbiota between the pre- and posttransplant periods. The results are shown in Table 3.
| Phylum | Pre Tx cohort | Post Tx cohort |
| Firmicutes | 91.8% | 87.7% |
| Actinobacteria | 2.0% | 7.6% |
| Proteobacteria | 0.9% | 4.1% |
| Bacteroidetes | 2.8% | 0.6% |
| Order | ||
| Clostridiales | 64.8% | 64.3% |
| Lactobacillales | 19.1% | 12.0% |
| Erysipelotrichales | 5.6% | 10.2% |
| Bifidobacteriales | 1.6% | 6.6% |
| Enterobacteriales | 0.4% | 3.9% |
| Bacteroidales | 2.8% | 0.6% |
Firmicutes were the most abundant bacteria detected pre- and posttransplantation, but their quantity posttransplantation was lower than in healthy subjects[80]. The same study reported posttransplantation an increase in the abundance of Bacteroides that included infective pathogens such as E. coli and Klebsiella pneumoniae[81].
Overall, the study by Lee and colleagues documented a dysbiosis that was later confirmed by other studies. A recent review from Xiao et al[82] on microbiota modifications in response to solid organ transplantation highlighted an increase in the abundance of pathogenic Proteobacteria, which might represent the cause of infectious diseases occurring after transplantation.
These data were confirmed by a recent study by Swarte et al[83] that confirmed a reduction in the abundance of Firmicutes with variability among the species. The most significant reduction was observed for Streptococcus thermophilus and Blautiawexlerae.
Overall these authors observed an increase in the abundance of Proteobacteria (E. coli) and a decrease in the abundance of Actinobacteria posttransplantation. The increase in Proteobacteria has already been proposed as a marker of dysbiosis[84]. Additionally, the same study observed a reduction in SFCAs producing bacteria after transplantation. In particular, reductions in the abundance of Eubacterium rectale, Coprococcuscatus and Roseburia were observed. All these bacteria produce SCFAs[85] that exert beneficial effects on the kidney and increase the number of Tregs, reducing systemic inflammation[86,87]. The use of proton pump inhibitors, of MMF and aging were the prevalent determinants of this form of dysbiosis[88,89].
Another study[90] analyzed the gut microbiota in 142 kidney transplant recipients. The authors detected potential pathogens, such as Clostridium difficile and E. coli in 30% of patients. These pathogens were not associated with diarrhea, as expected.
A different study[91] observed that major changes in the microbioma occur in the first month after transplantation, with substantial differences among patients. The authors concluded that longitudinal analyses should be performed to provide more information.
In conclusion, dysbiosis after renal transplantation is related to an imbalance between the indigenous microbiota and the pathobionts. This imbalance is related principally to the need for immunosuppressant and prophylactic and therapeutic antimicrobial agents[92].
The metabolic and clinical consequences of dysbiosis are represented by a higher incidence of acute rejections, acute infections, interstitial fibrosis, posttransplant diarrhea, reduced production of protective agents such as SCFAs by the gut microbiota, and modification of immunosuppressant levels in the blood.
Several experimental studies conducted in animals have documented en effect of the gut microbiota on immune responses that lead to transplant rejection[93].
Few studies have been conducted in the humans on this topic.
In the aforementioned study by Lee et al[79], the differences in the fecal bacteria composition of patients with and without rejection are shown in Table 4.
| Phylum | No AR cohort | AR cohort | P value |
| Firmicutes | 91.4% | 76.6% | 0.40 |
| Actinobacteria | 3.7% | 8.2% | 0.60 |
| Proteobacteria | 1.3% | 15.2% | 0.33 |
| Bacteroidetes | 3.1% | 0.02% | 0.03 |
| Order | |||
| Clostridiales | 63.1% | 16.9% | 0.01 |
| Lactobacillales | 12.7% | 49.9% | 0.04 |
| Erysipelotrichales | 13.3% | 9.2% | 0.32 |
| Bifidobacteriales | 3.1% | 7.9% | 0.44 |
| Enterobacteriales | 1.0% | 14.7% | 0.17 |
| Bacteroidales | 3.1% | 0.02% | 0.03 |
In one recent study[84], the microbiota was evaluated pre- and posttransplant in 60 patients who received a renal transplant.
Samples from urine, oral swabs, rectal swabs and blood were evaluated for up to 6 mo after transplantation.
In the study, the most relevant changes in the microbiota principally verified in the first month after transplant, when the immunosuppressive treatment was heavier because of the induction therapy. Further modifications in the microbiota were verified in the first six months after transplantation. In urine samples and in oral swab samples, changes were verified principally in the phylum Proteobacteria. In the rectal swab samples, Firmicutes were the bacteria whose composition changed more frequently.
Significant changes in Leptotrichia, Neisseria and Actinobacteria were observed in five patients who experienced acute rejection. Four patients experienced late acute rejection and displayed significant changes in Anaerotruncus, Coprobacillus and Coprococcus.
The same authors of the study on acute rejection[94] documented that similar changes in the microbiota were also associated with a higher incidence of urinary tract infections.
In particular, in four patients with posttransplant infections, the abundance of the genus Anaerotruncus of Firmicutes was markedly decreased compared to the other patients.
Several factors may cooperate with dysbiosis to generate infections, as shown in Table 5. This higher incidence of both urinary and gastrointestinal infections was also reported in the aforementioned studies by Lee et al[79] and Chan et al[95].
| Risk factors | Microbiota changes | Consequences | Interventions |
| Dietary patterns | Increase in bacteria translocation | Gastrointestinal upset e.g., diarrhea | Diet |
| Changes to colonic and bowel transit time | Increase in metabolic endotoxemia | Urinary tract infections | Prebiotics |
| Immunosuppression | Increase in gut-derived microbial toxin formation | Other infections not yet explored | Probiotics |
| Antibiotics | Synbiotics | ||
| Lifestyle (sedentary, smoking, alcohol) |
In a recent study[96], a transplant patient with recurrent urinary infections recovered after fecal microbiota transplantation (FMT), which induced a marked decrease in the abundance of E. coli in the urinary microbiota.
In conclusion, according to these studies, some microbial species may exert a protective effect on the mucosal surface under normal conditions, and when the microbiota changes, pathobionts and aggressive phenotypes appear to induce renal dysfunction.
The hypothesis that urinary dysbiosis is principally responsible for the development of interstitial fibrosis of the graft was based on the findings that patients affected by interstitial fibrosis/tubular atrophy (IF/TA) had abnormalities in the urinary microbiota with appearance of pathobionts and, consequently, in the immune response. Two studies, conducted in humans[97,98] detected antibodies directed against E. coli LPS, a powerful activator of the immune system via TLR4 receptor in the biopsies of patients affected by IF/TA.
In a recent study of transplant patients, Modena et al[99] collected urine samples from 25 patients at two time points after kidney transplantation (approximately 1 mo and 6 mo after transplantation). All these patients demonstrated developed IF/TA in surveillance biopsies collected 6 mo after transplantation.
These samples were compared with 23 patients with normal surveillance biopsies and stable renal function at 6 mo after transplantation.
At six months after transplantation, patients affected by IF/TA displayed decreased abundances in the Lactobacillus and Streptococcus genera along with an increase in the abundance of no dominant species.
The authors concluded that the urinary microbiota, modified posttransplantation, may contribute to IF/TA development by altering the host immune response.
IF/TA is associated with a loss of the indigenous dominant resident urinary microbiota and an increase in the abundance of pathobionts or nonresident, pathogenic bacteria.
The phenomenon of IF/TA may be mediated by myofibroblasts, as has already been documented in the gut, where gut dysbiosis potentially leads to intestinal fibrosis[100]. Myofibroblasts may be derived from transdifferentiation processes such as the epithelial to mesenchymal transition or endothelial to mesenchymal transition. These processes may be induced and aggravated by modifications in the indigenous microbiota.
In conclusion, myofibroblasts may play a relevant role in inducing IF/TA either at the gut or renal level, and the indigenous microbiota might have regulatory and protective functions under normal conditions.
Diarrhea represents a severe complication after kidney transplantation, affecting approximately 20% of patients[101], and it represents an important cause of graft loss and death[102]. However, its etiology is still being discussed, and a clear diagnosis not available for approximately 85% of transplanted patients affected by diarrhea. With the exception of the few cases that are ascribed to a specific infection and the presence of pathogens, the diarrhea etiology is often ascribed to the use of immuno-suppressants, in particular MMF. However, a reduction in the MMF dose is dangerous and may lead to an increased risk of allograft rejection[103].
In the pilot study by Lee et al[79], the authors observed a reduction in the commensal indigenous microbiota, such as Ruminococcus, Dorea and Coprococcus, in 26 renal transplant patients affected by diarrhea. In addition, they did not detect pathogens such as Clostridium difficile or norovirus in fecal specimens. These findings prompted the hypothesis that in the majority of patients, gut dysbiosis rather than the presence of pathogens may represent an important cause of posttransplant diarrhea. In a recent study by Lee et al[104], fecal specimens from 25 patients presenting diarrhea in the first three months after transplantation were compared with 46 patients who did not develop diarrhea. In the diarrhea group, the abundance of the genera Eubacterium, Anaerostipes, Coprococcus, Romboutsia, Ruminococcus, Dorea, and Faecalibacterium were significantly decreased, while the abundance of the genera Lachnoclostridium, Escherichia and Enterococcus were significantly increased. Table 6 provides a detailed description of the data. Many of the bacteria that were present at lower abundance in the diarrhea group belong to the Lachnospiraceae and Ruminococcaceae families[105] and contribute to metabolic functions essential for gut health[106]. Utilizing the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States Analysis[107], 9 metabolism-related pathways were decreased in the diarrhea group. The decrease in the abundance of these indigenous microbiota bacteria in the subjects affected by diarrhea contributes to the development of an abnormal metabolic status, which might lead to diarrhea.
| Bacterial Taxonomy Genus | Median relative abundance in the diarrhea group | Median relative abundance in the no diarrhea group | P value |
| Eubacterium | 0.002 | 0.017 | 1.5E-09 |
| Anaerostipes | 0.000 | 0.005 | 2.7E-08 |
| Coprococcus | 0.000 | 0.004 | 3.0E-08 |
| Romboutsia | 0.000 | 0.014 | 4.2E-06 |
| Ruminococcus | 0.007 | 0.025 | 8.3E-06 |
| Dorea | 0.000 | 0.007 | 3.4E-05 |
| Enterococcus | 0.002 | 0.000 | 1.3E-04 |
| Faecalibacterium | 0.000 | 0.019 | 1.4E-04 |
| Fusicatenibacter | 0.000 | 0.006 | 0.001 |
| Oscillibacter | 0.001 | 0.008 | 0.001 |
| Ruminiclostridium | 0.005 | 0.021 | 0.002 |
Interestingly, a similar decrease in the abundance of protective bacteria was also observed in nontransplant patients affected by diarrhea[108].
Notably, the specimens from transplanted patients with diarrhea were negative for known bacterial and protozoan pathogens that cause diarrhea.
Finally, two transplanted patients affected by persistent diarrhea underwent FMT from allogeneic donors. Diarrhea resolved in the first month after FMT, and the abundances of 13 protective bacteria taxa increased with a simultaneous decrease in the abundances of the 3 identified pathobionts or pathogenic bacterial taxa[96,108].
SCFAs are produced in the gut by the indigenous microbiota and have a trophyc action on the gut epithelium. In addition, these substances exert an anti-inflammatory effect on the whole body and regulate immune cells.
Ninety-five percent of SFCAs are represented by acetic acid, propionic acid, butyric acid and valeric acid, all of which are derived from saccharolytic fermentation. Under normal conditions with a microbiota producing normal quantities of SCFAs, several beneficial effects have been documented after transplantation both in animals and in humans.
In humans, SCFAs increase the expression of antimicrobial peptides secreted to the external surface by epithelial cells[109]. Studies in vitro or in animals documented that SCFAs modulate the production of immune mediators, including IL-18 and other cytokines and chemokines[110], regulate the differentiation, recruitment and activation of immune cells, including neutrophils[111], DCs, macrophages[112] and T lympho-cytes[113].
Finally, Wu et al[114] documented, in a murine kidney transplantation model, that SCFAs are able to induce donor-specific tolerance by inducing the production of T regulatory cells[114].
Andrade-Oliveira et al[115] evaluated the effect of SFCAs on a mouse model of ischemia-reperfusion[115].
In the animals, the treatment with SCFAs improved renal function after ischemia-reperfusion injury, reduced the apoptosis, inhibited NFkB activation and nitric oxide production and reactive oxygen species production. All these actions of SCFAs are summarized in Table 7.
| Actions |
| SCFAs improve renal function |
| SCFAs decrease apoptosis and increase tubular proliferating cells |
| SCFAs decrease activation of bone marrow derived dendritic cells and inhibit their function as antigen presenting cells |
| SCFAs inhibit NFkB activation and nitric oxide production |
| SFCAs inhibit ROS production |
In mice, SCFAs decrease the activation of bone marrow-derived DCs and inhibit their function as antigen presenting cells[115].
In conclusion, the authors showed that SFCA supplementation reduces inflammation in their model and improves ischemia-reperfusion injury.
To our knowledge, few studies have been conducted in humans. A recent study by Lee et al[116] studied 168 kidney transplant recipients and divided the patients according to whether they had higher levels of butyrate-producing bacteria (BPG) or low levels of BPG. The posttransplant administration of antibiotics was associated with a decrease in BPG levels. These patients have a higher incidence of respiratory tract infections.
For the first time, the clinically beneficial effects of higher butyrate levels and posttransplant-induced dysbiosis were documented in transplanted men and may induce higher infection rates.
Similarly, in another study on transplanted humans, 51 renal transplanted recipients have been followed up to 12 mo after transplantation to study the serum levels of uremic toxins as p cresyl sulfate, p cresyl glucoronide, indoxyl sulfate, TMAO and phenylacetylglutamine. The results were compared with CKD control patients with similar renal function. The study documented that after transplantation the colonic microbiota derived uremic retention solutes decreases. As the urinary excretion is lower in transplanted patients, this fact suggests an independent effect after transplantation on intestinal uptake and a different colonic microbial metabolism and absorption[117].
The aforementioned hypothesis that gut microbioma metabolites such as SCFAs could induce donor-specific tolerance through the induction of regulatory T cell differentiations[114], introduces the chapter on the relationship between microbiota and tolerance.
This relationship is well known in the development of immune tolerance in children. Indeed, in the first 1000 d of life, the early exposure of food allergens to indigenous intestinal microbiota induces tolerance through activation of Tregs and subsequent production of TGFβ and IL-10[118].
In a recent study, Colas et al[119] examined the urinary microbiota of 86 renal transplant patients. Patients were divided into 3 groups: Normally immuno-suppressed with stable renal function, minimally immunosuppressed, and spontaneously tolerant patients. Differences in microbiota profiles were observed, and a unique and specific urinary microbiota was detected in patients with spontaneous tolerance characterized by a clear Proteobacteria profile. The profile was different in patients stratified according to gender (higher in males) and inversely correlated with the quantity of immunosuppressive drugs.
The Proteobacteria detected in tolerant subjects included Janthinobacterium, Clostridia and Firmicutes. Janthinobacterium is known to produce an indole-derived peptide with antiproliferative and anti-inflammatory activities[120,121]. Clostridia exert an anti-inflammatory effect by producing SCFAs[122]. Firmicutes produce indole derivatives[123] and polyphosphate[124] with anti-inflammatory activities.
In conclusion, the indigenous microbiota may favor the induction of tolerance, but the use of immunosuppressants modifying the microbiota may represent an obstacle to the development of the tolerance state.
Bilateral actions between the microbiota and immunosuppressive drugs have been identified. On one hand, the microbiota may modify the absorption and the meta-bolism of immunosuppressants; on the other hand, immunosuppressants may modify the indigenous microbiota.
The vast majority of studies on this issue have been conducted on calcineurine inhibitors.
Several studies have extensively documented that factors such as age, gender, race and CYP3A5 polymorphisms influence the absorption and metabolism of immuno-suppressants and account for interindividual variability such that the individual dosing is not the same for all patients.
Recently, the gut indigenous microbiota or the pathobionts have been suspected to exert a powerful effect, justifying the different metabolism from one patient to another and in the same subject.
The assumption of other drugs, such as antibiotics, modifying the indigenous microbiota may account for this variability[125-128].
Lee et al[129] examined the microbiota in the fecal specimens of 19 patients who received a kidney transplant and were on tacrolimus (TAC) as the principal immuno-suppressive therapy. All patients received the same prophylactic antibiotic therapy to avoid biases. Patients were divided into two groups according to the need to receive increasing TAC doses (Dose Escalation Group) or not (Dose Stable Group). By examining the microbiota, the authors found a significantly higher level of Faecalibacterium prausnitzii in patients from the Dose Escalation Group than in patients from the Dose Stable Group. In addition, Faecalibacterium prausnitzii was the most significant factor justifying the need to increase the TAC dose. Even if a large quantity of TAC is absorbed by the small intestine, it may also be absorbed in the colon[130]. Although the Lee’s study is a pilot one, the results raise the question of the relevance of microbiota and of Faecalibacterium prausnitzii, particularly on TAC trough levels, which are also important due to the narrow therapeutic index of TAC.
In a different study, Guo et al[131] incubated Faecalibacterium prausnitzii cells in vitro with TAC. The authors detected a compound named M1 that is a cheto-produced metabolite of TAC with a less powerful immunosuppressant. The authors measured a large quantity of M1 in the stool samples of patients with a larger quantity of Faecalibacterium prausnitzii in the stool.
In addition, the same study documented that other bacteria, such as Clostridia and Bacteroidales, are able to convert TAC into M1 metabolites. The authors conclude that several commensal microbiota may metabolize TAC in the gut to less powerful compounds, explaining the differences in TAC exposure in transplant recipients.
On one hand, the microbiota may alter the metabolism of immunosuppressants; on the other hand, immunosuppressants may alter the gut indigenous microbiota. The study by Gibson et al[132] reviewed this topic extensively. Unfortunately the vast majority of studies have been conducted on calcineurine inhibitors and very few have examined renal transplantation.
The studies by Zhang et al[133] and by Lee et al[129] documented the effect of TAC on the gut microbiota in renal transplant recipients. Other studies[134] analyzed the same phenomenon in liver transplant recipients. Zaza et al[135] examined the microbiota in patients receiving TAC + MMF or everolimus + MMF, but they did not observe any difference.
In the pilot study by Lee et al [79], patients with early corticosteroid withdrawal had fewer Clostridiales and Erysipelotrichaeles in the microbiota, but the difference was not statistically significant.
Finally, a recent study[136] documenting that encapsulated cyclosporine A does not change the composition of the human indigenous microbiota is worth mentioning.
The treatment of gut dysbiosis may be divided into probiotics, smart bacteria, prebiotics, a high-fiber diet and fecal microbiota transplantation.
Several of these therapies have been used in patients affected by chronic kidney disease.
Probiotics are defined by the World Health Organization as live organisms that, when administered in adequate amounts, confer a health benefit to the host[137]. Probiotics such as Lactobacilli and Streptococci[138,139] have been used to treat CKD. They are able to enhance gut barriers, improve mucosal immunity and modulate the host signaling pathways by reducing the activation of NFkB and the MAPK[140,141]. Smart bacteria are genetically modified bacteria that are able to remove toxic molecules in animal studies[142,143].
Prebiotics are nonviable food components that confer health benefits to the host associated with the modulation of the microbiota[144]. A prebiotic must be resistant to gastric acid and digestive enzymes, allowing it to reach the small and the large intestines to stimulate the activity of beneficial microbes. To date, only insulin and trans-galacto-oligosaccharides have these characteristics and may be considered prebiotics[145].
The principal mechanisms of action of prebiotics are to increase the production of SCFAs and to decrease the intestinal pH[146].
Unfortunately, the vast majority of studies using these therapies have been conducted in animal models of CKD.
Few studies have assess probiotics in humans, particularly kidney transplant recipients and most studies were conducted in liver transplant patients[5,95].
Currently, the most effective treatment for renal transplant recipients appears to be FMT, principally in patients affected by infection and/or diarrhea due to resistant Clostridium difficile or E. coli[79,96].
Two main issues are involved in the search for new perspectives: the search for new therapies and an improved knowledge of gut microbiota and pathobionts.
New therapies: Potential benefits of nutritional and supplementation approaches may target microbiota in CKD patients. In CKD, nutritional management and supplementation, including salt and protein restriction, vegetable intakes, and the use of pro-, pre-, and synbiotics, has several benefits. Modulate gut microbiota dysbiosis, decrease colonic production of proteolytic derived uremic toxins and reduce inflammation and oxidative stress[147].
Strategies targeting the microbial source of immune regulation are also promising. The presence of Lactobacillales in the gut microbiota promotes Treg cells and suppresses Th17 in the kidney. The oral administration of Lacidophilus ATCC4356 in the animals attenuates atherosclerotic progression[148].
Lubiprostone, a synthetic derivative of prostaglandin, in a rat model of CKD is associated with reduction of kidney inflammation and improvement of microbioma profile with proliferation of saccarolytic bacteria.
Similarly, the trimethylamine inhibitor 3,3-dimethyl-1-butanol inhibits the atherosclerotic lesions in mice[149].
The identification of causative bacteria in the context of kidney disease and the distinction of indigenous microbioma from pathobionts is a technical challenge.
Sequencing techniques and a wide application of metabolomics allowed us for an improved understanding of microbioma in health and diseases.
The National Institute of Diabetes and Digestive and Kidney Diseases is conducting a study (ClinicalTrials.gov Identifier: NCT02572882)[150] aimed to Characterize the Gut Microbiome of Individuals With End-stage Renal Disease Treated With Maintenance Hemodialysis, and to Explore Effects of P-inulin on the Gut Microbiome.
Future studies should explore the interaction of microbioma with human genoma and how the microbioma should be treated in the case of renal disease and renal transplantation[137].
In the last decade, relevant importance in conditioning both the healthy status and several diseases has been assumed by the microbiota. The microbiota is defined as the microorganisms that live in our body.
Gut microbiota has an important function because can metabolize food and produce substances as SCFAs extremely useful for the body. In addition, the microbiota has important relationship with the immune system and, when modified may induce abnormal activation of the immunity that may cause disease.
Renal diseases may be induced by dysbiosis both for the activation of the immune system and for the production of an excess of uremic system.
In several renal diseases and in particular in the case of end stage renal disease the normal microbiota changes with development of pathobionts and the consequent dysbiosis is responsible for the further deterioration of the renal function.
In the case of renal transplantation, the microbiota has a relevant function.
After transplantation, because of the assumption of immunosuppressive drugs and of prophylactic antibiosis, the gut indigenous microbiota profile modifies, particularly in the first month after transplantation. This modification may influence the graft outcomes causing acute rejection, infections, renal fibrosis and modifications of the drug metabolism, immunosuppressants included. It is possible to modify an abnormal microbiota with the use of prebiotics, probiotics and diet modification.
It should be highlighted that all the studies referring to the microbiota in renal transplantation are few, refer to small number of patients, often retrospectives. In addition, many of these studies have been conducted in animals. Because of this fact the microbiota in general and in solid organ transplantation in particular may be considered a new frontier in medical studies.
Manuscript source: Invited manuscript
Specialty type: Transplantation
Country/Territory of origin: Italy
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Sales-Campos H, Zhu Y S-Editor: Zhang L L-Editor: A P-Editor: Yuan YY
| 1. | Yamaguchi H, Goto S, Takahashi N, Tsuchida M, Watanabe H, Yamamoto S, Kaneko Y, Higashi K, Mori H, Nakamura Y, Horii A, Kurokawa K, Narita I. Aberrant mucosal immunoreaction to tonsillar microbiota in immunoglobulin A nephropathy. Nephrol Dial Transplant. 2021;36:75-86. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 2. | Coppo R. The Gut-Renal Connection in IgA Nephropathy. Semin Nephrol. 2018;38:504-512. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 59] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 3. | Rollino C. Up-to date of glomerular disease. J Nephrol. 2016;29:461-462. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 4. | Mu Q, Zhang H, Liao X, Lin K, Liu H, Edwards MR, Ahmed SA, Yuan R, Li L, Cecere TE, Branson DB, Kirby JL, Goswami P, Leeth CM, Read KA, Oestreich KJ, Vieson MD, Reilly CM, Luo XM. Control of lupus nephritis by changes of gut microbiota. Microbiome. 2017;5:73. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 189] [Cited by in F6Publishing: 203] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 5. | Ardalan M, Vahed SZ. Gut microbiota and renal transplant outcome. Biomed Pharmacother. 2017;90:229-236. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 6. | Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016;14:e1002533. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2328] [Cited by in F6Publishing: 2502] [Article Influence: 312.8] [Reference Citation Analysis (0)] |
| 7. | Scholtens PA, Oozeer R, Martin R, Amor KB, Knol J. The early settlers: intestinal microbiology in early life. Annu Rev Food Sci Technol. 2012;3:425-447. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 129] [Cited by in F6Publishing: 114] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 8. | Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci. 2011;108 Suppl 1:4578-4585. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1666] [Cited by in F6Publishing: 1619] [Article Influence: 115.6] [Reference Citation Analysis (0)] |
| 9. | Rajilić-Stojanović M, Heilig HG, Molenaar D, Kajander K, Surakka A, Smidt H, de Vos WM. Development and application of the human intestinal tract chip, a phylogenetic microarray: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ Microbiol. 2009;11:1736-1751. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 366] [Cited by in F6Publishing: 385] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 10. | Chow J, Tang H, Mazmanian SK. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr Opin Immunol. 2011;23:473-480. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 284] [Cited by in F6Publishing: 289] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 11. | Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207-214. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8203] [Cited by in F6Publishing: 7225] [Article Influence: 602.1] [Reference Citation Analysis (2)] |
| 12. | Gevers D, Knight R, Petrosino JF, Huang K, McGuire AL, Birren BW, Nelson KE, White O, Methé BA, Huttenhower C. The Human Microbiome Project: a community resource for the healthy human microbiome. PLoS Biol. 2012;10:e1001377. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 284] [Cited by in F6Publishing: 279] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 13. | NIH HMP Working Group. , Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, Baker CC, Di Francesco V, Howcroft TK, Karp RW, Lunsford RD, Wellington CR, Belachew T, Wright M, Giblin C, David H, Mills M, Salomon R, Mullins C, Akolkar B, Begg L, Davis C, Grandison L, Humble M, Khalsa J, Little AR, Peavy H, Pontzer C, Portnoy M, Sayre MH, Starke-Reed P, Zakhari S, Read J, Watson B, Guyer M. The NIH Human Microbiome Project. Genome Res. 2009;19:2317-2323. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1410] [Cited by in F6Publishing: 1285] [Article Influence: 85.7] [Reference Citation Analysis (0)] |
| 14. | Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Jian M, Zhou Y, Li Y, Zhang X, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J; MetaHIT Consortium; Bork P, Ehrlich SD. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59-65. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7052] [Cited by in F6Publishing: 7123] [Article Influence: 508.8] [Reference Citation Analysis (3)] |
| 15. | Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, Khan MT, Zhang J, Li J, Xiao L, Al-Aama J, Zhang D, Lee YS, Kotowska D, Colding C, Tremaroli V, Yin Y, Bergman S, Xu X, Madsen L, Kristiansen K, Dahlgren J, Wang J. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe. 2015;17:852. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 374] [Cited by in F6Publishing: 384] [Article Influence: 42.7] [Reference Citation Analysis (0)] |
| 16. | Anderson JW, Baird P, Davis RH Jr, Ferreri S, Knudtson M, Koraym A, Waters V, Williams CL. Health benefits of dietary fiber. Nutr Rev. 2009;67:188-205. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1158] [Cited by in F6Publishing: 1051] [Article Influence: 70.1] [Reference Citation Analysis (2)] |
| 17. | Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490-D495. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4260] [Cited by in F6Publishing: 4253] [Article Influence: 386.6] [Reference Citation Analysis (0)] |
| 18. | Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294:1-8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1125] [Cited by in F6Publishing: 1267] [Article Influence: 84.5] [Reference Citation Analysis (0)] |
| 19. | Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262-1267. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2838] [Cited by in F6Publishing: 2916] [Article Influence: 243.0] [Reference Citation Analysis (0)] |
| 20. | Rawlings ND, Barrett AJ, Thomas PD, Huang X, Bateman A, Finn RD. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018;46:D624-D632. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 782] [Cited by in F6Publishing: 917] [Article Influence: 183.4] [Reference Citation Analysis (0)] |
| 21. | Portune KJ, Benítez-Páez A, Del Pulgar EM, Cerrudo V, Sanz Y. Gut microbiota, diet, and obesity-related disorders-The good, the bad, and the future challenges. Mol Nutr Food Res. 2017;61. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 112] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 22. | Adak A, Khan MR. An insight into gut microbiota and its functionalities. Cell Mol Life Sci. 2019;76:473-493. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 211] [Cited by in F6Publishing: 236] [Article Influence: 39.3] [Reference Citation Analysis (0)] |
| 23. | Saitoh Y, Suzuki H, Tani K, Nishikawa K, Irie K, Ogura Y, Tamura A, Tsukita S, Fujiyoshi Y. Tight junctions. Structural insight into tight junction disassembly by Clostridium perfringens enterotoxin. Science. 2015;347:775-778. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 142] [Cited by in F6Publishing: 147] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 24. | Yuhan R, Koutsouris A, Savkovic SD, Hecht G. Enteropathogenic Escherichia coli-induced myosin light chain phosphorylation alters intestinal epithelial permeability. Gastroenterology. 1997;113:1873-1882. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 183] [Cited by in F6Publishing: 189] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 25. | Hecht G, Koutsouris A, Pothoulakis C, LaMont JT, Madara JL. Clostridium difficile toxin B disrupts the barrier function of T84 monolayers. Gastroenterology. 1992;102:416-423. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 145] [Cited by in F6Publishing: 128] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 26. | Lee SH. Intestinal permeability regulation by tight junction: implication on inflammatory bowel diseases. Intest Res. 2015;13:11-18. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 408] [Cited by in F6Publishing: 475] [Article Influence: 52.8] [Reference Citation Analysis (0)] |
| 27. | Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313-323. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3154] [Cited by in F6Publishing: 3177] [Article Influence: 211.8] [Reference Citation Analysis (0)] |
| 28. | Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci. 2008;105:15064-15069. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1326] [Cited by in F6Publishing: 1388] [Article Influence: 86.8] [Reference Citation Analysis (1)] |
| 29. | Wilson CL, Ouellette AJ, Satchell DP, Ayabe T, López-Boado YS, Stratman JL, Hultgren SJ, Matrisian LM, Parks WC. Regulation of intestinal alpha-defensin activation by the metalloproteinase matrilysin in innate host defense. Science. 1999;286:113-117. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 799] [Cited by in F6Publishing: 786] [Article Influence: 31.4] [Reference Citation Analysis (0)] |
| 30. | Boneca IG, Dussurget O, Cabanes D, Nahori MA, Sousa S, Lecuit M, Psylinakis E, Bouriotis V, Hugot JP, Giovannini M, Coyle A, Bertin J, Namane A, Rousselle JC, Cayet N, Prévost MC, Balloy V, Chignard M, Philpott DJ, Cossart P, Girardin SE. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc Natl Acad Sci. 2007;104:997-1002. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 296] [Cited by in F6Publishing: 272] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 31. | Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126-1130. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1111] [Cited by in F6Publishing: 1008] [Article Influence: 56.0] [Reference Citation Analysis (0)] |
| 32. | Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662-1665. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1095] [Cited by in F6Publishing: 1056] [Article Influence: 52.8] [Reference Citation Analysis (0)] |
| 33. | Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, Granucci F, Kraehenbuhl JP, Ricciardi-Castagnoli P. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol. 2001;2:361-367. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1870] [Cited by in F6Publishing: 1767] [Article Influence: 76.8] [Reference Citation Analysis (0)] |
| 34. | Rahmel T. [SSC International Guideline 2016 - Management of Sepsis and Septic Shock]. Anasthesiol Intensivmed Notfallmed Schmerzther. 2018;53:142-148. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 35. | Belkaid Y, Harrison OJ. Homeostatic Immunity and the Microbiota. Immunity. 2017;46:562-576. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 591] [Cited by in F6Publishing: 664] [Article Influence: 94.9] [Reference Citation Analysis (0)] |
| 36. | Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283-307. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1139] [Cited by in F6Publishing: 1022] [Article Influence: 46.5] [Reference Citation Analysis (0)] |
| 37. | Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337-349. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1232] [Cited by in F6Publishing: 1305] [Article Influence: 81.6] [Reference Citation Analysis (0)] |
| 38. | Arrieta MC, Stiemsma LT, Amenyogbe N, Brown EM, Finlay B. The intestinal microbiome in early life: health and disease. Front Immunol. 2014;5:427. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 497] [Cited by in F6Publishing: 543] [Article Influence: 54.3] [Reference Citation Analysis (0)] |
| 39. | Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, Xavier RJ, Teixeira MM, Mackay CR. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282-1286. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2007] [Cited by in F6Publishing: 2169] [Article Influence: 144.6] [Reference Citation Analysis (0)] |
| 40. | Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16:341-352. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1428] [Cited by in F6Publishing: 1832] [Article Influence: 261.7] [Reference Citation Analysis (0)] |
| 41. | Kim YG, Udayanga KG, Totsuka N, Weinberg JB, Núñez G, Shibuya A. Gut dysbiosis promotes M2 macrophage polarization and allergic airway inflammation via fungi-induced PGE₂. Cell Host Microbe. 2014;15:95-102. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 225] [Cited by in F6Publishing: 254] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 42. | Zhang D, Chen G, Manwani D, Mortha A, Xu C, Faith JJ, Burk RD, Kunisaki Y, Jang JE, Scheiermann C, Merad M, Frenette PS. Neutrophil ageing is regulated by the microbiome. Nature. 2015;525:528-532. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 465] [Cited by in F6Publishing: 542] [Article Influence: 60.2] [Reference Citation Analysis (0)] |
| 43. | Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science. 2016;352:539-544. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 951] [Cited by in F6Publishing: 1097] [Article Influence: 137.1] [Reference Citation Analysis (0)] |
| 44. | Duan J, Chung H, Troy E, Kasper DL. Microbial colonization drives expansion of IL-1 receptor 1-expressing and IL-17-producing gamma/delta T cells. Cell Host Microbe. 2010;7:140-150. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 157] [Cited by in F6Publishing: 165] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 45. | Paget C, Chow MT, Gherardin NA, Beavis PA, Uldrich AP, Duret H, Hassane M, Souza-Fonseca-Guimaraes F, Mogilenko DA, Staumont-Sallé D, Escalante NK, Hill GR, Neeson P, Ritchie DS, Dombrowicz D, Mallevaey T, Trottein F, Belz GT, Godfrey DI, Smyth MJ. CD3bright signals on γδ T cells identify IL-17A-producing Vγ6Vδ1+ T cells. Immunol Cell Biol. 2015;93:198-212. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 57] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 46. | Bukina Y, Thyhonovska M, Koval M, Marushchak M, Krynytska I, Kamyshnyi A. The effect of immunoregulatory bacteria on the transcriptional activity of Foxp3 and RORyt genes in the gut-associated lymphoid tissue with Salmonella-induced inflammation in the presence of vancomycin and Bacteroides fragilis. Iran J Microbiol. 2020;12:231-241. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 47. | Yang BH, Hagemann S, Mamareli P, Lauer U, Hoffmann U, Beckstette M, Föhse L, Prinz I, Pezoldt J, Suerbaum S, Sparwasser T, Hamann A, Floess S, Huehn J, Lochner M. Foxp3(+) T cells expressing RORγt represent a stable regulatory T-cell effector lineage with enhanced suppressive capacity during intestinal inflammation. Mucosal Immunol. 2016;9:444-457. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 249] [Cited by in F6Publishing: 288] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 48. | Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485-498. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3529] [Cited by in F6Publishing: 3256] [Article Influence: 217.1] [Reference Citation Analysis (0)] |
| 49. | Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337-341. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2568] [Cited by in F6Publishing: 2647] [Article Influence: 189.1] [Reference Citation Analysis (0)] |
| 50. | Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620-625. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1625] [Cited by in F6Publishing: 1637] [Article Influence: 102.3] [Reference Citation Analysis (0)] |
| 51. | Kitching AR, Holdsworth SR. The emergence of TH17 cells as effectors of renal injury. J Am Soc Nephrol. 2011;22:235-238. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 86] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 52. | Krebs CF, Paust HJ, Krohn S, Koyro T, Brix SR, Riedel JH, Bartsch P, Wiech T, Meyer-Schwesinger C, Huang J, Fischer N, Busch P, Mittrücker HW, Steinhoff U, Stockinger B, Perez LG, Wenzel UO, Janneck M, Steinmetz OM, Gagliani N, Stahl RAK, Huber S, Turner JE, Panzer U. Autoimmune Renal Disease Is Exacerbated by S1P-Receptor-1-Dependent Intestinal Th17 Cell Migration to the Kidney. Immunity. 2016;45:1078-1092. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 126] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 53. | Krebs CF, Kapffer S, Paust HJ, Schmidt T, Bennstein SB, Peters A, Stege G, Brix SR, Meyer-Schwesinger C, Müller RU, Turner JE, Steinmetz OM, Wolf G, Stahl RA, Panzer U. MicroRNA-155 drives TH17 immune response and tissue injury in experimental crescentic GN. J Am Soc Nephrol. 2013;24:1955-1965. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 34] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 54. | Velden J, Paust HJ, Hoxha E, Turner JE, Steinmetz OM, Wolf G, Jabs WJ, Özcan F, Beige J, Heering PJ, Schröder S, Kneißler U, Disteldorf E, Mittrücker HW, Stahl RA, Helmchen U, Panzer U. Renal IL-17 expression in human ANCA-associated glomerulonephritis. Am J Physiol Renal Physiol. 2012;302:F1663-F1673. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 48] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 55. | Yang J, Kim CJ, Go YS, Lee HY, Kim MG, Oh SW, Cho WY, Im SH, Jo SK. Intestinal microbiota control acute kidney injury severity by immune modulation. Kidney Int. 2020;98:932-946. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 34] [Article Influence: 8.5] [Reference Citation Analysis (1)] |
| 56. | Rowland I, Gibson G, Heinken A, Scott K, Swann J, Thiele I, Tuohy K. Gut microbiota functions: metabolism of nutrients and other food components. Eur J Nutr. 2018;57:1-24. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 993] [Cited by in F6Publishing: 1253] [Article Influence: 179.0] [Reference Citation Analysis (0)] |
| 57. | Lin L, Zhang J. Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. BMC Immunol. 2017;18:2. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 325] [Cited by in F6Publishing: 377] [Article Influence: 53.9] [Reference Citation Analysis (0)] |
| 58. | Inoue D, Tsujimoto G, Kimura I. Regulation of Energy Homeostasis by GPR41. Front Endocrinol (Lausanne). 2014;5:81. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 66] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 59. | Tolhurst G, Heffron H, Lam YS, Parker HE, Habib AM, Diakogiannaki E, Cameron J, Grosse J, Reimann F, Gribble FM. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes. 2012;61:364-371. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1524] [Cited by in F6Publishing: 1411] [Article Influence: 117.6] [Reference Citation Analysis (0)] |
| 60. | Lukasova M, Malaval C, Gille A, Kero J, Offermanns S. Nicotinic acid inhibits progression of atherosclerosis in mice through its receptor GPR109A expressed by immune cells. J Clin Invest. 2011;121:1163-1173. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 177] [Cited by in F6Publishing: 195] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 61. | Pluznick J. A novel SCFA receptor, the microbiota, and blood pressure regulation. Gut Microbes. 2014;5:202-207. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 232] [Cited by in F6Publishing: 231] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 62. | Singh N, Thangaraju M, Prasad PD, Martin PM, Lambert NA, Boettger T, Offermanns S, Ganapathy V. Blockade of dendritic cell development by bacterial fermentation products butyrate and propionate through a transporter (Slc5a8)-dependent inhibition of histone deacetylases. J Biol Chem. 2010;285:27601-27608. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 164] [Cited by in F6Publishing: 192] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 63. | Andrade-Oliveira V, Foresto-Neto O, Watanabe IKM, Zatz R, Câmara NOS. Inflammation in Renal Diseases: New and Old Players. Front Pharmacol. 2019;10:1192. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 123] [Cited by in F6Publishing: 176] [Article Influence: 35.2] [Reference Citation Analysis (0)] |
| 64. | Matsumoto T, Kojima M, Takayanagi K, Taguchi K, Kobayashi T. Role of S-Equol, Indoxyl Sulfate, and Trimethylamine N-Oxide on Vascular Function. Am J Hypertens. 2020;33:793-803. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 65. | Tang WH, Wang Z, Kennedy DJ, Wu Y, Buffa JA, Agatisa-Boyle B, Li XS, Levison BS, Hazen SL. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ Res. 2015;116:448-455. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 666] [Cited by in F6Publishing: 785] [Article Influence: 78.5] [Reference Citation Analysis (0)] |
| 66. | Satoh M, Hayashi H, Watanabe M, Ueda K, Yamato H, Yoshioka T, Motojima M. Uremic toxins overload accelerates renal damage in a rat model of chronic renal failure. Nephron Exp Nephrol. 2003;95:e111-e118. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 84] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 67. | Noel S, Martina-Lingua MN, Bandapalle S, Pluznick J, Hamad AR, Peterson DA, Rabb H. Intestinal microbiota-kidney cross talk in acute kidney injury and chronic kidney disease. Nephron Clin Pract. 2014;127:139-143. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 68. | Vaziri ND, Wong J, Pahl M, Piceno YM, Yuan J, DeSantis TZ, Ni Z, Nguyen TH, Andersen GL. Chronic kidney disease alters intestinal microbial flora. Kidney Int. 2013;83:308-315. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 636] [Cited by in F6Publishing: 726] [Article Influence: 60.5] [Reference Citation Analysis (0)] |
| 69. | Cigarran Guldris S, González Parra E, Cases Amenós A. Gut microbiota in chronic kidney disease. Nefrologia. 2017;37:9-19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 62] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 70. | Aron-Wisnewsky J, Clément K. The gut microbiome, diet, and links to cardiometabolic and chronic disorders. Nat Rev Nephrol. 2016;12:169-181. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 197] [Cited by in F6Publishing: 210] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 71. | Sabatino A, Regolisti G, Brusasco I, Cabassi A, Morabito S, Fiaccadori E. Alterations of intestinal barrier and microbiota in chronic kidney disease. Nephrol Dial Transplant. 2015;30:924-933. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 138] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 72. | Guo TL, Chen Y, Xu HS, McDonough CM, Huang G. Gut microbiome in neuroendocrine and neuroimmune interactions: The case of genistein. Toxicol Appl Pharmacol. 2020;402:115130. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 73. | Anders HJ, Andersen K, Stecher B. The intestinal microbiota, a leaky gut, and abnormal immunity in kidney disease. Kidney Int. 2013;83:1010-1016. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 292] [Cited by in F6Publishing: 302] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 74. | Ramezani A, Raj DS. The gut microbiome, kidney disease, and targeted interventions. J Am Soc Nephrol. 2014;25:657-670. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 423] [Cited by in F6Publishing: 470] [Article Influence: 42.7] [Reference Citation Analysis (0)] |
| 75. | Evenepoel P, Poesen R, Meijers B. The gut-kidney axis. Pediatr Nephrol. 2017;32:2005-2014. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 162] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 76. | Koppe L, Mafra D, Fouque D. Probiotics and chronic kidney disease. Kidney Int. 2015;88:958-966. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 130] [Cited by in F6Publishing: 142] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 77. | Ramezani A, Massy ZA, Meijers B, Evenepoel P, Vanholder R, Raj DS. Role of the Gut Microbiome in Uremia: A Potential Therapeutic Target. Am J Kidney Dis. 2016;67:483-498. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 198] [Cited by in F6Publishing: 240] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 78. | Rapone B, Ferrara E, Santacroce L, Cesarano F, Arazzi M, Liberato LD, Scacco S, Grassi R, Grassi FR, Gnoni A, Nardi GM. Periodontal Microbiological Status Influences the Occurrence of Cyclosporine-A and Tacrolimus-Induced Gingival Overgrowth. Antibiotics (Basel). 2019;8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 79. | Lee JR, Muthukumar T, Dadhania D, Toussaint NC, Ling L, Pamer E, Suthanthiran M. Gut microbial community structure and complications after kidney transplantation: a pilot study. Transplantation. 2014;98:697-705. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 103] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 80. | Integrative HMP (iHMP) Research Network Consortium. The Integrative Human Microbiome Project. Nature. 2019;569:641-648. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 669] [Cited by in F6Publishing: 585] [Article Influence: 117.0] [Reference Citation Analysis (0)] |
| 81. | Taur Y, Xavier JB, Lipuma L, Ubeda C, Goldberg J, Gobourne A, Lee YJ, Dubin KA, Socci ND, Viale A, Perales MA, Jenq RR, van den Brink MR, Pamer EG. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis. 2012;55:905-914. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 609] [Cited by in F6Publishing: 662] [Article Influence: 55.2] [Reference Citation Analysis (0)] |
| 82. | Xiao J, Peng Z, Liao Y, Sun H, Chen W, Chen X, Wei Z, Yang C, Nüssler AK, Liu J, Yang W. Organ transplantation and gut microbiota: current reviews and future challenges. Am J Transl Res. 2018;10:3330-3344. [PubMed] [Cited in This Article: ] |
| 83. | Swarte JC, Douwes RM, Hu S, Vich Vila A, Eisenga MF, van Londen M, Gomes-Neto AW, Weersma RK, Harmsen HJM, Bakker SJL. Characteristics and Dysbiosis of the Gut Microbiome in Renal Transplant Recipients. J Clin Med. 2020;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 47] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 84. | Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33:496-503. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1476] [Cited by in F6Publishing: 1933] [Article Influence: 214.8] [Reference Citation Analysis (0)] |
| 85. | Flint HJ, Duncan SH, Scott KP, Louis P. Links between diet, gut microbiota composition and gut metabolism. Proc Nutr Soc. 2015;74:13-22. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 512] [Cited by in F6Publishing: 558] [Article Influence: 55.8] [Reference Citation Analysis (0)] |
| 86. | Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446-450. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2951] [Cited by in F6Publishing: 3272] [Article Influence: 297.5] [Reference Citation Analysis (0)] |
| 87. | Sampaio-Maia B, Simões-Silva L, Pestana M, Araujo R, Soares-Silva IJ. The Role of the Gut Microbiome on Chronic Kidney Disease. Adv Appl Microbiol. 2016;96:65-94. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 73] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 88. | Imhann F, Bonder MJ, Vich Vila A, Fu J, Mujagic Z, Vork L, Tigchelaar EF, Jankipersadsing SA, Cenit MC, Harmsen HJ, Dijkstra G, Franke L, Xavier RJ, Jonkers D, Wijmenga C, Weersma RK, Zhernakova A. Proton pump inhibitors affect the gut microbiome. Gut. 2016;65:740-748. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 780] [Cited by in F6Publishing: 770] [Article Influence: 96.3] [Reference Citation Analysis (0)] |
| 89. | Spasić A, Catić-Đorđević A, Veličković-Radovanović R, Stefanović N, Džodić P, Cvetković T. Adverse effects of mycophenolic acid in renal transplant recipients: gender differences. Int J Clin Pharm. 2019;41:776-784. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 90. | Westblade LF, Satlin MJ, Albakry S, Botticelli B, Robertson A, Alston T, Magruder M, Zhang LT, Edusei E, Chan K, Lubetzky M, Dadhania DM, Pamer EG, Suthanthiran M, Lee JR. Gastrointestinal pathogen colonization and the microbiome in asymptomatic kidney transplant recipients. Transpl Infect Dis. 2019;21:e13167. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 91. | Wang W, Xu S, Ren Z, Jiang J, Zheng S. Gut microbiota and allogeneic transplantation. J Transl Med. 2015;13:275. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 58] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 92. | Ahmad S, Bromberg JS. Current status of the microbiome in renal transplantation. Curr Opin Nephrol Hypertens. 2016;25:570-576. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 93. | Lei YM, Chen L, Wang Y, Stefka AT, Molinero LL, Theriault B, Aquino-Michaels K, Sivan AS, Nagler CR, Gajewski TF, Chong AS, Bartman C, Alegre ML. The composition of the microbiota modulates allograft rejection. J Clin Invest. 2016;126:2736-2744. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 73] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 94. | Fricke WF, Maddox C, Song Y, Bromberg JS. Human microbiota characterization in the course of renal transplantation. Am J Transplant. 2014;14:416-427. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 114] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 95. | Chan S, Hawley CM, Campbell KL, Morrison M, Campbell SB, Isbel NM, Francis RS, Playford EG, Johnson DW. Transplant associated infections-The role of the gastrointestinal microbiota and potential therapeutic options. Nephrology (Carlton). 2020;25:5-13. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 96. | Biehl LM, Cruz Aguilar R, Farowski F, Hahn W, Nowag A, Wisplinghoff H, Vehreschild MJGT. Fecal microbiota transplantation in a kidney transplant recipient with recurrent urinary tract infection. Infection. 2018;46:871-874. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 97. | Cheng J, Torkamani A, Grover RK, Jones TM, Ruiz DI, Schork NJ, Quigley MM, Hall FW, Salomon DR, Lerner RA. Ectopic B-cell clusters that infiltrate transplanted human kidneys are clonal. Proc Natl Acad Sci U S A. 2011;108:5560-5565. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 44] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 98. | Grover RK, Cheng J, Peng Y, Jones TM, Ruiz DI, Ulevitch RJ, Glass JI, Dennis EA, Salomon DR, Lerner RA. The costimulatory immunogen LPS induces the B-Cell clones that infiltrate transplanted human kidneys. Proc Natl Acad Sci U S A. 2012;109:6036-6041. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 99. | Modena BD, Milam R, Harrison F, Cheeseman JA, Abecassis MM, Friedewald JJ, Kirk AD, Salomon DR. Changes in Urinary Microbiome Populations Correlate in Kidney Transplants With Interstitial Fibrosis and Tubular Atrophy Documented in Early Surveillance Biopsies. Am J Transplant. 2017;17:712-723. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 100. | Rieder F. The gut microbiome in intestinal fibrosis: environmental protector or provocateur? Sci Transl Med. 2013;5:190ps10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 48] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 101. | mofetil in renal transplantation: 3-year results from the placebo-controlled trial. European Mycophenolate Mofetil Cooperative Study Group. Transplantation. 1999;68:391-396. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 189] [Cited by in F6Publishing: 189] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 102. | Bunnapradist S, Neri L, Wong W, Lentine KL, Burroughs TE, Pinsky BW, Takemoto SK, Schnitzler MA. Incidence and risk factors for diarrhea following kidney transplantation and association with graft loss and mortality. Am J Kidney Dis. 2008;51:478-486. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 112] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 103. | Bunnapradist S, Lentine KL, Burroughs TE, Pinsky BW, Hardinger KL, Brennan DC, Schnitzler MA. Mycophenolate mofetil dose reductions and discontinuations after gastrointestinal complications are associated with renal transplant graft failure. Transplantation. 2006;82:102-107. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 127] [Cited by in F6Publishing: 131] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 104. | Lee JR, Magruder M, Zhang L, Westblade LF, Satlin MJ, Robertson A, Edusei E, Crawford C, Ling L, Taur Y, Schluter J, Lubetzky M, Dadhania D, Pamer E, Suthanthiran M. Gut microbiota dysbiosis and diarrhea in kidney transplant recipients. Am J Transplant. 2019;19:488-500. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 59] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 105. | Lopetuso LR, Scaldaferri F, Petito V, Gasbarrini A. Commensal Clostridia: leading players in the maintenance of gut homeostasis. Gut Pathog. 2013;5:23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 445] [Cited by in F6Publishing: 489] [Article Influence: 44.5] [Reference Citation Analysis (0)] |
| 106. | Rajilić-Stojanović M, de Vos WM. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev. 2014;38:996-1047. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 765] [Cited by in F6Publishing: 693] [Article Influence: 69.3] [Reference Citation Analysis (0)] |
| 107. | Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814-821. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5694] [Cited by in F6Publishing: 5765] [Article Influence: 524.1] [Reference Citation Analysis (0)] |
| 108. | Antharam VC, Li EC, Ishmael A, Sharma A, Mai V, Rand KH, Wang GP. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J Clin Microbiol. 2013;51:2884-2892. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 341] [Cited by in F6Publishing: 349] [Article Influence: 31.7] [Reference Citation Analysis (0)] |
| 109. | Raqib R, Sarker P, Bergman P, Ara G, Lindh M, Sack DA, Nasirul Islam KM, Gudmundsson GH, Andersson J, Agerberth B. Improved outcome in shigellosis associated with butyrate induction of an endogenous peptide antibiotic. Proc Natl Acad Sci. 2006;103:9178-9183. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 201] [Cited by in F6Publishing: 220] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 110. | Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, Bayless AJ, Wilson KE, Glover LE, Kominsky DJ, Magnuson A, Weir TL, Ehrentraut SF, Pickel C, Kuhn KA, Lanis JM, Nguyen V, Taylor CT, Colgan SP. Crosstalk between Microbiota-Derived Short-Chain Fatty Acids and Intestinal Epithelial HIF Augments Tissue Barrier Function. Cell Host Microbe. 2015;17:662-671. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 807] [Cited by in F6Publishing: 986] [Article Influence: 109.6] [Reference Citation Analysis (0)] |
| 111. | Rodrigues HG, Takeo Sato F, Curi R, Vinolo MAR. Fatty acids as modulators of neutrophil recruitment, function and survival. Eur J Pharmacol. 2016;785:50-58. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 59] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 112. | Millard AL, Mertes PM, Ittelet D, Villard F, Jeannesson P, Bernard J. Butyrate affects differentiation, maturation and function of human monocyte-derived dendritic cells and macrophages. Clin Exp Immunol. 2002;130:245-255. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 150] [Cited by in F6Publishing: 147] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 113. | Park J, Kim M, Kang SG, Jannasch AH, Cooper B, Patterson J, Kim CH. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol. 2015;8:80-93. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 575] [Cited by in F6Publishing: 693] [Article Influence: 77.0] [Reference Citation Analysis (0)] |
| 114. | Wu H, Singer J, Kwan TK, Loh YW, Wang C, Tan J, Li YJ, Lai SWC, Macia L, Alexander SI, Chadban SJ. Gut Microbial Metabolites Induce Donor-Specific Tolerance of Kidney Allografts through Induction of T Regulatory Cells by Short-Chain Fatty Acids. J Am Soc Nephrol. 2020;31:1445-1461. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 40] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 115. | Andrade-Oliveira V, Amano MT, Correa-Costa M, Castoldi A, Felizardo RJ, de Almeida DC, Bassi EJ, Moraes-Vieira PM, Hiyane MI, Rodas AC, Peron JP, Aguiar CF, Reis MA, Ribeiro WR, Valduga CJ, Curi R, Vinolo MA, Ferreira CM, Câmara NO. Gut Bacteria Products Prevent AKI Induced by Ischemia-Reperfusion. J Am Soc Nephrol. 2015;26:1877-1888. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 269] [Cited by in F6Publishing: 333] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 116. | Lee JR, Huang J, Magruder M, Zhang LT, Gong C, Sholi AN, Albakry S, Edusei E, Muthukumar T, Lubetzky M, Dadhania DM, Taur Y, Pamer EG, Suthanthiran M. Butyrate-producing gut bacteria and viral infections in kidney transplant recipients: A pilot study. Transpl Infect Dis. 2019;21:e13180. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 117. | Poesen R, Evenepoel P, de Loor H, Bammens B, Claes K, Sprangers B, Naesens M, Kuypers D, Augustijns P, Meijers B. The influence of renal transplantation on retained microbial-human co-metabolites. Nephrol Dial Transplant. 2016;31:1721-1729. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 118. | Vitetta L, Vitetta G, Hall S. Immunological Tolerance and Function: Associations Between Intestinal Bacteria, Probiotics, Prebiotics, and Phages. Front Immunol. 2018;9:2240. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 95] [Cited by in F6Publishing: 80] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 119. | Colas L, Mongodin EF, Montassier E, Chesneau M, Guerif P, Hittle L, Giral M, Bromberg JS, Brouard S; DIVAT Consortium. Unique and specific Proteobacteria diversity in urinary microbiota of tolerant kidney transplanted recipients. Am J Transplant. 2020;20:145-158. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 120. | Masuelli L, Pantanella F, La Regina G, Benvenuto M, Fantini M, Mattera R, Di Stefano E, Mattei M, Silvestri R, Schippa S, Manzari V, Modesti A, Bei R. Violacein, an indole-derived purple-colored natural pigment produced by Janthinobacterium lividum, inhibits the growth of head and neck carcinoma cell lines both in vitro and in vivo. Tumour Biol. 2016;37:3705-3717. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 121. | Verinaud L, Lopes SC, Prado IC, Zanucoli F, Alves da Costa T, Di Gangi R, Issayama LK, Carvalho AC, Bonfanti AP, Niederauer GF, Duran N, Costa FT, Oliveira AL, Höfling MA, Machado DR, Thomé R. Violacein Treatment Modulates Acute and Chronic Inflammation through the Suppression of Cytokine Production and Induction of Regulatory T Cells. PLoS One. 2015;10:e0125409. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 122. | Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, Kim S, Fritz JV, Wilmes P, Ueha S, Matsushima K, Ohno H, Olle B, Sakaguchi S, Taniguchi T, Morita H, Hattori M, Honda K. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232-236. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1875] [Cited by in F6Publishing: 1929] [Article Influence: 175.4] [Reference Citation Analysis (2)] |
| 123. | Etienne-Mesmin L, Chassaing B, Gewirtz AT. Tryptophan: A gut microbiota-derived metabolites regulating inflammation. World J Gastrointest Pharmacol Ther. 2017;8:7-9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 47] [Cited by in F6Publishing: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 124. | Segawa S, Fujiya M, Konishi H, Ueno N, Kobayashi N, Shigyo T, Kohgo Y. Probiotic-derived polyphosphate enhances the epithelial barrier function and maintains intestinal homeostasis through integrin-p38 MAPK pathway. PLoS One. 2011;6:e23278. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 145] [Cited by in F6Publishing: 141] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 125. | Teuteberg JJ, Shullo MA, Zomak R, Toyoda Y, McNamara DM, Bermudez C, Kormos RL, McCurry KR. Alemtuzumab induction prior to cardiac transplantation with lower intensity maintenance immunosuppression: one-year outcomes. Am J Transplant. 2010;10:382-388. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 126. | Federico S, Carrano R, Capone D, Gentile A, Palmiero G, Basile V. Pharmacokinetic interaction between levofloxacin and ciclosporin or tacrolimus in kidney transplant recipients: ciclosporin, tacrolimus and levofloxacin in renal transplantation. Clin Pharmacokinet. 2006;45:169-175. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 30] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 127. | Page RL 2nd, Klem PM, Rogers C. Potential elevation of tacrolimus trough concentrations with concomitant metronidazole therapy. Ann Pharmacother. 2005;39:1109-1113. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 31] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 128. | Zheng Y, Masand A, Wagner M, Kapur S, Dadhania D, Lubetzky M, Lee JR. Identification of Antibiotic Administration as a Potentially Novel Factor Associated With Tacrolimus Trough Variability in Kidney Transplant Recipients: A Preliminary Study. Transplant Direct. 2019;5:e485. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 129. | Lee JR, Muthukumar T, Dadhania D, Taur Y, Jenq RR, Toussaint NC, Ling L, Pamer E, Suthanthiran M. Gut microbiota and tacrolimus dosing in kidney transplantation. PLoS One. 2015;10:e0122399. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 107] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 130. | Tsunashima D, Kawamura A, Murakami M, Sawamoto T, Undre N, Brown M, Groenewoud A, Keirns JJ, Holman J, Connor A, Wylde H, Wilding I, Ogawara K, Sako K, Higaki K, First R. Assessment of tacrolimus absorption from the human intestinal tract: open-label, randomized, 4-way crossover study. Clin Ther. 2014;36:748-759. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 41] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 131. | Guo Y, Crnkovic CM, Won KJ, Yang X, Lee JR, Orjala J, Lee H, Jeong H. Commensal Gut Bacteria Convert the Immunosuppressant Tacrolimus to Less Potent Metabolites. Drug Metab Dispos. 2019;47:194-202. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 67] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 132. | Gibson CM, Childs-Kean LM, Naziruddin Z, Howell CK. The alteration of the gut microbiome by immunosuppressive agents used in solid organ transplantation. Transpl Infect Dis. 2020: e13397. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 133. | Zhang Z, Liu L, Tang H, Jiao W, Zeng S, Xu Y, Zhang Q, Sun Z, Mukherjee A, Zhang X, Hu X. Immunosuppressive effect of the gut microbiome altered by high-dose tacrolimus in mice. Am J Transplant. 2018;18:1646-1656. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 62] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 134. | Lu H, He J, Wu Z, Xu W, Zhang H, Ye P, Yang J, Zhen S, Li L. Assessment of microbiome variation during the perioperative period in liver transplant patients: a retrospective analysis. Microb Ecol. 2013;65:781-791. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 59] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 135. | Zaza G, Dalla Gassa A, Felis G, Granata S, Torriani S, Lupo A. Impact of maintenance immunosuppressive therapy on the fecal microbiome of renal transplant recipients: Comparison between an everolimus- and a standard tacrolimus-based regimen. PLoS One. 2017;12:e0178228. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 136. | O'Reilly C, O'Sullivan Ó, Cotter PD, O'Connor PM, Shanahan F, Cullen A, Rea MC, Hill C, Coulter I, Ross RP. Encapsulated cyclosporine does not change the composition of the human microbiota when assessed ex vivo and in vivo. J Med Microbiol. 2020;69:854-863. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 137. | Nallu A, Sharma S, Ramezani A, Muralidharan J, Raj D. Gut microbiome in chronic kidney disease: challenges and opportunities. Transl Res. 2017;179:24-37. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 145] [Cited by in F6Publishing: 152] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 138. | Konstantinov SR, Smidt H, de Vos WM, Bruijns SC, Singh SK, Valence F, Molle D, Lortal S, Altermann E, Klaenhammer TR, van Kooyk Y. S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc Natl Acad Sci. 2008;105:19474-19479. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 395] [Cited by in F6Publishing: 404] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 139. | van Baarlen P, Troost FJ, van Hemert S, van der Meer C, de Vos WM, de Groot PJ, Hooiveld GJ, Brummer RJ, Kleerebezem M. Differential NF-kappaB pathways induction by Lactobacillus plantarum in the duodenum of healthy humans correlating with immune tolerance. Proc Natl Acad Sci. 2009;106:2371-2376. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 304] [Cited by in F6Publishing: 298] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 140. | Maudet C, Mano M, Eulalio A. MicroRNAs in the interaction between host and bacterial pathogens. FEBS Lett. 2014;588:4140-4147. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 110] [Cited by in F6Publishing: 120] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 141. | Thomas CM, Versalovic J. Probiotics-host communication: Modulation of signaling pathways in the intestine. Gut Microbes. 2010;1:148-163. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 283] [Cited by in F6Publishing: 272] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 142. | Prakash S, Chang TM. Microencapsulated genetically engineered live E. coli DH5 cells administered orally to maintain normal plasma urea level in uremic rats. Nat Med. 1996;2:883-887. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 143] [Cited by in F6Publishing: 148] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 143. | Piñero-Lambea C, Ruano-Gallego D, Fernández LÁ. Engineered bacteria as therapeutic agents. Curr Opin Biotechnol. 2015;35:94-102. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 52] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 144. | Roberfroid M. Prebiotics: the concept revisited. J Nutr. 2007;137:830S-837S. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 920] [Cited by in F6Publishing: 733] [Article Influence: 43.1] [Reference Citation Analysis (0)] |
| 145. | Slavin J. Fiber and prebiotics: mechanisms and health benefits. Nutrients. 2013;5:1417-1435. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1380] [Cited by in F6Publishing: 1050] [Article Influence: 95.5] [Reference Citation Analysis (0)] |
| 146. | de Vrese M, Marteau PR. Probiotics and prebiotics: effects on diarrhea. J Nutr. 2007;137:803S-811S. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 181] [Cited by in F6Publishing: 189] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 147. | Cosola C, Rocchetti MT, Cupisti A, Gesualdo L. Microbiota metabolites: Pivotal players of cardiovascular damage in chronic kidney disease. Pharmacol Res. 2018;130:132-142. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 66] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 148. | Gong J, Noel S, Pluznick JL, Hamad ARA, Rabb H. Gut Microbiota-Kidney Cross-Talk in Acute Kidney Injury. Semin Nephrol. 2019;39:107-116. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 60] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 149. | Wang Z, Roberts AB, Buffa JA, Levison BS, Zhu W, Org E, Gu X, Huang Y, Zamanian-Daryoush M, Culley MK, DiDonato AJ, Fu X, Hazen JE, Krajcik D, DiDonato JA, Lusis AJ, Hazen SL. Non-lethal Inhibition of Gut Microbial Trimethylamine Production for the Treatment of Atherosclerosis. Cell. 2015;163:1585-1595. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 733] [Cited by in F6Publishing: 852] [Article Influence: 106.5] [Reference Citation Analysis (0)] |
| 150. | University of Pennsylvania. Gut Microbiome and p-Inulin in Hemodialysis. [accessed 2021 January 5]. In: ClinicalTrials. gov [Internet]. Bethesda (MD): U.S. National Library of Medicine. Available from: https://clinicaltrials.gov/ct2/show/NCT02572882. . [Cited in This Article: ] |