Published online Dec 24, 2021. doi: 10.5306/wjco.v12.i12.1215
Peer-review started: April 28, 2021
First decision: June 13, 2021
Revised: June 26, 2021
Accepted: November 28, 2021
Article in press: November 28, 2021
Published online: December 24, 2021
The mutation-based analysis of circulating tumor DNA (ctDNA) is a promising diagnostic tool for clinical oncology. However, it has low success rate because many cancer patients do not have detectable ctDNA in the bloodstream.
To evaluate whether preoperative tumor irradiation results in a transient increase of plasma ctDNA concentration due to the induction of apoptosis in radiation-exposed cells.
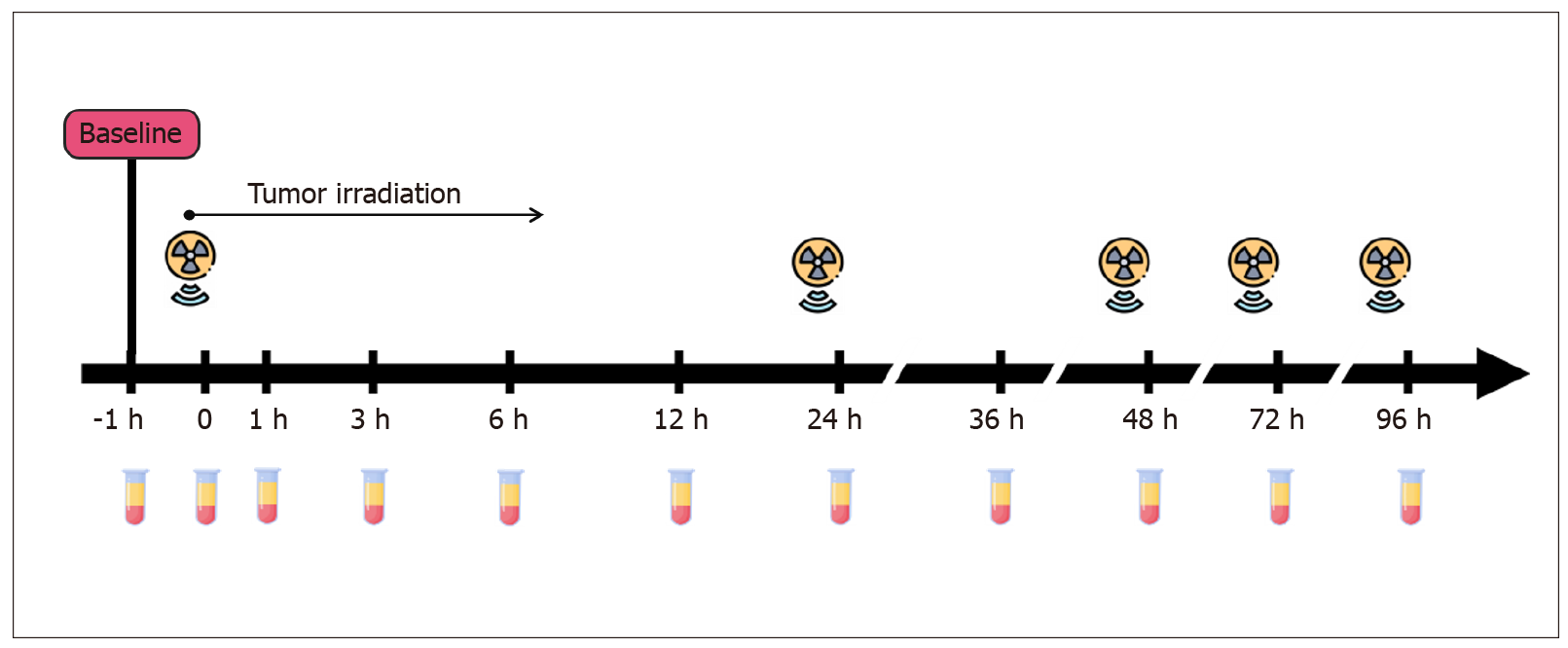
This study focused on patients with locally advanced rectal cancer, because preoperative tumor irradiation is a part of their standard treatment plan. Nine subjects, whose tumors contained KRAS, NRAS or BRAF mutations, donated serial blood samples 1 h prior to the first fraction of irradiation (at baseline), immediately after the first fraction (time 0), and 1, 3, 6, 12, 24, 36, 48, 72 and 96 h after the first fraction. The amount of mutated gene copies was measured by droplet digital PCR.
Five out of nine patients were mutation-negative by ctDNA test at baseline; two of these subjects demonstrated an emergence of the mutated DNA copies in the bloodstream within the follow-up period. There were 4 patients, who had detectable ctDNA in the plasma at the start of the experiment; three of them showed an evident treatment-induced increase of the content of mutated RAS/RAF alleles.
Local tumor irradiation may facilitate the detection of tumor-specific DNA in the bloodstream. These data justify further assessment of the clinical feasibility of irradiation-assisted liquid biopsy.
Core Tip: The detection of circulating tumor DNA (ctDNA) in cancer patients is compromised by the low sensitivity of this assay. We hypothesized that tumor irradiation may lead to the transient increase of ctDNA content due to induction of cell death. Nine patients with locally advanced RAS/RAF-mutated rectal cancer provided serial blood samples at baseline and during the first 96 h after the first dose of tumor irradiation. Treatment-induced elevation of the concentration of mutated RAS/RAF alleles in the blood was revealed in five of these subjects. In conclusion, local tumor irradiation may facilitate the detection of plasma ctDNA and thus improve the efficacy of liquid biopsy.
- Citation: Kuligina E, Moiseyenko F, Belukhin S, Stepanova E, Zakharova M, Chernobrivtseva V, Aliev I, Sharabura T, Moiseyenko V, Aleksakhina S, Laidus T, Martianov A, Kholmatov M, Whitehead A, Yanus G, Imyanitov E. Tumor irradiation may facilitate the detection of tumor-specific mutations in plasma. World J Clin Oncol 2021; 12(12): 1215-1226
- URL: https://www.wjgnet.com/2218-4333/full/v12/i12/1215.htm
- DOI: https://dx.doi.org/10.5306/wjco.v12.i12.1215
“Liquid biopsy” is a popular diagnostic tool, which is based on the identification of tumor-specific markers in plasma or other body fluids. The analysis of several proteins, e.g., prostate-specific antigen, carcinoembryonic antigen, CA-125 etc. has been utilized for years, however, these assays have significant limitations with regard to specificity and sensitivity[1,2]. The examination of tumor-derived mutations in circulating DNA (ctDNA) is considered to be more promising[3]. Indeed, some methods of genetic testing, for instance, droplet digital PCR (ddPCR) or next-generation sequencing allow the detection of a single mutated allele within a huge excess of wild-type nucleic acids[4,5]. In addition, while the protein-based liquid biopsy is not truly cancer-specific but rather tissue-specific, oncogenic mutations are strongly associated with the malignant phenotype. For the time being, clinical use of ctDNA tests is largely limited to the analysis of secondary mutations emerging during targeted therapy, as these assays may help to identify mechanisms of acquired drug resistance and therefore guide the subsequent treatment choice. It is anticipated that in the near future liquid biopsy will support other components of cancer care, such as screening, early diagnosis, analysis of treatment outcome and monitoring of relapse of tumor disease[6-11].
ctDNA-based liquid biopsy may have unacceptably low sensitivity. It appears that many categories of neoplasms (medulloblastomas; gliomas; cancers of kidney, thyroid, breast, etc.) are composed of relatively well-preserved cells, which do not shed DNA in the bloodstream, at least when the tumor is small[12,13]. Consequently, the analysis of ctDNA cannot substitute the detection of mutations in tumors tissue, i.e., tissue biopsy cannot be easily replaced by liquid biopsy. The development of tools, which allow non-invasive examination of tumor characteristics, is of great value. One of the options may involve utilization of various agents, which induce tumor cell death and thus facilitate DNA shedding. In particular, tumor irradiation may increase tumor-specific ctDNA level due to the involvement of the above mechanism[14-16].
While considering the appropriate model for the validation of this assumption, we found it reasonable to focus on rectal cancer. Approximately 40%-50% of rectal carcinomas contain missense mutations in KRAS, NRAS or BRAF oncogenes, which can be used for ctDNA assays[17-19]. Furthermore, many rectal cancer patients undergo preoperative radiotherapy (RT) as a part of the treatment plan. Here we present the results of the study, which involved consecutive patients with mutation-positive rectal cancer. We demonstrate, that tumor irradiation indeed results in a transient increase of concentration of tumor-derived DNA and thus can be considered as a liquid biopsy supporting tool.
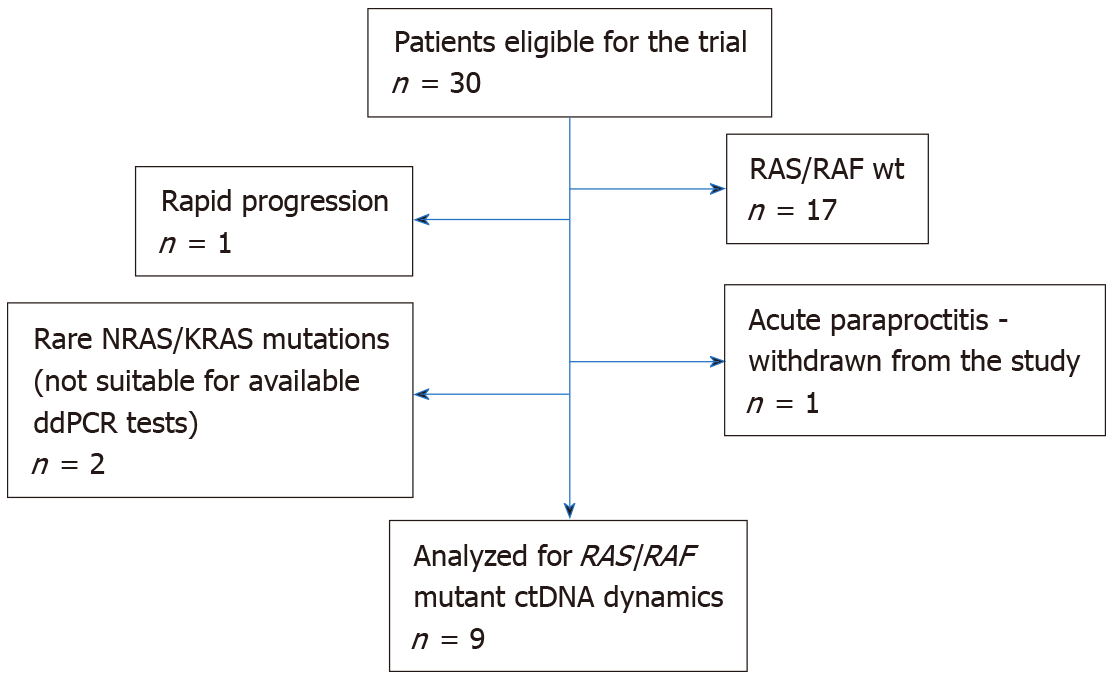
The study considered treatment-naive patients with histologically verified locally advanced rectal cancer (T1-2/N1-2/M0, T3-4/N0-2/M0), who were referred to the St.-Petersburg City Cancer Center between February 2019 and April 2020 and who planned to undergo preoperative RT. The study was approved by the local Ethics Committee. Thirty patients provided informed consent and underwent RAS/RAF mutation testing (Figure 1). Thirteen analyzed tumors carried nucleotide substitutions in the mentioned genes. Four subjects failed to participate in the study due to various reasons (two tumors contained “rare” RAS mutations (KRAS A59G and NRAS G12C), which could not be detected by available ddPCR assays; 1 patient experienced rapid disease progression and was not subjected to RT; 1 patient developed acute par
RT was performed according to routine procedures either with 45–50 Gy in 25–28 fractions or short-course radiation therapy (25 Gy in 5 fractions) with or without concurrent fluoropyrimidine-based chemotherapy. Chemotherapy was delivered according to standard regimens (capecitabine 825 mg/m2 twice daily given within 5 d per week for 6-8 weeks). After RT patients were restaged with magnetic resonance imaging and the response was evaluated according to the TRG (tumor regression grade) system[20]. When the patients were surgically treated, the pathologic response was evaluated according to Mandard criteria[21]. The treatment results summary is presented in Supplementary Table 2.
Patients provided blood at 11 different time points: 1 h before the first fraction of radiation (at baseline), immediately after the first fraction (time 0), and 1, 3, 6, 12, 24, 36, 48, 72 and 96 h after the first fraction (Figure 2). Ten milliliters of blood were collected into PAXgene Blood ccfDNA Tubes (Qiagen) or cf-DNA/cf-RNA Preservative Tubes (Norgen). Plasma samples were separated from the cellular fraction within 2-8 h after the blood-draw by two-step centrifugation (400 g for 10 min at room temperature followed by 14400 g for 10 min at 4 °C). The supernatants were aliquoted into 2 mL tubes and stored at −70 °C until further use. Cell-free DNA was extracted with the QIAmp Circulating Nucleic Acid Kit (Qiagen) as recommended by Diefenbach et al[22]. Isolated DNA was subsequently diluted in sterile distilled water and frozen at −24 °C until further analysis.
The fractions of KRAS/NRAS mutations in codons 12, 13, 61 or BRAF V600E allele were measured by ddPCR using the QX100 Bio-Rad System[23]. ddPCR reactions were performed in triplicate. These reactions contained 2X ddPCR Supermix for Probes (no UTP, Bio-Rad), mutation-specific oligonucleotides (see Supplementary Tables 3 and 4) and 2-3 μL of the template DNA in a total reaction volume of 22-23 μL. Data analyses were performed with the QuantaSoft Software version 1.7.4 as recommended by the manufacturer. All ddPCR reactions, which yielded 10 or more droplets with the target DNA molecule, were considered informative. The absolute number of tumor-derived “mutated” DNA copies in 1 mL of plasma (Cmut) was calculated according to the formula:
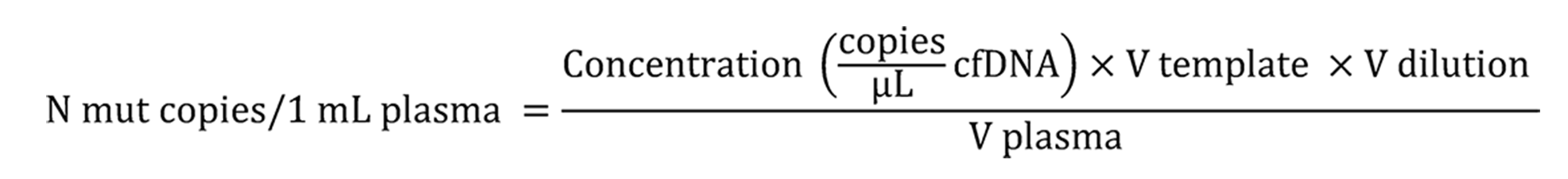
Where: Concentration – number of «mutated» droplets per 1 μL of ddPCR reaction; Vtemplate – volume of ctDNA aliquot taken into ddPCR, μL; Vdilution – total volume of diluted ctDNA sample collected from the plasma, μL; V plasma – volume of processed plasma, mL.
The change of ctDNA content after tumor irradiation was evaluated according to the following formula:
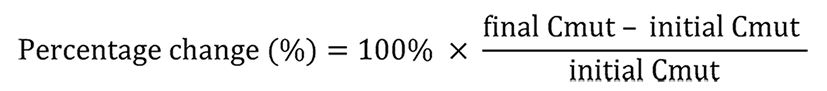
Quantitative data were present as a median values/range or means ± 95% confidence interval (1.960σx). The non-parametric Wilcoxon Signed Rank Test and Mann–Whitney U test were utilized to compare the medians. P value of < 0.05 was considered statistically significant. All calculations were performed using IBM SPSS v.23 software package.
Nine rectal patients were included in the study of ctDNA fluctuations occurring within the first hours after RT. Individual characteristics of the patients are given in Table 1. Four out of nine (44%) analyzed subjects had detectable RAS/RAF mutations in plasma DNA at baseline (at least 5 mutation-specific signals per reaction). The probability of detecting ctDNA in plasma did not correlate with any clinical characteristics, e.g. age, gender, mutation type, T/N stages, tumor grade, tumor location within the rectum, extramural venous invasion, circumferential resection margin, tumor response to treatment or PFS (statistical data not shown).
| Patient ID | Gender | Age | сТ | сN | Stage | CRM | EMV | Tumor location | RAS/RAFstatus | Total RT dose, Gy | Chemosensibiliza | RECIST | Surgery | MRI TRG | Mandard TRG | ypT | уN | Progression2
| Follow-up, mo | ctDNA positive at baseline |
| Yes/no | ||||||||||||||||||||
| ArAS | M | 44 | 3 | 1 | 3 | No | No | L | KRAS G12S | 50 | Yes | SD | Yes | III | NA | 3 | 0 | No | 6.90 | No |
| GaZM | F | 66 | 3 | 2 | 3 | Yes | No | U-M | KRAS G13D | 46 | Yes | PR | Yes | IV | 4 | 4 | 1 | No | 4.03 | No |
| DaKS | M | 73 | 4 | 1 | 3 | Yes | No | L | KRAS G12A | 25 (short course) | No | PD | No | IV | NA | NA | NA | Yes | 4.73 | Yes |
| ArTP | F | 81 | 4 | 1 | 3 | Yes | Yes | L | KRAS Q61L | 50 | No | SD | Yes | III | 3 | 3 | 0 | Nd | 5.80 | Yes |
| MaLI | F | 78 | 3 | 1 | 3 | No | No | L | NRAS G12D | 25(short course) | No | PD | No | IV | NA | NA | NA | Yes | 3.97 | Yes |
| MaNK | F | 48 | 3 | 2 | 3 | No | Yes | M | KRAS G12D | 50 | Yes | SD | Yes | III | 2 | 3 | 2 | No | 6.57 | Yes |
| ZuNM | F | 63 | 3 | 2 | 3 | No | No | L-M | BRAF V600E | 44 | Yes | PR | Yes | II | 2 | 3 | 0 | No | 5.13 | No |
| MiMF | F | 74 | 3 | 1 | 3 | No | Yes | M | NRAS G12D | 25(short course) | No | PR | No | III | NA | NA | NA | No | 3.13 | No |
| SaVV | M | 65 | 3 | 1 | 3 | Yes | No | L-M | NRAS Q61R | 50 | Yes | SD | No | III | NA | NA | NA | Nd | 1.13 | No |
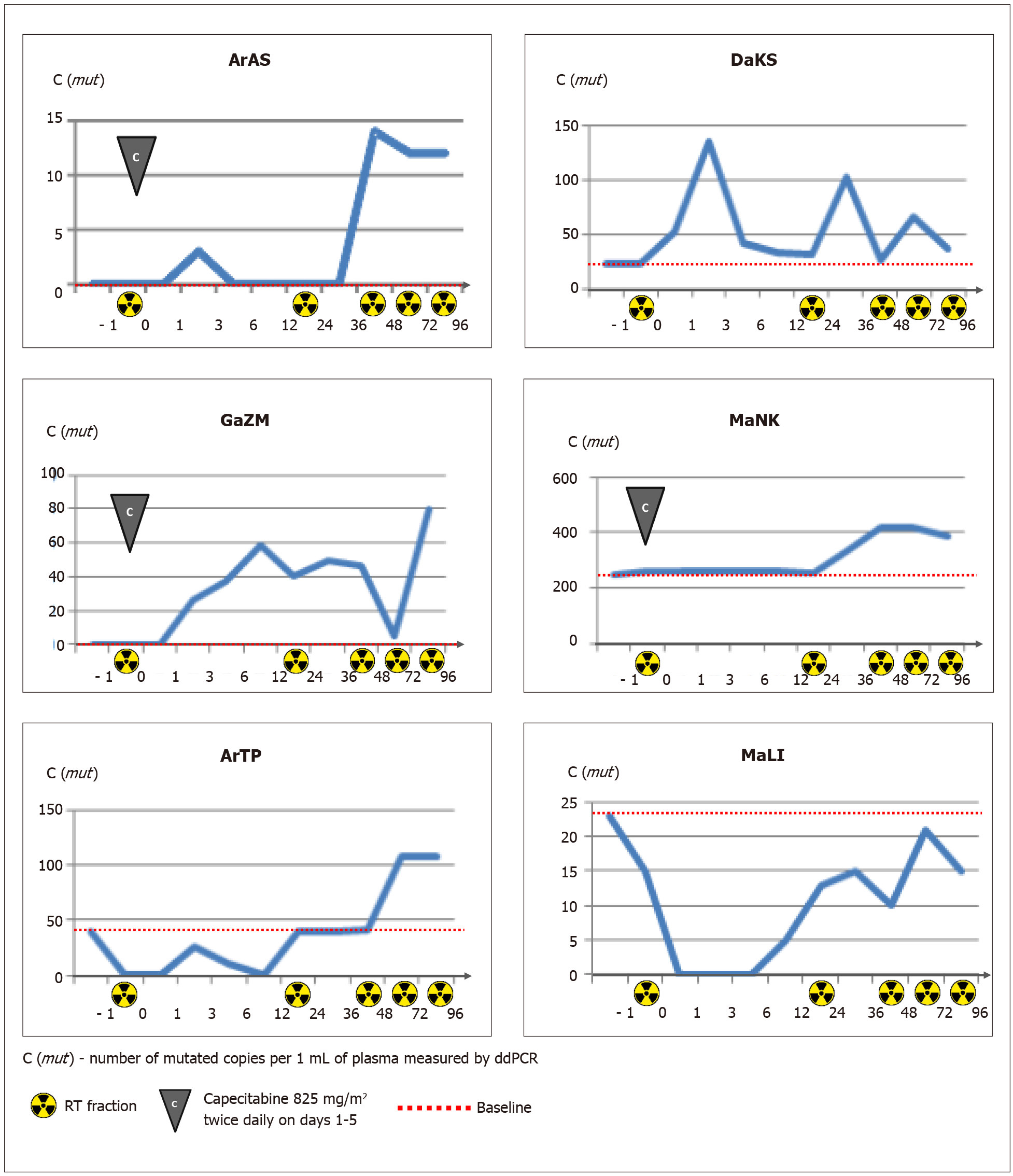
Three of 5 patients, who were negative for plasma RAS/RAF-mutated DNA at baseline, did not show the presence of mutated copies in subsequent serial samples obtained after tumor irradiation. The remaining two patients (#ArAS and #GaZM) demonstrated an appearance of the mutated DNA copies within the follow-up period (Table 2 and Figure 3).
| Patient ID | Mutation | Baseline mutated ctDNA | ctDNA analysis | -1 h | 0 | 1 h | 3 h | 6 h | 12 h | 24 h | 36 h | 48 h | 72 h | 96 h | Radiation dose per day / number of fractions / |
| ArAS | KRAS G12S | Neg | C (mut)1 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 14 | 12 | nd | 2.0 Gy/4 fr/8 Gy + capecitabine3 |
| C (mut + wt)2 | 642 | 1006 | 1337 | 1963 | 633 | 906 | 829 | 762 | 4298 | 3088 | nd | ||||
| VAF, % | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.3 | 0.4 | nd | ||||
| GaZM | KRAS G13D | Neg | C (mut) | 0 | 0 | 0 | 26 | 37 | 59 | 40 | 50 | 46 | 5 | 80 | 2.0 Gy/5 fr/10 Gy + capecitabine3 |
| C (mut + wt) | 6 | 7 | 8 | 866 | 808 | 1450 | 604 | 441 | 506 | 112 | 1219 | ||||
| VAF, % | 0.0 | 0.0 | 0.0 | 3.0 | 4.6 | 4.1 | 6.6 | 11.3 | 9.1 | 4.5 | 6.6 | ||||
| DaKS | KRAS G12A | Pos | C(mut) | 22 | 22 | 52 | 134 | 42 | 33 | 32 | 102 | 27 | 65 | 36 | 5.0 Gy/5fr/25 Gy |
| C (mut + wt) | 295 | 118 | 655 | 1802 | 588 | 292 | 170 | 549 | 359 | 391 | 331 | ||||
| VAF, % | 7.5 | 18.6 | 7.9 | 7.4 | 7.1 | 11.3 | 18.8 | 18.6 | 7.5 | 16.6 | 10.9 | ||||
| ArTP | KRAS Q61L | Pos | C (mut) | 39 | 0 | 0 | 26 | 10 | 0 | 39 | nd | 42 | 107 | nd | 2.0 Gy/4 fr/8 Gy |
| C (mut + wt) | 7606 | 9430 | 4036 | 5028 | 3187 | 3033 | 12574 | nd | 10622 | 14443 | nd | ||||
| VAF, % | 0.5 | 0.0 | 0.0 | 0.5 | 0.3 | 0.0 | 0.3 | nd | 0.4 | 0.7 | nd | ||||
| MaLI | NRAS G12D | Pos | C (mut) | 23 | 15 | 0 | 0 | 0 | 5 | 13 | 15 | 10 | 21 | 15 | 5.0 Gy/5 fr/25 Gy |
| C (mut + wt) | 981 | 1011 | 1073 | 1303 | 1309 | 1160 | 1010 | 724 | 815 | 673 | 1088 | ||||
| VAF, % | 2.3 | 1.5 | 0.0 | 0.0 | 0.0 | 0.0 | 1.3 | 2.1 | 1.2 | 3.1 | 1.4 | ||||
| MaNK | KRAS G12D | Pos | C (mut) | 244 | 257 | nd | nd | nd | Nd | 254 | 335 | 415 | 418 | 387 | 2.0 Gy/5 fr/10 Gy + capecitabine3 |
| C (mut + wt) | 897 | 1091 | nd | nd | nd | Nd | 1424 | 1832 | 2241 | 1719 | 1955 | ||||
| VAF, % | 27.2 | 23.6 | nd | nd | nd | Nd | 17.8 | 18.0 | 18.5 | 24.3 | 19.8 | ||||
| ZuNM | BRAF V600E | Neg | C (mut) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.0 Gy/5 fr/10 Gy + capecitabine3 |
| C (mut + wt) | 1652 | 372 | 522 | 522 | 522 | 522 | 671 | 837 | 1004 | 791 | 2805 | ||||
| VAF, % | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| MiMF | NRAS G12D | Neg | C (mut) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5.0 Gy/5 fr/25 Gy |
| C (mut + wt) | 594 | 500 | 740 | 2294 | 804 | 681 | 681 | 386 | 644 | 2891 | 1536 | ||||
| VAF, % | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| SaVV | NRAS Q61R | Neg | C (mut) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.0 Gy/5 fr/10 Gy + capecitabine3 |
| C (mut + wt) | 196 | 237 | 541 | 840 | 621 | 748 | 353 | 372 | 272 | 261 | 249 | ||||
| VAF, % | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Four patients were recognized as “plasma-positive” at baseline. The mean concentration of mutated copies in plasma samples was 82 copies per 1 mL (22, 23, 39, and 244, respectively). The variant allele frequency (VAF) of circulating mutations ranged from 0.5% to 27.2%. The analysis of changes in mutated ctDNA concentration occurring within the first 96 h of treatment revealed a pronounced increase in the number of circulating RAS/RAF mutant copies in patients #DaKS, #ArTP, #MaNK (with the maximum percent changes equal to 509%, 174% and 71%, respectively). Patient #MaLI showed less consistent variations in ctDNA content, with a maximal concentration at the start (23 mutant copies per 1 mL plasma, VAF 2.3%) and a number of subsequent spikes and drops (Figure 3).
There was no correlation between the content of mutated ctDNA and the total irradiation dose accumulated during the blood collection time (r = -0.400; P (2-tailed) = 0.253, Spearman’s Rho).
Patients with locally advanced rectal cancer provide a good opportunity for the analysis of RT-induced changes in the ctDNA level, as these malignancies frequently contain RAS/RAF mutations and the tumor irradiation is a part of routine clinical management of this disease[21]. The data obtained within this study are consistent with prior investigations, which were performed on lung cancer patients and demonstrated that radiotherapeutic or chemoradiotherapeutic intervention may result in a transient increase of the level of ctDNA in some cases[14-16]. As compared to published reports[14-16], our study considered multiple evenly distributed time points within the first day after tumor irradiation. We anticipated, that this additional effort may help us to identify a time interval characterized by maximal RT-induced ctDNA release. However, there was a significant interpatient variability with regard to the timing of ctDNA concentration peaks (Table 2 and Figure 3).
There are several limitations of this investigation. Human studies involving multiple serial blood takes are logistically complicated and need to be well balanced with ethical issues, therefore it is understandable that our study and similar reports[14-16] are of limited size. Furthermore, the range of “natural” variations of ctDNA measurements occurring due to imperfect reproducibility of laboratory protocols or physiological fluctuations of ctDNA content is largely unknown. Therefore, although our study demonstrated a trend towards the RT-induced increase of ctDNA concentration in some rectal cancer patients, it is not clear how these observations need to be adjusted for the described above confounding factors. This limitation is also applicable to other published data sets[14-16].
The analysis of tumor-specific mutations at the initial diagnostic work-up is usually not complicated, given that the management of cancer patients always requires morphological visualization of transformed cells and thus implies the availability of malignant tissue. However, the detection of actionable mutations acquired during the course of therapy presents a challenge. For example, the management of lung cancer patients, whose tumors progressed during gefitinib, erlotinib or afatinib treatment, involves the analysis of EGFR T790M mutation. The presence of this mutation justifies the administration of osimertinib, while the absence of this substitution calls for other treatment options. Re-biopsy of multiple visceral tumor lumps is often not feasible; therefore, the analysis of EGFR T790M mutation usually relies on liquid biopsy[24,25]. Clinical studies demonstrate that the detection of EGFR T790M mutation in plasma is seriously compromised by the low sensitivity of the test, especially in patients with limited tumor burden[26,27].
This study utilized patients with localized rectal cancer, who had a moderate volume of tumor masses. It is therefore explainable that only 4 out of 9 patients had detectable ctDNA at baseline. These data are comparable with the results obtained in other studies[12,28]. We deliberately focused on rectal cancer disease, as these patients often receive irradiation during the standard preoperative treatment, so no additional interventions were involved within this investigation. We demonstrated that two out of five subjects, who were initially ctDNA-negative, showed the presence of mutated DNA copies in the plasma after the start of the therapy. In addition, 3 out of 4 initially ctDNA-positive subjects experienced a RT-related increase of ctDNA content. The obtained data look promising, so further studies, which involve tumor irradiation not as a part of regular treatment plan, but as an additional intervention aimed to support ctDNA analysis, appear to be justified.
The clinical utility of this approach deserves to be evaluated in lung cancer patients, who demonstrate the disease progression during the therapy by first- or second-generation EGFR inhibitors and therefore require the diagnostic detection of EGFR T790M substitution. It is feasible to organize a prospective study, where the tumor lumps observed in these patients will be subjected to irradiation in order to provoke the release of tumor DNA in the bloodstream. It is essential to minimize the risks of this procedure by considering the anatomic location of targeted tumor foci (particularly, the vicinity of large blood vessels), ensuring a highly precise topical delivery of the irradiation dose and accounting for potentially significant comorbidities. If this intervention was to increase the rate of EGFR T790M allele detection in the plasma while being sufficiently safe, the proposed approach would have significant potential for clinical use.
Local tumor irradiation may facilitate the detection of plasma ctDNA. This study calls for a comprehensive evaluation of the clinical feasibility of irradiation-assisted liquid biopsy.
The detection of circulating tumor DNA (ctDNA) is a valuable diagnostic tool, however many cancer patients do not have detectable amount of ctDNA in their plasma.
We evaluated whether tumor irradiation may provoke the release of tumor DNA in the bloodstream and thus improve the efficiency of liquid biopsy.
We have chosen for the study patients with locally advanced rectal cancer as they usually receive preoperative tumor irradiation as a part of standard treatment plan.
The study included 9 patients with RAF/RAF mutations. Multiple serial blood draws were taken within first 96 h after the first fraction of radiotherapy. The amount of mutated RAF/RAF copies in the plasma was quantified by the droplet digital PCR.
Five out of nine patients demonstrated increased ctDNA content at least at some plasma samples obtained after the beginning of radiotherapy.
Radiotherapy is a promising tool for the improvement of performance of liquid biopsy.
It is feasible to extend this study to lung cancer patients, who receive tyrosine kinase inhibitors and may experience acquired tumor resistance due to the gain of secondary mutation.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: Russia
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Chen SY, Suzuki R S-Editor: Liu M L-Editor: A P-Editor: Liu M
| 1. | Locker GY, Hamilton S, Harris J, Jessup JM, Kemeny N, Macdonald JS, Somerfield MR, Hayes DF, Bast RC Jr; ASCO. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol. 2006;24:5313-5327. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1057] [Cited by in F6Publishing: 1042] [Article Influence: 57.9] [Reference Citation Analysis (0)] |
| 2. | Lakemeyer L, Sander S, Wittau M, Henne-Bruns D, Kornmann M, Lemke J. Diagnostic and Prognostic Value of CEA and CA19-9 in Colorectal Cancer. Diseases. 2021;9:21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 43] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 3. | Cheng ML, Pectasides E, Hanna GJ, Parsons HA, Choudhury AD, Oxnard GR. Circulating tumor DNA in advanced solid tumors: Clinical relevance and future directions. CA Cancer J Clin. 2021;71:176-190. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 50] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 4. | Zhang H, Liu R, Yan C, Liu L, Tong Z, Jiang W, Yao M, Fang W, Chen Z. Advantage of Next-Generation Sequencing in Dynamic Monitoring of Circulating Tumor DNA over Droplet Digital PCR in Cetuximab Treated Colorectal Cancer Patients. Transl Oncol. 2019;12:426-431. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 5. | Suzuki T, Suzuki T, Yoshimura Y, Yahata M, Yew PY, Nakamura T, Nakamura Y, Park JH, Matsuo R. Detection of circulating tumor DNA in patients of operative colorectal and gastric cancers. Oncotarget. 2020;11:3198-3207. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 6. | Liu L, Toung JM, Jassowicz AF, Vijayaraghavan R, Kang H, Zhang R, Kruglyak KM, Huang HJ, Hinoue T, Shen H, Salathia NS, Hong DS, Naing A, Subbiah V, Piha-Paul SA, Bibikova M, Granger G, Barnes B, Shen R, Gutekunst K, Fu S, Tsimberidou AM, Lu C, Eng C, Moulder SL, Kopetz ES, Amaria RN, Meric-Bernstam F, Laird PW, Fan JB, Janku F. Targeted methylation sequencing of plasma cell-free DNA for cancer detection and classification. Ann Oncol. 2018;29:1445-1453. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 82] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 7. | Reece M, Saluja H, Hollington P, Karapetis CS, Vatandoust S, Young GP, Symonds EL. The Use of Circulating Tumor DNA to Monitor and Predict Response to Treatment in Colorectal Cancer. Front Genet. 2019;10:1118. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 8. | Tie J, Cohen JD, Wang Y, Christie M, Simons K, Lee M, Wong R, Kosmider S, Ananda S, McKendrick J, Lee B, Cho JH, Faragher I, Jones IT, Ptak J, Schaeffer MJ, Silliman N, Dobbyn L, Li L, Tomasetti C, Papadopoulos N, Kinzler KW, Vogelstein B, Gibbs P. Circulating Tumor DNA Analyses as Markers of Recurrence Risk and Benefit of Adjuvant Therapy for Stage III Colon Cancer. JAMA Oncol. 2019;5:1710-1717. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 209] [Cited by in F6Publishing: 329] [Article Influence: 109.7] [Reference Citation Analysis (0)] |
| 9. | Formica V, Lucchetti J, Doldo E, Riondino S, Morelli C, Argirò R, Renzi N, Nitti D, Nardecchia A, Dell'Aquila E, Ferroni P, Guadagni F, Palmieri G, Orlandi A, Roselli M. Clinical Utility of Plasma KRAS, NRAS and BRAF Mutational Analysis with Real Time PCR in Metastatic Colorectal Cancer Patients-The Importance of Tissue/Plasma Discordant Cases. J Clin Med. 2020;10:87. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 10. | Naidoo M, Gibbs P, Tie J. ctDNA and Adjuvant Therapy for Colorectal Cancer: Time to Re-Invent Our Treatment Paradigm. Cancers (Basel). 2021;13:346. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 36] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 11. | Rodriguez-Casanova A, Costa-Fraga N, Bao-Caamano A, López-López R, Muinelo-Romay L, Diaz-Lagares A. Epigenetic Landscape of Liquid Biopsy in Colorectal Cancer. Front Cell Dev Biol. 2021;9:622459. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 27] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 12. | Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, Antonarakis ES, Azad NS, Bardelli A, Brem H, Cameron JL, Lee CC, Fecher LA, Gallia GL, Gibbs P, Le D, Giuntoli RL, Goggins M, Hogarty MD, Holdhoff M, Hong SM, Jiao Y, Juhl HH, Kim JJ, Siravegna G, Laheru DA, Lauricella C, Lim M, Lipson EJ, Marie SK, Netto GJ, Oliner KS, Olivi A, Olsson L, Riggins GJ, Sartore-Bianchi A, Schmidt K, Shih lM, Oba-Shinjo SM, Siena S, Theodorescu D, Tie J, Harkins TT, Veronese S, Wang TL, Weingart JD, Wolfgang CL, Wood LD, Xing D, Hruban RH, Wu J, Allen PJ, Schmidt CM, Choti MA, Velculescu VE, Kinzler KW, Vogelstein B, Papadopoulos N, Diaz LA Jr. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2770] [Cited by in F6Publishing: 3117] [Article Influence: 311.7] [Reference Citation Analysis (0)] |
| 13. | Zhang Y, Yao Y, Xu Y, Li L, Gong Y, Zhang K, Zhang M, Guan Y, Chang L, Xia X, Jia S, Zeng Q. Pan-cancer circulating tumor DNA detection in over 10,000 Chinese patients. Nat Commun. 2021;12:11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 93] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 14. | Kageyama SI, Nihei K, Karasawa K, Sawada T, Koizumi F, Yamaguchi S, Kato S, Hojo H, Motegi A, Tsuchihara K, Akimoto T. Radiotherapy increases plasma levels of tumoral cell-free DNA in non-small cell lung cancer patients. Oncotarget. 2018;9:19368-19378. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 15. | Nygård L, Ahlborn LB, Persson GF, Chandrananda D, Langer JW, Fischer BM, Langer SW, Gabrielaite M, Kjær A, Rosenfeld N, Mouliere F, Østrup O, Vogelius IR, Bentzen SM. Circulating cell free DNA during definitive chemo-radiotherapy in non-small cell lung cancer patients - initial observations. PLoS One. 2020;15:e0231884. [PubMed] [Cited in This Article: ] |
| 16. | Walls GM, McConnell L, McAleese J, Murray P, Lynch TB, Savage K, Hanna GG, de Castro DG. Early circulating tumour DNA kinetics measured by ultra-deep next-generation sequencing during radical radiotherapy for non-small cell lung cancer: a feasibility study. Radiat Oncol. 2020;15:132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 17. | Vaughn CP, Zobell SD, Furtado LV, Baker CL, Samowitz WS. Frequency of KRAS, BRAF, and NRAS mutations in colorectal cancer. Genes Chromosomes Cancer. 2011;50:307-312. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 262] [Cited by in F6Publishing: 280] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 18. | Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330-337. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5743] [Cited by in F6Publishing: 6144] [Article Influence: 512.0] [Reference Citation Analysis (0)] |
| 19. | Yanus GA, Belyaeva AV, Ivantsov AO, Kuligina ESh, Suspitsin EN, Mitiushkina NV, Aleksakhina SN, Iyevleva AG, Zaitseva OA, Yatsuk OS, Gorodnova TV, Strelkova TN, Efremova SA, Lepenchuk AY, Ochir-Garyaev AN, Paneyah MB, Matsko DE, Togo AV, Imyanitov EN. Pattern of clinically relevant mutations in consecutive series of Russian colorectal cancer patients. Med Oncol. 2013;30:686. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 20. | Patel UB, Brown G, Rutten H, West N, Sebag-Montefiore D, Glynne-Jones R, Rullier E, Peeters M, Van Cutsem E, Ricci S, Van de Velde C, Kjell P, Quirke P. Comparison of magnetic resonance imaging and histopathological response to chemoradiotherapy in locally advanced rectal cancer. Ann Surg Oncol. 2012;19:2842-2852. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 150] [Cited by in F6Publishing: 159] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 21. | Mandard AM, Dalibard F, Mandard JC, Marnay J, Henry-Amar M, Petiot JF, Roussel A, Jacob JH, Segol P, Samama G. Pathologic assessment of tumor regression after preoperative chemoradiotherapy of esophageal carcinoma. Clinicopathologic correlations. Cancer. 1994;73:2680-2686. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 16] [Reference Citation Analysis (0)] |
| 22. | Diefenbach RJ, Lee JH, Kefford RF, Rizos H. Evaluation of commercial kits for purification of circulating free DNA. Cancer Genet. 2018;228-229:21-27. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 66] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 23. | Sacher AG, Paweletz C, Dahlberg SE, Alden RS, O'Connell A, Feeney N, Mach SL, Jänne PA, Oxnard GR. Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer. JAMA Oncol. 2016;2:1014-1022. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 379] [Cited by in F6Publishing: 436] [Article Influence: 62.3] [Reference Citation Analysis (0)] |
| 24. | Jenkins S, Yang JC, Ramalingam SS, Yu K, Patel S, Weston S, Hodge R, Cantarini M, Jänne PA, Mitsudomi T, Goss GD. Plasma ctDNA Analysis for Detection of the EGFR T790M Mutation in Patients with Advanced Non-Small Cell Lung Cancer. J Thorac Oncol. 2017;12:1061-1070. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 170] [Cited by in F6Publishing: 204] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 25. | Akhoundova D, Mosquera Martinez J, Musmann LE, Britschgi C, Rütsche C, Rechsteiner M, Nadal E, Garcia Campelo MR, Curioni-Fontecedro A. The Role of the Liquid Biopsy in Decision-Making for Patients with Non-Small Cell Lung Cancer. J Clin Med. 2020;9:3674. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 26. | Passiglia F, Rizzo S, Di Maio M, Galvano A, Badalamenti G, Listì A, Gulotta L, Castiglia M, Fulfaro F, Bazan V, Russo A. The diagnostic accuracy of circulating tumor DNA for the detection of EGFR-T790M mutation in NSCLC: a systematic review and meta-analysis. Sci Rep. 2018;8:13379. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 60] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 27. | Cortiula F, Pasello G, Follador A, Nardo G, Polo V, Scquizzato E, Del Conte A, Miorin M, Giovanis P, D'Urso A, Girlando S, Settanni G, Picece V, Veccia A, Corvaja C, Indraccolo S, De Maglio G. A Multi-Center, Real-Life Experience on Liquid Biopsy Practice for EGFR Testing in Non-Small Cell Lung Cancer (NSCLC) Patients. Diagnostics (Basel). 2020;10:765. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 28. | Sclafani F, Chau I, Cunningham D, Hahne JC, Vlachogiannis G, Eltahir Z, Lampis A, Braconi C, Kalaitzaki E, De Castro DG, Wotherspoon A, Capdevila J, Glimelius B, Tarazona N, Begum R, Lote H, Hulkki Wilson S, Mentrasti G, Brown G, Tait D, Oates J, Valeri N. KRAS and BRAF mutations in circulating tumour DNA from locally advanced rectal cancer. Sci Rep. 2018;8:1445. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |