Abstract
MicroRNAs (miRNAs) are a class of small RNAs of 19-23 nucleotides that regulate gene expression through target mRNA degradation or translational gene silencing. The miRNAs are reported to be involved in many biological processes, and the discovery of miRNAs has been provided great impacts on computational biology as well as traditional biology. Most miRNA-associated computational methods comprise the prediction of miRNA genes and their targets, and increasing numbers of computational algorithms and web-based resources are being developed to fulfill the need of scientists performing miRNA research. Here we summarize the rules to predict miRNA targets and introduce some computational algorithms that have been developed for miRNA target prediction and the application of the methods. In addition, the issue of target gene validation in an experimental way will be discussed.
Similar content being viewed by others
Introduction
MicroRNAs (miRNAs) are a class of small, non-coding regulatory RNAs that are important in post-transcriptional gene silencing (Bartel, 2004). They regulate gene expression by binding to 3' untranslated region (UTR) of their target mRNAs for cleavage or translational repression and play important roles in many biological processes including cell proliferation, cell death, hematopoiesis, and oncogenesis.
In the canonical pathway of miRNA biogenesis, mature miRNAs arise from long primary miRNA transcripts (pri-miRNAs) that are transcribed from non-protein-coding genes in the nucleus (Figure 1; Lodish et al., 2008). The pri-miRNAs are then cleaved by the RNase III enzyme Drosha to liberate ~70 nucleotide (nt) precursor miRNAs (pre-mRNAs) which are subsequently transported into the cytoplasm by Exportin-5, a Ran-GTP-dependent nuclear export factor. In the cytoplasm, the pre-miRNAs are processed by RNase III-like nuclease Dicer (animals) or DICER-LIKE1 (DCL1 [plants]) to generate ~21 to 22 nucleotide duplexes. The functional mature miRNA strand is then selectively incorporated into RISC (RNA-induced silencing complex) effector complex to regulate specific target mRNAs. In general, plant miRNAs interact with their targets through near-perfect base-paring, resulting in target degradation, whereas animal miRNAs form imprecise base-pairing and cause translational repression.
MicroRNA biogenesis and function in animal cells (Lodish et al., 2008). miRNAs are transcribed as long primary transcripts (pri-miRNAs) in the nucleus. The pri-miRNAs are then processed by the RNase III-type Drosha, yielding pre-miRNAs of ~70 nucleotide (nt). Subsequently, the pre-miRNAs are exported to the cytoplasm by exportin-5, and further cleaved into ~21 to 22 nucleotide miRNA duplex by another RNase III enzyme Dicer. The less stable strand of the miRNA duplex is then incorporated into a multiple protein nuclease complex, the RISC, and regulates protein expression.
Since the discovery of the very first miRNAs, computational approaches have been invaluable tools in understanding the biology of miRNAs (Bentwich, 2005; Rajewsky, 2006). Web-based-miRNA databases have been constructed and provided not only thousands of published miRNA sequences and annotation (e.g. miRBase Sequences database; Griffiths-Jones et al., 2008) but also potential miRNA target genes (e.g. miRBase Targets database; Griffiths-Jones et al., 2008). Many pri-miRNA transcripts are computationally predicted to undergo folding into elaborate stem-loop structures. In addition, computational algorithms have been developed to predict pre-miRNAs (Huang et al., 2007) and to search for homologous conserved miRNA genes in several animal species. However, most computational approaches associated with miRNA research are about miRNA gene detection and miRNA target prediction.
Researchers initially determined miRNA targets through experiments. The first miRNAs and their target genes were identified through classical genetic techniques (Lee et al., 1993). However, due to the laborious nature of experiments and the absence of high-throughput experimental methods, it is inevitable to develop computational techniques to determine miRNA targets. In this review, we summarize the principles to predict miRNA targets and discuss some currently available computational methods that have been developed for miRNA targets prediction and the application of these methods.
Principles of miRNA target recognition
Target prediction and its biological validation have been major obstacles to miRNA researchers. Because miRNAs are short, and animal miRNAs have limited sequence complementarity to their targets, it is a challenging task to predict animal miRNA targets with high specificity. However, target prediction in plants is relatively uncomplicated, because plant miRNAs bind to their target mRNAs with perfect or nearly perfect complementarity for target cleavage.
In order to develop computational algorithms identifying miRNA target genes, principles of miRNA target recognition are often established based on empirical evidences. For example, the importance of base pairing between miRNAs and their targets was suspected according to the observation that the 'target site' of the lin-14 UTR is complementary to the 5' region of the lin-4 miRNA (Lee et al., 1993). Some features used by the mammalian target prediction programs are described below.
-
1
Base pairing pattern
-
2
Thermodynamic stability of miRNA-mRNA hybrid
-
3
Comparative sequence analysis to check conservation
-
4
Examination of the presence of multiple target sites
Base pairing pattern
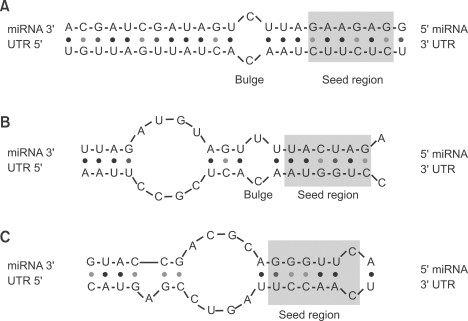
In the first step, target prediction programs identify potential binding sites according to specific pairing patterns. The binding sites can be classified into 3 categories (Maziere and Enright, 2007): (i) 5'-dominant canonical, (ii) 5'-dominant seed only and (iii) 3'-compensatory (Figure 2). MiRNA seed is defined as the consecutive 7 to 8-nucleotide sequence starting from either the first or second base at the 5' end of an miRNA (Lewis et al., 2003). The 5'-dominant canonical sites have perfect base paring to the 5' end seed region and extensive base pairing to the 3' end of the miRNA with a characteristic bulge in the middle. The seed-only sites have perfect base pairing to the seed region and imperfect base pairing to the 3' end of the miRNA. The 3'-compensatory sites have a mismatch or wobble in the seed region of the miRNA, but have long stretch of base pairing to the 3' end of the miRNA to make up for the weak binding at the 5' seed (Brennecke et al., 2005).
Approximate secondary structures of the three main types of target site duplex. (A) Canonical sites have perfect base paring in seed region, a bulge in the middle and extensive base pairing in the 3' end of the miRNA. (B) Dominant seed sites form perfect complementarity in the seed, but poor complementarity in the 3' end of the miRNA. (C) Compensatory sites have a mismatch or G:U wobble in the seed region, but have extensive base pairing to the 3' end of the miRNA (Maziere and Enright, 2007).
Thermodynamic analysis of miRNA-mRNA hybrid
The thermodynamic properties of miRNA-mRNA duplexes are assessed by calculating free-energy (ΔG) of the putative binding. The approximate free energy and secondary structure of the miRNA-mRNA duplex can be calculated by RNA folding program such as the Vienna package (Wuchty et al., 1999), RNAfold (Hofacker, 2003) and Mfold (Mathews et al., 1999). A threshold of free-energy of binding is then calculated in accordance with specificity and sensitivity. However, since data sets of identified miRNA-mRNA duplexes are very restricted, and a low free energy of hybridization does not guarantee accurate prediction of miRNA target genes (Watanabe et al., 2007), it is complicated to resolve appropriate thresholds of free energy. Thus, it is inevitable to consider additional features such as conservation analysis for reliable prediction of target transcripts. For example, a recent study by Lewis et al. (2005) has shown that thermodynamics can be removed without lowering the specificity of the algorithm by incorporating evolutionary conservation derived from multiple sequence alignments.
Comparative sequence analysis
Comparative sequence analysis within related species is performed to check if target sequences are evolutionarily conserved across species (Watanabe et al., 2007). In order to reduce the number of false positives, many target prediction algorithms identify orthologous 3' UTR sequences and then perform conservation analysis across related species. However, there are issues related to conservation analysis. For instance, given that transcripts between humans and chimpanzees are highly conserved, it might not be meaningful to search for conserved targets between humans and chimpanzees (Maziere and Enright, 2007). Instead, other organisms such as rats and dogs might be more appropriate for conservation analysis with human transcripts, but genomes might not be sequenced along with their evolutionary distance. As a result, the use of conservation filter has a risk of increasing false negatives while it decreases false positives.
Examination of multiple target sites per target transcript
Previous studies have shown that multiple miRNAs are co-expressed and are likely to regulate the same mRNA coordinately (Rajewsky, 2006). Multiple target sites in the same 3' UTR can potentially increase the degree of translational suppression and enhance specificity of gene regulation. Thus, some algorithms check the presence of multiple target sites and take the number of target sites into account for prediction (Stark et al., 2005).
Programs for miRNA target recognition
Tens of different methods have been developed for computational target prediction. The programs based on base pairing pattern (Lewis et al., 2003) are most common, and other features including evolutionary conservation (Lewis et al., 2003; Grün et al., 2005), secondary structure of target transcript (Kertesz et al., 2007; Long et al., 2007), and nucleotide composition of target sequences (Grimson et al., 2007) are often added to increase accuracy. Currently available target production methods are described in Table 1, and some of them are reviewed below in more detail.
TargetScan and TargetScanS
TargetScan is an algorithm developed by Lewis et al. (2003) to identify the targets of vertebrate miRNAs. The program integrates thermodynamics-based modeling of miRNA-mRNA interactions and comparative sequence analysis to predict miRNA targets conserved across multiple genomes such as human, mouse, rat, and pufferfish.
The 'miRNA seed' is a 7-nucleotide sequence at base 2 to 8 in the 5' end of the miRNAs. It forms perfect Watson-Crick base pairing complementary to 'seed matches' which refers to the 3' UTR heptamer in the target mRNA. TargetScan searches for seed matches in the first organism such as human and expand each seed match with additional base pairings to the miRNA. The algorithm then calculates the thermodynamic free energy of the binding between the putative miRNA target and extended seed sequences by using the RNAFold package (Hofacker, 2003) and assigns a score to each UTR. Then, it repeats the process for the sets of UTRs from other organisms including mouse, rat, and pufferfish for phylogenic analysis. The estimated false-positive rate varies between 22% and 31%, and the method was shown to predict not only known miRNA binding sites but also 451 novel potential sites. In addition, by using luciferase reporter constructs, 11 out of the 15 tested sites were experimentally validated.
TargetScanS simplified the TargetScan method and improved the target prediction fidelity (Lewis et al., 2005). TargetScanS requires a six-nucleotide seed (position 2 to 7) followed by an additional 3' match of adenosines surrounding the miRNA seed (It was found that the immediate downstream position of the seed match is highly conserved and is often an adenosine). The method is independent of thermodynamic stability or multiple target sites, but two more species (dog and chicken) were added for conservation analysis. As a result, the estimated false-positive rate was reduced to 22% in mammals, and all known miRNA-target interactions were successfully predicted.
Although TargetScan and TargetScanS efficiently reduced false positive rates, there is a concern about using conservation analysis and complementarity in the seed region. As shown in Figure 2C, the 3' compensatory site has a mismatch or GU wobble in the seed region and does not form a perfect Watson-Crick base pairing. Therefore, some targets having the 3' compensatory site cannot be detected. In addition, as mentioned earlier, if targets are loosely conserved, they will not be picked by TargetScan and TargetScanS resulting in an increase of false negatives.
PicTar
Contrary to TargetScan and TargetScanS that require a seed match at exactly corresponding positions in a cross-species UTR alignment, PicTar requires binding sites that are coregulated by multiple miRNAs across species (Grün et al., 2005). PicTar checks the alignments of 3' UTRs for those displaying seed matches to miRNAs, filters the retained alignments based on their thermodynamic stability, and computes a hidden Markov model (HMM) maximum likelihood score (PicTar score) for each predicted target. To filter out false positives, PicTar used statistical tests based on genome-wide alignments of eight vertebrate genomes, and considered clustering co-expressed miRNAs and matching miRNAs with putative targets that are expressed in the same context (Yoon and De Micheli, 2006). This algorithm was able to correctly identify some known miRNA targets and its false positive rate was estimated to be around 30%.
By using PicTar, Krek et al. (2005) suggested that each vertebrate miRNA has approximately 200 target transcripts on average. In addition, they experimentally validated 7 out of 13 predicted targets and 8 out of 9 previously known targets, demonstrating the efficiency of the algorithm. Furthermore, Grün et al. (2005) performed cross-species comparison and predicted that about 54 genes are regulated by a given miRNA. PicTar was also used for genome-wide search of miRNA targets in C. elegans (Lall et al., 2006). By using a new version of PicTar and sequence alignments of three nematodes, the authors reported that at least 10% of C. elegans genes are predicted miRNA targets, and a number of nematode miRNAs are likely to control biological processes by targeting functionally related genes.
miRanda
This method was originally developed to predict miRNA target genes in D. melanogaster (Enright et al., 2003), but was also used to predict human miRNA targets. For each miRNA, miRanda selected target genes on the basis of three properties: sequence complementarity using a position-weighted local alignment algorithm, free energies of RNA-RNA duplexes, and conservation of target sites in related genomes. miRanda was able to correctly identify 9 out of 10 currently characterized target genes, and its false-positive rate was around 24%. When targets of all miRNAs were analyzed for the distribution of functional annotation using GO terms, the functions of the predicted target genes were found to be enriched in the components of the ubiquitin machinery, transcription factors, components of miRNA machinery, and translational regulation.
John et al. (2004) improved the method by implementing a strict model for the binding sites that require almost perfect complementarity in the seed region allowing a single wobble pairing. The authors reported about 2000 human genes with miRNA target sites conserved in mammals and about 250 human genes conserved between mammals and fish. Their analysis also suggests that miRNA genes, which comprise around 1% of the human genome, control the production of protein for 10% or more of all human genes.
DIANA-microT/DIANA-micro T web server
Kiriakidou et al. (2004) developed DIANA-microT by combining computational and experimental approaches. In order to identify putative miRNA-recognition elements (MREs), this method uses a window of 38 nucleotide that progressively goes through a 3' UTR of potential target. Using dynamic programming, the minimum binding energy between the miRNAs and sequences in the human 3' UTR database is calculated at each step and is compared with the outcomes obtained from scrambled sequences with the same dinucleotide content as real 3' UTRs. In contrast to TargetScan/TargetScanS or PicTar, DIANA-microT method allows a weak binding at 5' seed, involving six consecutively paired nucleotides or G:U wobble pairs, if there exists additional base paring between the miRNA 3' end and target gene.
This algorithm successfully identified all currently known C. elegans miRNA target sites, and 7 predicted mammalian miRNA target genes were experimentally validated. Moreover, this method was reported to show the precision levels of 66%, which is the highest among several prediction programs, when their performance were assessed through microarray and the pulsed stable isotope labeling with amino acids in cell culture (pSILAC) method that measures changes in the synthesis of thousands of protein in response to miRNA transfection or endogenous miRNA knockdown (Selbach et al., 2008).
The DIANA-microT web server (Maragkakis et al., 2009) is the user interface to DIANA-microT, providing extensive connectivity to online biological resources as well as information on predicted miRNA:target gene interactions. The server contains links to UniProt for protein information, iHOP for functional and bibliographic information (iHOP), miRBase for miRNA information, and KEGG pathway for pathway analysis.
RNAHybrid
RNAHybrid is an extension of classical RNA secondary structure prediction software tools such as RNAfold (Hofacker, 2003) and Mfold (Mathews et al., 1999). The classical methods were designed for single-sequence folding, and therefore require an artificial linker between an miRNA and its potential binding site. However, there are some issues about using these methods (Stark et al., 2003). The short artificial linker sequence might lead to artifacts in the prediction, and intramolecular hybridizations (hybridization between target nucleotides or between miRNA sequences), or hybridization of the target and miRNA with the linker, can happen. An additional problem is that the prospective binding sites should be excised and folded separately for prediction of multiple bindings in one target. However, RNAHybrid finds the energetically most favorable hybridization sites of a small RNA within a large target RNA sequence, and base pairings between target nucleotides or between miRNA nucleotides are not allowed (Rehmsmeier et al., 2004).
Beyond prediction: extension of target prediction resources
While algorithms for target prediction remain mainstream, many web-based servers have been developed by combining new features to existing prediction programs. These servers mostly integrate multiple established prediction programs and include functional annotations that are exhaustively linked to many miRNA, gene, protein or biological pathway resources such as miRBase, Ensembl, Swiss-Prot, UCSC genome browser, KEGG pathway, and other databases. For target prediction, TargetScan, miRanda, and PicTar are the most frequently adopted, and RNAhybrid, DIANA-micro T, and TarBase are also widely used. A list of the web servers is shown in Table 2, and some of them are described in more detail as follows:
TarBase and miRecords
TarBase5.0 (Sethupathy et al., 2006; Papadopoulos et al., 2009) is a database that is built by extracting miRNA targets that are experimentally validated from 203 scientific reports. In 2008, the database included over 1300 records with information on miRNAs, target genes, and experimental conditions used for target support. It is also functionally linked to other databases including Ensembl, Swiss-Prot, Hugo and HGNC to extend information about miRNAs and their target genes.
MiRecords (Xiao et al., 2009) is another database curating predicted targets generated by 11 miRNA target prediction programs as well as 1135 entries of validated targets (as of August, 2008). For each query of miRNA-target interaction, miRecords presents validated targets plus prediction results from different prediction programs, which meets the need of researchers who want to run several programs and find their intersections at a time. Especially, the predicted targets component of miRecords integrates putative targets produced by 11 programs providing thorough prediction results, while most other miRNA target resources integrate 3-4 prediction programs.
With the expansion of knowledge on miRNAs and target interactions, construction of centralized archive that holds experimentally confirmed, comprehensive, and up-to-date information is of necessity, and in that sense, TarBase and miRecords are welcome to the miRNA research community.
miRGator
miRGator (Nam et al., 2008) is a database that integrates target prediction, functional analysis, gene expression data and genome annotation. For target prediction, it uses TargetScan, miRanda, and PicTar and combines their results in a Boolean logic.
Given that functional relationships between target genes may provide a critical clue for elucidating functional significance of each miRNA, it implements a number of functional categories such as the GO, KEGG/GenMAPP/BioCarta pathways, and disease classification by using Ingenuity pathway analysis. Furthermore, in order to assess the quality of target prediction, expression correlation analysis between miRNA and target mRNA or target protein is performed. Since reciprocal expression pattern is expected for genuine miRNA:target pairs, and proportional expression with high correlation represents miRNA: non-target pairs, correlation coefficients can be informative for evaluating candidate target genes. It also contains an miRNA expression profiling module from the gene expression omnibus (GEO) database supporting differentially regulated miRNAs in 24 tissues/organs and 28 cell types.
GOmir
GOmir (Roubelakis et al., 2009) was established for human miRNA target prediction and ontology clustering. It consists of two JAVA modules, namely JTarget and TAGGO. JTarget integrates putative targets obtained by 4 prediction softwares (TargetScan, miRanda, RNAHybrid, PicTar-4 way, PicTar-5 way) and an experimental database TarBase to find common targets and provides detailed information about gene description and function. TAGGO then performs GO clustering with the common genes obtained from JTarget and analyzes how many target proteins share a common GO category.
MirZ
Hausser et al. from M. Zavolan group developed a web-server called mirZ (Hausser et al., 2009), incorporating the smiRNAdb miRNA expression atlas (Landgraf et al., 2007) and the ElMMo miRNA target prediction algorithm (Gaidatzis et al., 2007) that were both developed by the Zavolan group. The smiRNAdb is a web-accessible resource of miRNA profiles determined by sequencing 250 small RNA libraries from 26 different organ systems and cell types in human and rodents. It also has features of an extended repertoire of on-line analyses such as visualization and hierarchical clustering of miRNA expression profiles, principal component analysis, and comparison of miRNA expression between two samples. The ElMMo is a miRNA target prediction method based on a Bayesian probabilistic model that uses comparative genomics information. The prediction results are composed of two sections: 1) an miRNA-centric summary showing the smiRNAdb tissues where the selected miRNA is mostly expressed, and 2) a target mRNA-centric summary with the putative target site locations in the 3'-UTR region. With the idea that miRNAs that are most strongly expressed within a given tissue have the largest impact on mRNA targets, MirZ tries to find the mRNA that is most likely to be affected by the changes in miRNA expression.
MMIA (miRNA and mRNA integrated analysis)
MMIA (Nam et al., 2009) is a web-server developed to integrate miRNA and mRNA expression data for target prediction. Based on the fact that the expressions of miRNA and its target mRNA are reciprocal, the MMIA finds target mRNAs by combining computational prediction results and expression data analysis. In more detail, the MMIA selects up- or down-regulated miRNAs from input data and predicts their target mRNAs using TargetScan, PicTar or Probability of Interaction by Target Accessibility (PITA; Kertesz et al., 2007). It then identifies the mRNAs whose expressions are inversely correlated from microarray input and performs gene set analysis to find intersection of predicted mRNA and inversely correlated mRNA. The final output includes optional information about diseases associated with miRNAs, transcription factors enriched in the promoters of miRNAs as well as resources from GO, KEGG pathways and MIT MSigDB (Subramanian et al., 2005).
miRDB
This is an online database system for target prediction and functional annotation (Wang, 2008). What is unique for miRDB is that it uses a wiki interface for community editing, which has been proven to be successful as shown from an example of Wikipedia, and thus, anyone with internet access can freely write miRNA functional annotations. In addition to wiki annotations, it also has uneditable sections containing officially adopted information such as miRNA names, Sanger accessions, sequences and genomic locations, and target predictions are made through its own algorithm named MirTarget2 (Wang and El Naqa, 2008).
What to choose?: comparison of target prediction algorithms
Tens of target prediction programs have been developed and they are all freely available. Now an issue to the researchers is to choose the appropriate tool for each one's unique situation. For that purpose, groups of researchers tried to evaluate the performance of target prediction programs experimentally (Baek et al., 2008; Selbach et al., 2008) or computationally with an experimentally-verified miRNA target dataset (Alexiou et al., 2009). Selbach et al. (2008) reported that 3 programs such as TargetScanS, Pictar and DIANA-microT have precision levels (the fraction of the predicted targets that were actually downregulated) >60%, and Alexiou et al. found that 5 programs (DIANA-microT, TargetScan, TargetScanS, Pictar, and ElMMO) have a precision ~50% with a sensitivity raging from 6 to 12%. Accordingly, these 3-5 programs would be the good ones to start with if your target organisms are vertebrates, flies or worms. However, if your model system is virus, slim mold or some unique organisms, you have a choice of DIANA-microT among the five popular ones plus miTarget, rna22, and RNAhybrid.
For the researchers who want to run multiple algorithms and choose all possible union or intersection of the combinations, the methods that provide prediction by using multiple algorithms might be attracting. In fact, several methods such as GOmir, miRecords, and miTarget employ the integration of as many as 11 algorithms to meet such a need. However, it turns out that many of the combinatorial predictions perform worse than the prediction by one accurate algorithm, because of the trade-off between specificity and sensitivity (Alexiou et al., 2009).
Since existing target prediction algorithms rely on different assumptions and models for prediction, it would be wise to check the underlying assumptions and limitations first before employing a target prediction tool. Combining results from multiple tools seems to be a common practice and is often encouraged in order to reduce the probability of introducing false positives and/or negatives as much as possible.
Discussion
Most computational algorithms for target prediction combine 5' seed matches, thermodynamic stability and conservation analysis in order to maximize specificity. However, there exist some exceptions to these generalized rules, and target selection mechanisms need to be adjusted in a species specific manner (Watanabe et al., 2007).
Although the rule of seed pairing has been successfully used to predict target sites with statistical support, the seed matches are not always sufficient for repression, implying that additional features would require for reliable target selection (Grimson et al., 2007). Through the combination of computational and experimental approaches, Grimson et al. (2007) revealed five general characteristics of site milieu that increase effectiveness: 1) high local density of AU nucleotides, 2) closeness of sites for co-expression of multiple miRNAs leading to synergistic activity, 3) additional base-paring at the 12-17 nucleotide region of miRNA, most especially at the 13-16, 4) site location in the 3' UTR at least 15 nucleotide away from the stop codon, and 5) extensive and contiguous 3' pairing. Thus, in designing an algorithm, those five features as well as the rule of seed match should be considered.
Another problem of using 5' dominant site is that 3' compensatory site containing a mismatch or wobble in the seed region cannot be detected by most target prediction methods. Although miRanda is sensitive for such targets (Sethupathy et al., 2006), it is of necessity to develop more computational algorithms to identify those 3' compensatory target sites with accuracy.
Evolutionary conservation is another important factor to filter out false positive targets and increase specificity. It helps to predict only the target sites which are under selective pressure to preserve their sequence and presumably functionality, across evolution (Sethupathy et al., 2006). However, Farh et al. (2005) demonstrated that many of the non-conserved target sites, which outnumber the conserved sites 10 to 1, are also functional and mediate repression. Thus, the presence of those non-conserved target sites should not be overlooked when designing an algorithm for target prediction.
Once miRNA targets are predicted with a fair degree of accuracy, the next step is to validate the miRNA-target interaction experimentally. Since computational methods are not perfect, and there is a risk of false-positive prediction, target validation in biological system is inevitable to complete the study of target prediction. A reporter assay is the most common method to check the interaction between miRNA and its target mRNA directly. In a standard reporter assay, the putative target sites are fused to a reporter construct (e.g. luciferase, green fluorescence protein or yellow fluorescence protein), and reporter expression is measured in the absence and presence of the cognate miRNA. Additionally, northern blot analysis, quantitative real-time PCR (qRT-PCR), ribonuclease protection assay, or in situ hybridization is often performed to examine the reciprocal expression of predicted miRNA and mRNA target genes. Levels of protein are often measured by western blot or immunocytochemistry to compare protein expression given the presence and absence of the miRNA.
For comprehensive study, biological function can be examined through miRNA overexpression or knockout experiment under in vitro or in vivo conditions. Overexpression of miRNA can be accomplished by constructing an expression vector containing mature miRNA, precursor (hairpin) miRNA, or the pri-miRNA sequence followed by transfection. miRNA overexpression may also be indirectly induced by using the DNA methylating agent 5-aza-deoxycytidine (Lujambio et al., 2007) or histone deacetylase inhibitor phenylbutyrate (Saito et al., 2006), while these methods are not miRNA sequence specific and are not common. To silence a specific miRNA, chemically modified oligonucleotides that are perfectly complimentary to the mature miRNA are introduced. Such antisense modified oligonucleotides are morpholinos, antagomir, locked nucleic acids (LNA), or 2'-O-methyl oligonucleotides. The technique of siRNAs is also applied to knock-down miRNA gene as it has been done for silencing regular genes (Kim et al., 2008).
Although much work has been done on target validation, and even a couple of databases of validated targets have been constructed, those wet lab experiments (even the reporter assay) are still too lengthy and laborious to simultaneously deal with many pairs of miRNA and its targets. Worse, it is more difficult to catch up with the expanding numbers of new miRNAs and their targets that are computationally predicted. Therefore, the development of high-throughput experimental strategies is inevitable for large-scale analysis of miRNA targets and their biological function. Microarrays and pSILAC are greatly useful to measure global changes in the transcriptome (Lim et al., 2005) or proteome (Selbach et al., 2008) following overexpression or silencing of miRNA. However, these methods cannot distinguish direct targets from indirect targets and only give indirect evidence about miRNA-target interactions. Degradome analysis (Addo-Quaye et al., 2008; German et al., 2008) is also available, but it only works in a system where a miRNA induces RISC-mediated mRNA cleavage, and thus, its usage is limited mostly in plants.
Since the discovery of the first miRNAs and their target genes in 1993, there has been a dramatic growth in the number of annotated miRNAs and their validated or putative targets which are supported by a number of computational algorithms. Although these algorithms are still lack sensitivity and specificity, they are able to provide valuable help to researchers investigating new miRNA targets. In addition, ample web-based resources on miRNA expression profiles, gene function, gene ontology, transcriptional regulatory interactions, signaling pathways, and other functional genomic data are easily accessible for the study of miRNA-target interactions at a system-wide level. By integrating such genome-wide computational and experimental approaches, research on miRNA will be prosperous than ever.
Abbreviations
- GEO:
-
gene expression omnibus
- GO:
-
gene ontology
- LNA:
-
locked nucleic acids
- miRNA:
-
microRNA
- MREs:
-
miRNA-recognition elements
- HMM:
-
hidden Markov model
- pre-mRNAs:
-
precursor miRNAs
- pri-miRNAs:
-
primary miRNA transcripts
- pSILAC:
-
pulsed stable isotope labeling with amino acids in cell culture
- qRT-PCR:
-
quantitative real-time PCR
- RISC:
-
RNA-induced silencing complex
- UTR:
-
untranslated region
References
Alexiou P, Maragkakis M, Papadopoulos GL, Reczko M, Hatzigeorgiou AG . Lost in translation: an assessment and perspective for computational microRNA target identification . Bioinformatics 2009 ; 25 : 3049 - 3055
Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ . Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome . Curr Biol 2008 ; 18 : 758 - 762
Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP . The impact of microRNAs on protein output . Nature 2008 ; 455 : 64 - 71
Bartel DP . MicroRNAs: genomics, biogenesis, mechanism, and function . Cell 2004 ; 116 : 281 - 297
Bentwich I . Prediction and validation of microRNAs and their targets . FEBS Lett 2005 ; 579 : 5904 - 5910
Brennecke J, Stark A, Russell RB, Cohen SM . Principles of microRNA-target recognition . PLoS Biol 2005 ; 3 : e85 -
Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS . MicroRNA targets in Drosophila . Genome Biol 2003 ; 5 : R1 -
Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP . The widespread impact of mammalian MicroRNAs on mRNA repression and evolution . Science 2005 ; 310 : 1817 - 1821
Gaidatzis D, van Nimwegen E, Hausser J, Zavolan M . Inference of miRNA targets using evolutionary conservation and pathway analysis . BMC Bioinformatics 2007 ; 8 : 69 -
Gerlach W, Giegerich R . GUUGle: a utility for fast exact matching under RNA complementary rules including G-U base pairing . Bioinformatics 2006 ; 22 : 762 - 764
German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R . Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends . Nat Biotechnol 2008 ; 26 : 941 - 946
Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ . miRBase: tools for microRNA genomics . Nucleic Acids Res 2008 ; 36 : D154 - D158
Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP . MicroRNA targeting specificity in mammals:Determinants beyond seed pairing . Mol Cell 2007 ; 27 : 91 - 105
Grün D, Wang YL, Langenberger D, Gunsalus KC, Rajewsky N . MicroRNA target predictions across seven Drosophila species and comparison to mammalian targets . PLoS Comput Biol 2005 ; 1 : e13 -
Hausser J, Berninger P, Rodak C, Jantscher Y, Wirth S, Zavolan M . MirZ: an integrated microRNA expression atlas and target prediction resource . Nucleic Acids Res 2009 ; 36 : W266 - W272
Hofacker IL . Vienna RNA secondary structure server . Nucleic Acids Res 2003 ; 31 : 3429 - 3431
Hsu SD, Chu CH, Tsou AP, Chen SJ, Chen HC, Hsu PW, Wong YH, Chen YH, Chen GH, Huang HD . miRNAMap 2.0: genomic maps of microRNAs in metazoan genomes . Nucleic Acids Res 2008 ; 36 : D165 - D169
Huang TH, Fan B, Rothschild MF, Hu ZL, Li K, Zhao SH . miRFinder: an improved approach and software implementation for genome-wide fast microRNA precursor scans . BMC Bioinformatics 2007 ; 8 : 341 -
John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS . Human MicroRNA targets . PLos Biol 2004 ; 2 : e363 -
Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E . The role of site accessibility in miRNA target recognition . Nat Genet 2007 ; 39 : 1278 - 1284
Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N . Combinatorial microRNA target predictions . Nat Genet 2005 ; 37 : 495 - 500
Kim B-Y, Kim H, Cho E-J, Youn H-D . Nurr77 upregulates HIF-α by inhibiting pVHL-mediated degradation . Exp Mol Med 2008 ; 40 : 71 - 83
Kim SK, Nam JW, Rhee JK, Lee WJ, Zhang BT . miTarget: microRNA target-gene prediction using a support vector machine . BMC Bioinformatics 2006 ; 7 : 411 -
Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z, Hatzigeorgiou A . A combined computational-experimental approach predicts human microRNA targets . Genes Dev 2004 ; 18 : 1165 - 1178
Lall S, Grün D, Krek A, Chen K, Wang YL, Dewey CN, Sood P, Colombo T, Bray N, Macmenamin P, Kao HL, Gunsalus KC, Pachter L, Piano F, Rajewsky N . A genome-wide map of conserved microRNA targets in C. elegans . Curr Biol 2006 ; 16 : 460 - 471
Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M . A mammalian microRNA expression atlas based on small rna library sequencing . Cell 2007 ; 129 : 1401 - 1414
Lee RC, Feinbaum RL, Ambros V . The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14 . Cell 1993 ; 75 : 843 - 854
Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB . Prediction of mammalian microRNA targets . Cell 2003 ; 115 : 787 - 798
Lewis BP, Burge CB, Bartel DP . Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets . Cell 2005 ; 120 : 15 - 20
Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM . Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs . Nature 2005 ; 433 : 769 - 773
Lodish HF, Zhou B, Liu G, Chen CZ . Micromanagement of the immune system by microRNAs . Nat Rev Immunol 2008 ; 8 : 120 - 130
Long D, Lee R, Williams P, Chan CY, Ambros V, Ding Y . Potent effect of target structure on microRNA function . Nat Struct Mol Biol 2007 ; 14 : 287 - 294
Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setien F, Casado S, Suarez-Gauthier A, Sanchez-Cespedes M, Gitt A, Spiteri I, Das PP, Caldas C, Miska E, Esteller M . Genetic unmasking of an epigenetically silenced microRNA in human cancer cells . Cancer Res 2007 ; 67 : 1424 - 1429
Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, Vergoulis T, Koziris N, Sellis T, Tsanakas P, Hatzigeorgiou AG . DIANA-microT web server: elucidating microRNA functions through target prediction . Nucleic Acids Res 2009 ; 37 : W273 - W276
Mathews DH, Sabina J, Zuker M, Turner DH . Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure . J Mol Biol 1999 ; 288 : 911 - 940
Maziere P, Enright AJ . Prediction of microRNA targets . Drug Discov Today 2007 ; 12 : 452 - 458
Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I . A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes . Cell 2006 ; 126 : 1203 - 1217
Nam S, Kim B, Shin S, Lee S . miRGator: an integrated system for functional annotation of microRNAs . Nucleic Acid Res 2008 ; 36 : D159 - D164
Nam S, Li M, Choi K, Balch C, Kim S, Nephew KP . MicroRNA and mRNA integrated analysis (MMIA): a web tool for examining biological functions of microRNA expression . Nucleic Acids Res 2009 ; 37 : W356 - W362
Papadopoulos GL, Reczko M, Simossis VA, Sethupathy P, Hatzigeorgiou AG . The database of experimentally supported targets: a functional update of TarBase . Nucleic Acids Res 2009 ; 37 : D155 - D158
Rajewsky N . microRNA target predictions in animals . Nat Genet 2006 ; 38 : S8 - S13
Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R . Fast and effective prediction of microRNA/target duplexes . RNA 2004 ; 10 : 1507 - 1517
Roubelakis MG, Zotos P, Papachristoudis G, Michalopoulos I, Pappa KI, Anagnou NP, Kossida S . Human microRNA target analysis and gene ontology clustering by GOmir, a novel stand-alone application . BMC Bioinformatics 2009 ; 10 : S20 -
Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA . Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells . Cancer Cell 2006 ; 9 : 435 - 443
Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N . Widespread changes in protein synthesis induced by microRNAs . Nature 2008 ; 455 : 58 - 63
Sethupathy P, Corda B, Hatzigeorgiou AG . TarBase: A comprehensive database of experimentally supported animal microRNA targets . RNA 2006 ; 12 : 192 - 197
Stark A, Brennecke J, Russell RB, Cohen SM . Identification of Drosophila MicroRNA targets . PLoS Biol 2003 ; 1 : E60 -
Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM . Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3'UTR evolution . Cell 2005 ; 123 : 1133 - 1146
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES . Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles . Proc Natl Acad Sci USA 2005 ; 102 : 15545 - 15550
Wang X . miRDB: A microRNA target prediction and functional annotation database with a wiki interface . RNA 2008 ; 14 : 1012 - 1017
Wang X, El Naqa IM . Prediction of both conserved and nonconserved microRNA targets in animals . Bioinformatics 2008 ; 24 : 325 - 332
Watanabe Y, Tomita M, Kanai A . Computational methods for microRNA target prediction . Methods Enzymol 2007 ; 427 : 65 - 86
Wuchty S, Fontana W, Hofacker IL, Schuster P . Complete suboptimal folding of RNA and the stability of secondary structures . Biopolymers 1999 ; 49 : 145 - 165
Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T . MiRecords: an integrated resource for microRNA-target interactions . Nucleic Acids Res 2009 ; 37 : D105 - D110
Yoon S, De Micheli G . Computational identification of microRNAs and their targets . Birth Defects Res C Embryo Today 2006 ; 78 : 118 - 128
Zuker M, Stiegler P . Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information . Nucleic Acids Res 1981 ; 9 : 133 - 148
Acknowledgements
This research was supported by the Chung-Ang University Research Grants in 2009.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Min, H., Yoon, S. Got target?: computational methods for microRNA target prediction and their extension. Exp Mol Med 42, 233–244 (2010). https://doi.org/10.3858/emm.2010.42.4.032
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3858/emm.2010.42.4.032
Keywords
This article is cited by
-
Comprehensive machine-learning-based analysis of microRNA–target interactions reveals variable transferability of interaction rules across species
BMC Bioinformatics (2021)
-
Overexpression of HOXA10 is associated with unfavorable prognosis of acute myeloid leukemia
BMC Cancer (2020)
-
LINC00649 underexpression is an adverse prognostic marker in acute myeloid leukemia
BMC Cancer (2020)
-
Using exosomal miRNAs extracted from porcine follicular fluid to investigate their role in oocyte development
BMC Veterinary Research (2020)
-
Polymorphisms in miRNA binding sites involved in metabolic diseases in mice and humans
Scientific Reports (2020)