Published online Oct 15, 2002. doi: 10.3748/wjg.v8.i5.857
Revised: April 15, 2002
Accepted: April 20, 2002
Published online: October 15, 2002
AIM: To determine the frequencies of HGV and TTV infections in serum and saliva samples of non-hepatitis patients with oral diseases in Hangzhou area, and to understand the correlation between detected results of HGV RNA and/or TTV DNA in sera and in saliva from the same patients.
METHODS: RT-nested PCR for HGV RNA detection and semi-nested PCR for TTV DNA detection were performed in the serum and saliva samples from 226 non-hepatitis patients with oral diseases, and nucleotide sequence analysis.
RESULTS: Twenty-seven (11.9%) and 21 (9.3%) of the 226 serum samples were only positive for HGV RNA and TTV DNA, respectively. 10 (4.4%) and 9 (3.9%) of the 226 saliva samples were only positive for HGV RNA and TTV DNA, respectively. And 7 (3.1%) of the serum samples and 2 (0.9%) of the saliva samples showed the positive amplification results for both HGV RNA and TTV DNA. 12 saliva samples from the 34 patients (35.3%) with HGV or HGV/TTV viremia and 11 saliva samples from the 28 patients (39.3%) with TTV or HGV/TTV viremia were HGV RNA detectable, respectively, including two patients positive for both HGV RNA and TTV DNA in serum and saliva samples. No saliva samples from the 226 patients were found to be HGV RNA or TTV DNA detectable while their serum samples were negative for HGV or TTV. Homologies of the nucleotide sequences of HGV and TTV amplification products from the serum and saliva samples of the two patients compared with the reported sequences were 88.65%-91.49% and 65.32%-66.67%, respectively. In comparison with the nucleotide sequences of amplification products between serum and from saliva sample from any one of the two patients, the homologies were 98.58% and 99.29% for HGV, and were 98.65% and 98.20% for TTV, respectively.
CONCLUSION: Relatively high carrying rates of HGV and/or TTV in the sera of non-hepatitis patients with oral diseases in Hangzhou area are demonstrated. Parts of the carriers are HGV and/or TTV positive in their saliva. The results of this study indicate that dentists may be one of the populations with high risk for HGV and/or TTV infection, and by way of saliva HGV and TTV may be transmitted among individuals.
- Citation: Yan J, Chen LL, Lou YL, Zhong XZ. Investigation of HGV and TTV infection in sera and saliva from non-hepatitis patients with oral diseases. World J Gastroenterol 2002; 8(5): 857-862
- URL: https://www.wjgnet.com/1007-9327/full/v8/i5/857.htm
- DOI: https://dx.doi.org/10.3748/wjg.v8.i5.857
Viral hepatitis is relatively common in China[1-13]. Except for human A-E hepatitis viruses, hepatitis G virus (HGV) and GB virus C (GBV-C) were recently identified as the causative agents of human non-A-E hepatitis[14,15]. Transfusion transmitted virus (TTV), another novel virus isolated from a patient suffering from non-A-G hepatitis, was considered to be responsible for human non-A-G hepatitis[16]. Since the nucleotide sequences and putative amino acid sequences of HGV and GBV-C genomes show high homologies of 85% and 100%, respectively, the two viral agents are considered to be the different isolates of the same virus belonging to Flaviviridae family[14,15]. TTV was proposed to be a member of a new virus family temporarily named Circinoviridae[17]. Although HGV is a positive-stranded RNA virus and TTV is a negative-stranded DNA virus, the two viruses are mainly transmitted through a hematogenous pathway[18-21], especially through blood transfusion[22-25]. Many investigation data demonstrated that HGV or TTV was able to cause a long-term asymptomatic viremia in non A-E hepatitis patients and only a few patients could suffer from hepatitis with mild liver in-jury[26-32]. On the contrary, a few references reported that HGV infection might accelerate the progression of chronic liver diseases and cause fulminant hepatic failure[33-37]. TTV was frequently found in non-A-G hepatitis patients with a single elevation of alanine aminotransferase (ALT)[38-40]. Recent published literatures revealed that HGV or TTV infection frequency in peripheral bloods of normal populations including healthy blood donors was found to be relatively high[41-50] and part of the infected individuals were HGV RNA or TTV DNA detectable not only in the serum samples but also in their saliva samples[51-55]. Therefore, dentists may be one of the populations at high risk of HGV and TTV infection, and may play an important role in HGV and TTV transmission among individuals. On the other hand, the possibility of HGV and TTV transmission through saliva is also an interesting subject for investigation.
To determine the frequency of HGV or TTV infection in non-hepatitis patients with oral diseases and to understand the correlation of HGV and TTV detection between in sera and in saliva from the same one patients, RT-nested PCR and semi-nested PCR were applied to detect HGV and TTV respectively in the serum and saliva samples from 226 non-hepatitis patients for oral treatment in Hangzhou area. The HGV RNA or TTV DNA positive amplification products from representatives of the serum and saliva samples were cloned and then sequenced. The results of this study may contribute to understand transmission mechanism of the two viruses and populations with high risk of HGV and TTV infection, and may be helpful for the control of the prevalence of human hepatitis associated with HGV or TTV.
Total 226 patients with tooth, pulp or periodontal diseases came from the oral departments of three hospitals in Hangzhou, Zhejiang Province of China, respectively. Each of the patients was told the aim and content of this study and all of them agreed to be the volunteers. A serum sample as well as a saliva sample from each of the patients was collected. These samples were immediately performed for HGV and TTV detection or temporarily stored at -20 °C. The patients were confirmed to have no any clinical signals of hepatitis and no elevation of ALT by conventional hepatic examinations, and the serum samples were negative for hepatitis A-C viruses by PCR and EIA. The reagents used in reverse transcription (RT) and PCR were purchased from Boehringer and BMI, and the other reagents used in this study came from Sigma.
Isolation of total RNA and DNA in sera Total RNA in 200 μL of each serum samples was prepared by Trizol method according to the manufacturer's instruction and then dissolved in 50 μL of DEPC treated super-purified water. 200 μL of each serum samples was mixed with 200 μL DNA extraction solution containing 20 mmol•L-1 EDTA, 0.5% (W/W) SDS, 10 mmol•L-1 Tris-HCl and 40 mg proteinase K (pH8.0) and was treated at 55 °C for 30 min. Total DNA in the mixture was extracted by phenol-chloroform method[56] and then dissolved in 50 μL of TE buffer (pH8.0).
Isolation of total RNA and DNA in saliva 200 μL of each saliva samples was mixed with 400 mL RNA extraction solution containing 4 mol•L-1 guanidine thiocyanate, 0.1 mol•L-1 2-mercaptoethanol, 0.5% (W/W) sodium N-lauroylsarcosine and 25 mmol•L-1 sodium citrate (pH7.0). Vortex the tube gently for 15 min and lay the tube at room temperature for 5 min. 100 mL 2 mol•L-1 sodium acetate (pH4.0), 600 mL TE buffer (pH8.0) and 200 mL phenol (pH4.5)-chloroform (5:1, V:V) were added into the tube and then mixed. Supernatant of the mixture after centrifugation was added with 400 mL 2-propanol (-20 °C) and then mixed. Lay the tube at -20 °C for 2 h to precipitate RNA. RNA pellet obtained by using centrifugation at 4 °C was washed with 75% ethanol and then dissolved in 50 μL of DEPC treated super-purified water[51,52]. 200 μL of each saliva samples was mixed with 200 mL DNA extraction solution containing 20 mmol•L-1 EDTA, 100 mmol•L-1 NaCl, 0.5% (W/W) SDS, 10 mmol•L-1 Tris-HCl (pH8.0) and 80 μg proteinase K. Incubate the tube at 55 °C for 60 min. Total DNA was obtained by phenol-chloroform method[56] and then dissolved in 50 μL of TE buffer (pH8.0)[53-55].
RT-nested PCR for HGV RNA detection Ten microliters of total RNA preparation was mixed with 10 μL RT master mixture containing 5 μmol•L-1 hexanucleotide as random primers, 12.5 mol•L-1 each of dNTP, 20 U M-MuLV-reverse transcriptase, 20 U RNase inhibitor and 4 μL of 5 × RT buffer (pH8.3). The steps of RT were described as the following: at 70 °C × 5 min for denaturation, at 42 °C × 60 min for cDNA synthesis, and at 70 °C × 10 min to stop the reaction.
Primers derived from HGV 5'-NCR were used in the RT-nested PCR for HGV detection[13,50]. The external primers: 5'-ATGACAGGGTTGGTAGGTCGTAAATC-3' (sense), 5'-CCCCACTGGTCCTTGTCAACTCGCCG-3' (antisense). The internal primers: 5'-TGGTAGCCACTATAGGTGGGTCTTAA-3' (sense), 5'-ACATTGAAGGGCGACGTGGACCGTAC-3' (antisense). For the first round PCR, 10 μL of RT product was mixed with 90 μL PCR master mixture containing 250 nmol•L-1 each of the external primers, 2.5 mol•L-1 each of dNTP, 25 mol•L-1 MgCl2, 2.5 U of Taq DNA polymerase and 10 μL 10 × PCR buffer (pH8.3). For the second round PCR, 5 μL product from the first round PCR was used as template, and the other reaction reagents were the same as that in the first round PCR except of the primers. DNA Thermal cycler 480 (Perkin Elmer) was used for amplification. The parameters for each of the PCR rounds were described as the following: 94 °C 5 min (× 1); 94 °C 1 min, 56 °C 1 min, 72 °C 1.5 min (× 35); 72 °C 7 min (× 1). The expected size of target fragments amplified from HGV 5'-NCR RNA was 193 bp.
Semi-nested PCR for TTV DNA detection The primers used in the semi-nested PCR for TTV DNA detection were the same described by Okamoto et al[57]. The external primers: 5'-ACAGACAGAGGAGAAGGCAACATG-3' (sense), 5'-CTGGCATTTTACCATTTCCAAAGTT-3' (antisense). The internal primers: 5'-GGCAACATGTTATGATAGACTGG-3' (sense), 5'-CTGGCATTTTACCATTTCCAAAGTT-3' (antisense). Except the primers, MgCl2 concentration (15 mol•L-1), total reaction volume (50 μL) and annealing temperature (60 °C), the compositions and concentrations of other reaction agents and the parameters for the semi-nested PCR was the same as that of the RT-nested PCR for HGV detection. The expected size of target fragments amplified from TTV DNA was 271 bp.
Examination of amplification products The results of amplification reactions were observed on UV light after electrophoresis of 20 g•L-1 agarose stained with ethidium bromide. 100 bp DNA ladder was used as a size marker to estimate the length of products.
Cloning and sequencing of PCR amplification products The target DNA fragments of HGV or TTV amplification products by PCR were cloned into plasmid pGEM-T vector by using T-A cloning kit according to the manufacturer’s instruction. The recombinant plasmid was amplified in E. coli strain DH5α and then recovered by Sambrook's method. A professional company (sangon) was responsible for the analysis of nucleotide sequences of the inserted fragments. Homology of the nucleotide sequences was compared with those reported respectively[14,56].
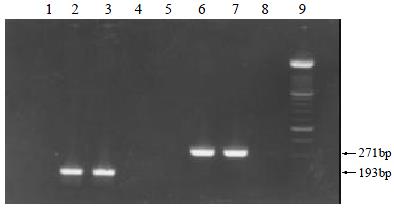
Twenty-seven (11.9%) and 21 (9.3%) of the 226 serum samples were only positive for HGV RNA and TTV DNA, respectively. 10 (4.4%) and 9 (3.9%) of the 226 saliva samples were only detectable for HGV RNA and TTV DNA, respectively. And 7 (3.1%) of the serum samples and 2 (0.9%) of the saliva samples showed the positive results for both HGV RNA and TTV DNA. 171 of the 226 serum samples (75.7%) and 205 of the 226 saliva samples (90.7%) were undetectable for both HGV RNA and TTV DNA. Representatives of the target fragments respectively amplified from HGV and TTV genomes in the serum and saliva samples were shown in Figure 1.
Twelve saliva samples from the 34 patients (35.3%) positive for HGV RNA and 11 saliva samples from the 28 patients (39.3%) positive for TTV DNA in their sera were HGV RNA and TTV DNA detectable respectively, including 2 patients with both HGV RNA and TTV in their serum and saliva samples. No saliva samples from the 226 patients were found to be positive for HGV RNA or TTV DNA but their serum samples were negative for HGV or TTV. Distribution of HGV and TTV detection results in serum and saliva samples of the 226 patients was shown in Table 1.
In comparison with the reported HGV[14] and TTV genotype-1a[57] nucleotide sequences, the homologies of HGV and TTV amplification products from 2 serum and 2 saliva samples from the two same patients (No.: 010906 and 011022) were 88.65%-91.49% and 65.32%-66.67%, respectively. When to compare the nucleotide sequences of HGV and TTV amplification products between the 2 serum samples and between the 2 sliva samples, the homologies were as high as 98.65% and 98.20%, 93.24% and 94.59%, respectively. The homology comparison mentioned above did not contain the primer sequences (Figure 2 and Figure 3).
Viral hepatitis is a common infectious disease of human beings, which is prevalent in all countries of the world and causes a serious healthy problem. Although hepatitis virus A-E have been demonstrated to be the causative agents of human hepatitis A-E respectively, approximately 20% of acute and 15% of chronic hepatitis were associated with unknown etiology[23,24]. HGV and TTV were recently identified as transfusion-transmitted viruses and were considered to be responsible for human non-A-E hepatitis[14-17]. Many published literatures reported that among the patients infected with HGV or TTV, a few of them showed mild liver injury, which resulting in a conclusion of a low clinical importance of HGV or TTV infection[26-32]. However, a wide variety of questions about the potential pathogenicity of HGV and TTV infection still remain unanswered[13,23]. So far the assays for HGV and TTV detection are not the routing laboratory examination items in many countries and areas. Not few of investigation data indicated that high frequencies of HGV or TTV infection were frequently found in different normal populations such as in healthy blood donors[41-50]. Some investigation results revealed that the genomic nuclear of HGV or TTV in saliva of partial infected individuals was also detectable[51-55], suggesting the possibility by way of saliva for transmission of the two viruses. Although HGV or TTV infection might be not clinically significant, the existence of extensive and potential infectious sources of the two viruses will still generate a great problem for human health.
In the present study, HGV and TTV infection rates in serum samples of the 226 non-hepatitis patients with oral diseases were 15.0% (34/226) and 12.4% (28/226) respectively, which indicated that HGV infection rate was obviously higher than that of the reported in other areas of China and abroad (1.3%-10.6%)[7,8], and TTV infection rate was higher than that of the reported abroad (< 5%)[41,43-47] but similar to the reported in China (9.3%-16.8%)[42,48-50]. Such high HGV and TTV infection frequencies in the non-hepatitis population in Hangzhou are probably due to geographic difference. In the previous study, we found 2.5% of healthy blood donors in Hangzhou coinfected with both HGV and TTV[50]. In this study, the serum samples from 7 of the 226 patients (3.1%) were positive for both HGV RNA and TTV DNA, which offered further evidence for the existence of coinfection of the two viruses. Therefore, it is valuable to make further studies to determine the pathological and clinical significance of the coinfection.
Since the patients with oral diseases in this study did not show any clinical symptoms and laboratory markers for hepatitis, which suggested that most of the patients infected with HGV and/or TTV are usually asymptomatic. However, these asymptomatic patients carrying HGV and/or TTV may be more risky and important for the source of infection. It was worthy to notice that 5.3% and 4.9% of the non-hepatitis patients with oral diseases were HGV RNA and/or TTV DNA detectable in the saliva, indicating dentists may be one of the populations at high risk of HGV and/or TTV infection except of blood precipitants and hemodialysed patients[18,25,31,32,57,58]. It was more important that HGV and TTV in saliva might be transmitted among the patients for oral treatment through dentists’ hands and therapeutic instruments. So it is necessary to take effective measures to block the possible transmission of HGV and TTV by way of saliva.
In this study, 2 serum samples and 2 saliva samples from the two patients showed positive results for both HGV RNA and TTV DNA. The nucleotide sequences of HGV amplification products from these samples are highly homologous (88.65%-91.49%) to the sequence reported by Linnen et al[14]. This result of sequence analysis indicates that the RT-nested PCR used in this study is reliable for HGV RNA detection. To analyze the details of the nucleotide sequencing data, it seems to show a HGV genotype different from the reported[59-62]. Lower homology (65.32%-66.67%) of the nucleotide sequences of TTV Semi-nested PCR products from these samples of the 2 patients compared with the reported TTV genotype 1a sequence[56] reveals the genomic heterogeneity of TTV isolates from different areas, and this finding accords with the conclusions of previously published literatures[31,56]. In this study, no saliva samples from the 226 patients were found to be positive for HGV RNA or TTV DNA while their serum samples were negative for HGV or TTV. Furthermore, the nucleotide sequence homology of HGV amplification products between the serum and saliva samples from any one of the 2 patients is 98.65% and 98.20%, respectively, and so is the nucleotide sequence homology of TTV amplification products (93.24% and 94.59%), which are much higher than that compared with the reported HGV and TTV sequences[14,56]. These data indicate a closely and directly correlation between HGV and TTV genomes detected from serum and saliva from the same one patients.
Edited by Zhao P
| 1. | Han FC, Hou Y, Yan XJ, Xiao LY, Guo YH. Dot immunogold filtration assay for rapid detection of anti-HAV IgM in Chinese. World J Gastroenterol. 2000;6:400-401. [PubMed] [Cited in This Article: ] |
| 2. | Zhao YL, Meng ZD, Xu ZY, Guo JJ, Chai SA, Duo CG, Wang XY, Yao JF, Liu HB, Qi SX. H2 strain attenuated live hepatitis A vaccines: protective efficacy in a hepatitis A outbreak. World J Gastroenterol. 2000;6:829-832. [PubMed] [Cited in This Article: ] |
| 3. | Fang JN, Jin CJ, Cui LH, Quan ZY, Choi BY, Ki M, Park HB. A comparative study on serologic profiles of virus hepatitis B. World J Gastroenterol. 2001;7:107-110. [PubMed] [Cited in This Article: ] |
| 4. | Tang RX, Gao FG, Zeng LY, Wang YW, Wang YL. Detection of HBV DNA and its existence status in liver tissues and peripheral blood lymphocytes from chronic hepatitis B patients. World J Gastroenterol. 1999;5:359-361. [PubMed] [Cited in This Article: ] |
| 5. | Wei J, Wang YQ, Lu ZM, Li GD, Wang Y, Zhang ZC. Detection of anti-preS1 antibodies for recovery of hepatitis B patients by immunoassay. World J Gastroenterol. 2002;8:276-281. [PubMed] [Cited in This Article: ] |
| 6. | Xiao LY, Yan XJ, Mi MR, Han FC, Hou Y. Preliminary study of a dot immunogold filtration assay for rapid detection of anti-HCV IgG. World J Gastroenterol. 1999;5:349-350. [PubMed] [Cited in This Article: ] |
| 7. | Song YH, Lin JS, Liu NZ, Kong XJ, Xie N, Wang NX, Jin YX, Liang KH. Anti-HBV hairpin ribozyme-mediated cleavage of target RNA in vitro. World J Gastroenterol. 2002;8:91-94. [PubMed] [Cited in This Article: ] |
| 8. | Li CP, Wang KX, Wang J, Pan BR. mIL-2R, T cell subsets & amp; hepatitis C. World J Gastroenterol. 2002;8:298-300. [PubMed] [Cited in This Article: ] |
| 9. | Yan FM, Chen AS, Hao F, Zhao XP, Gu CH, Zhao LB, Yang DL, Hao LJ. Hepatitis C virus may infect extrahepatic tissues in patients with hepatitis C. World J Gastroenterol. 2000;6:805-811. [PubMed] [Cited in This Article: ] |
| 10. | Chen MY, Huang ZQ, Chen LZ, Gao YB, Peng RY, Wang DW. Detection of hepatitis C virus NS5 protein and genome in Chinese carcinoma of the extrahepatic bile duct and its significance. World J Gastroenterol. 2000;6:800-804. [PubMed] [Cited in This Article: ] |
| 11. | Wang XZ, Jiang XR, Chen XC, Chen ZX, Li D, Lin JY, Tao QM. Seek protein which can interact with hepatitis B virus X protein from human liver cDNA library by yeast two-hybrid system. World J Gastroenterol. 2002;8:95-98. [PubMed] [Cited in This Article: ] |
| 12. | Wang NS, Liao LT, Zhu YJ, Pan W, Fang F. Follow-up study of hepatitis C virus infection in uremic patients on maintenance hemodialysis for 30 mo. World J Gastroenterol. 2000;6:888-892. [PubMed] [Cited in This Article: ] |
| 13. | Yan J, Dennin RH. A high frequency of GBV-C/HGV coinfection in hepatitis C patients in Germany. World J Gastroenterol. 2000;6:833-841. [PubMed] [Cited in This Article: ] |
| 14. | Linnen J, Wages J, Zhang-Keck ZY, Fry KE, Krawczynski KZ, Alter H, Koonin E, Gallagher M, Alter M, Hadziyannis S. Molecular cloning and disease association of hepatitis G virus: a transfusion-transmissible agent. Science. 1996;271:505-508. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 957] [Cited by in F6Publishing: 882] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 15. | Muerhoff AS, Leary TP, Simons JN, Pilot-Matias TJ, Dawson GJ, Erker JC, Chalmers ML, Schlauder GG, Desai SM, Mushahwar IK. Genomic organization of GB viruses A and B: two new members of the Flaviviridae associated with GB agent hepatitis. J Virol. 1995;69:5621-5630. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 10] [Reference Citation Analysis (0)] |
| 16. | Nishizawa T, Okamoto H, Konishi K, Yoshizawa H, Miyakawa Y, Mayumi M. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem Biophys Res Commun. 1997;241:92-97. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 856] [Cited by in F6Publishing: 822] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 17. | Mushahwar IK, Erker JC, Muerhoff AS, Leary TP, Simons JN, Birkenmeyer LG, Chalmers ML, Pilot-Matias TJ, Dexai SM. Molecular and biophysical characterization of TT virus: evidence for a new virus family infecting humans. Proc Natl Acad Sci USA. 1999;96:3177-3182. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 298] [Cited by in F6Publishing: 298] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 18. | Simmonds P, Smith DB. Structural constraints on RNA virus evolution. J Virol. 1999;73:5787-5794. [PubMed] [Cited in This Article: ] |
| 19. | Abe K, Inami T, Ishikawa K, Nakamura S, Goto S. TT virus infection in nonhuman primates and characterization of the viral genome: identification of simian TT virus isolates. J Virol. 2000;74:1549-1553. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 37] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 20. | Moaven LD, Tennakoon PS, Bowden DS, Locarnini SA. Mother-to-baby transmission of hepatitis G virus. Med J Aust. 1996;165:84-85. [PubMed] [Cited in This Article: ] |
| 21. | Okamoto H, Akahane Y, Ukita M, Fukuda M, Tsuda F, Miyakawa Y, Mayumi M. Fecal excretion of a nonenveloped DNA virus (TTV) associated with posttransfusion non-A-G hepatitis. J Med Virol. 1998;56:128-132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 6] [Reference Citation Analysis (0)] |
| 22. | Feucht HH, Zollner B, Polywka S, Laufs R. Vertical transmission of hepatitis G. Lancet. 1996;347:615-616. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 80] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 23. | Mortimer PP. HGV and the blood supply. J Infect. 1998;37:106-107. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.0] [Reference Citation Analysis (0)] |
| 24. | Tanaka H, Okamoto H, Luengrojanakul P, Chainuvati T, Tsuda F, Tanaka T, Miyakawa Y, Mayumi M. Infection with an unenveloped DNA virus (TTV) associated with posttransfusion non-A to G hepatitis in hepatitis patients and healthy blood donors in Thailand. J Med Virol. 1998;56:234-238. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 5] [Reference Citation Analysis (0)] |
| 25. | Yang SS, Wu CH, Chen TH, Huang YY, Huang CS. TT viral infection through blood transfusion: retrospective investigation on patients in a prospective study of post-transfusion hepatitis. World J Gastroenterol. 2000;6:70-73. [PubMed] [Cited in This Article: ] |
| 26. | Sarrazin C, Herrmann G, Roth WK, Lee JH, Marx S, Zeuzem S. Prevalence and clinical and histological manifestation of hepatitis G/GBV-C infections in patients with elevated aminotransferases of unknown etiology. J Hepatol. 1997;27:276-283. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 27. | Alter HJ. The cloning and clinical implications of HGV and HGBV-C. N Engl J Med. 1996;334:1536-1537. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 191] [Cited by in F6Publishing: 192] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 28. | Feucht HH, Zöllner B, Polywka S, Knödler B, Schröter M, Nolte H, Laufs R. Prevalence of hepatitis G viremia among healthy subjects, individuals with liver disease, and persons at risk for parenteral transmission. J Clin Microbiol. 1997;35:767-768. [PubMed] [Cited in This Article: ] |
| 29. | Naoumov NV, Petrova EP, Thomas MG, Williams R. Presence of a newly described human DNA virus (TTV) in patients with liver disease. Lancet. 1998;352:195-197. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 226] [Cited by in F6Publishing: 232] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 30. | Alter MJ, Gallagher M, Morris TT, Moyer LA, Meeks EL, Krawczynski K, Kim JP, Margolis HS. Acute non-A-E hepatitis in the United States and the role of hepatitis G virus infection. Sentinel Counties Viral Hepatitis Study Team. N Engl J Med. 1997;336:741-746. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 281] [Cited by in F6Publishing: 289] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 31. | Forns X, Hegerich P, Darnell A, Emerson SU, Purcell RH, Bukh J. High prevalence of TT virus (TTV) infection in patients on maintenance hemodialysis: frequent mixed infections with different genotypes and lack of evidence of associated liver disease. J Med Virol. 1999;59:313-317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 5] [Reference Citation Analysis (0)] |
| 32. | Kao JH, Chen W, Hsiang SC, Chen PJ, Lai MY, Chen DS. Prevalence and implication of TT virus infection: minimal role in patients with non-A-E hepatitis in Taiwan. J Med Virol. 1999;59:307-312. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 6] [Reference Citation Analysis (0)] |
| 33. | Fiordalisi G, Zanella I, Mantero G, Bettinardi A, Stellini R, Paraninfo G, Cadeo G, Primi D. High prevalence of GB virus C infection in a group of Italian patients with hepatitis of unknown etiology. J Infect Dis. 1996;174:181-183. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 104] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 34. | Sugai Y, Nakayama H, Fukuda M, Sawada N, Tanaka T, Tsuda F, Okamoto H, Miyakawa Y, Mayumi M. Infection with GB virus C in patients with chronic liver disease. J Med Virol. 1997;51:175-181. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 2] [Reference Citation Analysis (0)] |
| 35. | Yoshiba M, Okamoto H, Mishiro S. Detection of the GBV-C hepatitis virus genome in serum from patients with fulminant hepatitis of unknown aetiology. Lancet. 1995;346:1131-1132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 299] [Cited by in F6Publishing: 305] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 36. | Heringlake S, Osterkamp S, Trautwein C, Tillmann HL, Böker K, Muerhoff S, Mushahwar IK, Hunsmann G, Manns MP. Association between fulminant hepatic failure and a strain of GBV virus C. Lancet. 1996;348:1626-1629. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 141] [Cited by in F6Publishing: 146] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 37. | Tanaka T, Hess G, Tanaka S, Kohara M. The significance of hepatitis G virus infection in patients with non-A to C hepatic diseases. Hepatogastroenterology. 1999;46:1870-1873. [PubMed] [Cited in This Article: ] |
| 38. | Takayama S, Miura T, Matsuo S, Taki M, Sugii S. Prevalence and persistence of a novel DNA TT virus (TTV) infection in Japanese haemophiliacs. Br J Haematol. 1999;104:626-629. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 45] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 39. | Sugiyama T, Shimizu M, Yamauchi O, Kojima M. [Seroepidemiological survey of TT virus (TTV) infection in an endemic area for hepatitis C virus]. Nihon Rinsho. 1999;57:1402-1405. [PubMed] [Cited in This Article: ] |
| 40. | Cleavinger PJ, Persing DH, Li H, Moore SB, Charlton MR, Sievers C, Therneau TM, Zein NN. Prevalence of TT virus infection in blood donors with elevated ALT in the absence of known hepatitis markers. Am J Gastroenterol. 2000;95:772-776. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 17] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 41. | Park YM, Mizokami M, Nakano T, Choi JY, Cao K, Byun BH, Cho CH, Jung YT, Paik SY, Yoon SK. GB virus C/hepatitis G virus infection among Korean patients with liver diseases and general population. Virus Res. 1997;48:185-192. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 42. | Ling BH, Zhuang H, Cui YH, An WF, Li ZJ, Wang SP, Zhu WF. A cross-sectional study on HGV infection in a rural population. World J Gastroenterol. 1998;4:489-492. [PubMed] [Cited in This Article: ] |
| 43. | Casteling A, Song E, Sim J, Blaauw D, Heyns A, Schweizer R, Margolius L, Kuun E, Field S, Schoub B. GB virus C prevalence in blood donors and high risk groups for parenterally transmitted agents from Gauteng, South Africa. J Med Virol. 1998;55:103-108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 3] [Reference Citation Analysis (0)] |
| 44. | Blair CS, Davidson F, Lycett C, McDonald DM, Haydon GH, Yap PL, Hayes PC, Simmonds P, Gillon J. Prevalence, incidence, and clinical characteristics of hepatitis G virus/GB virus C infection in Scottish blood donors. J Infect Dis. 1998;178:1779-1782. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 27] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 45. | Goubau P, Andrade FB, Liu HF, Basilio FP, Croonen L, Barreto-Gomes VA. Prevalence of GB virus C/hepatitis G virus among blood donors in north-eastern Brazil. Trop Med Int Health. 1999;4:365-367. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 46. | Simmonds P, Davidson F, Lycett C, Prescott LE, MacDonald DM, Ellender J, Yap PL, Ludlam CA, Haydon GH, Gillon J. Detection of a novel DNA virus (TTV) in blood donors and blood products. Lancet. 1998;352:191-195. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 292] [Cited by in F6Publishing: 302] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 47. | Gad A, Tanaka E, Orii K, Kafumi T, Serwah AE, El-Sherif A, Nooman Z, Kiyosawa K. Clinical significance of TT virus infection in patients with chronic liver disease and volunteer blood donors in Egypt. J Med Virol. 2000;60:177-181. [PubMed] [Cited in This Article: ] |
| 48. | Chen YP, Liang WF, Zhang L, He HT, Luo KX. Transfusion transmitted virus infection in general populations and patients with various liver diseases in south China. World J Gastroenterol. 2000;6:738-741. [PubMed] [Cited in This Article: ] |
| 49. | Huang CH, Zhou YS, Chen RG, Xie CY, Wang HT. The prevalence of transfusion transmitted virus infection in blood donors. World J Gastroenterol. 2000;6:268-270. [PubMed] [Cited in This Article: ] |
| 50. | Yan J, Chen LL, Luo YH, Mao YF, He M. High frequencies of HGV and TTV infections in blood donors in Hangzhou. World J Gastroenterol. 2001;7:637-641. [PubMed] [Cited in This Article: ] |
| 51. | Chen M, Sönnerborg A, Johansson B, Sällberg M. Detection of hepatitis G virus (GB virus C) RNA in human saliva. J Clin Microbiol. 1997;35:973-975. [PubMed] [Cited in This Article: ] |
| 52. | Seemayer CA, Viazov S, Philipp T, Roggendorf M. Detection of GBV-C/HGV RNA in saliva and serum, but not in urine of infected patients. Infection. 1998;26:39-41. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 18] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 53. | Ross RS, Viazov S, Runde V, Schaefer UW, Roggendorf M. Detection of TT virus DNA in specimens other than blood. J Clin Virol. 1999;13:181-184. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 79] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 54. | Inami T, Konomi N, Arakawa Y, Abe K. High prevalence of TT virus DNA in human saliva and semen. J Clin Microbiol. 2000;38:2407-2408. [PubMed] [Cited in This Article: ] |
| 55. | Deng X, Terunuma H, Handema R, Sakamoto M, Kitamura T, Ito M, Akahane Y. Higher prevalence and viral load of TT virus in saliva than in the corresponding serum: another possible transmission route and replication site of TT virus. J Med Virol. 2000;62:531-537. [PubMed] [Cited in This Article: ] |
| 56. | Okamoto H, Takahashi M, Nishizawa T, Ukita M, Fukuda M, Tsuda F, Miyakawa Y, Mayumi M. Marked genomic heterogeneity and frequent mixed infection of TT virus demonstrated by PCR with primers from coding and noncoding regions. Virology. 1999;259:428-436. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 181] [Cited by in F6Publishing: 184] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 57. | Tian DY, Yang DF, Xia NS, Zhang ZG, Lei HB, Huang YC. The serological prevalence and risk factor analysis of hepatitis G virus infection in Hubei Province of China. World J Gastroenterol. 2000;6:585-587. [PubMed] [Cited in This Article: ] |
| 58. | Hu ZJ, Lang ZW, Zhou YS, Yan HP, Huang DZ, Chen WR, Luo ZX. Clinicopathological study on TTV infection in hepatitis of unknown etiology. World J Gastroenterol. 2002;8:288-293. [PubMed] [Cited in This Article: ] |
| 59. | Handajani R, Soetjipto MI, Suryohudoyo P, Adi P, Setiawan PB, Nidom CA, Soemarto R, Katayama Y, Fujii M, Hotta H. Prevalence of GB virus C/Hepatitis G virus infection among various populations in Surabaya, Indonesia, and identification of novel groups of sequence variants. J Clin Microbiol. 2000;38:662-668. [PubMed] [Cited in This Article: ] |
| 60. | Wang XT, Zhuang H, Song HB, Li HM, Zhang HY, Yu Y. Partial sequencing of 5' noncoding region of 7 HGV strains isolated from different areas of China. World J Gastroenterol. 1999;5:432-434. [PubMed] [Cited in This Article: ] |
| 61. | Smith DB, Cuceanu N, Davidson F, Jarvis LM, Mokili JL, Hamid S, Ludlam CA, Simmonds P. Discrimination of hepatitis G virus/GBV-C geographical variants by analysis of the 5' non-coding region. J Gen Virol. 1997;78:1533-1542. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 90] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 62. | López-Alcorocho JM, Castillo I, Tomás JF, Carreño V. Identification of a novel GB type C virus/hepatitis G virus subtype in patients with hematologic malignancies. J Med Virol. 1999;57:80-84. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 3] [Reference Citation Analysis (0)] |