Published online Feb 14, 2022. doi: 10.3748/wjg.v28.i6.653
Peer-review started: September 13, 2021
First decision: November 16, 2021
Revised: November 19, 2021
Accepted: January 19, 2022
Article in press: January 19, 2022
Published online: February 14, 2022
Gastric cancer (GC) is one of the most frequently diagnosed tumor globally. In most cases, GC develops in a stepwise manner from chronic gastritis or atrophic gastritis (AG) to cancer. One of the major issues in clinical settings of GC is diagnosis at advanced disease stages resulting in poor prognosis. MicroRNAs (miRNAs) are small noncoding molecules that play an essential role in a variety of fundamental biological processes. However, clinical potential of miRNA profiling in the gastric cancerogenesis, especially in premalignant GC cases, remains unclear.
To evaluate the AG and GC tissue miRNomes and identify specific miRNAs’ potential for clinical applications (e.g., non-invasive diagnostics).
Study included a total of 125 subjects: Controls (CON), AG, and GC patients. All study subjects were recruited at the Departments of Surgery or Gastroenterology, Hospital of Lithuanian University of Health Sciences and divided into the profiling (n = 60) and validation (n = 65) cohorts. Total RNA isolated from tissue samples was used for preparation of small RNA sequencing libraries and profiled using next-generation sequencing (NGS). Based on NGS data, deregulated miRNAs hsa-miR-129-1-3p and hsa-miR-196a-5p were analyzed in plasma samples of independent cohort consisting of CON, AG, and GC patients. Expression level of hsa-miR-129-1-3p and hsa-miR-196a-5p was determined using the quantitative real-time polymerase chain reaction and 2-ΔΔCt method.
Results of tissue analysis revealed 20 differentially expressed miRNAs in AG group compared to CON group, 129 deregulated miRNAs in GC compared to CON, and 99 altered miRNAs comparing GC and AG groups. Only 2 miRNAs (hsa-miR-129-1-3p and hsa-miR-196a-5p) were identified to be step-wise deregulated in healthy-premalignant-malignant sequence. Area under the curve (AUC)-receiver operating characteristic analysis revealed that expression level of hsa-miR-196a-5p is significant for discrimination of CON vs AG, CON vs GC and AG vs GC and resulted in AUCs: 88.0%, 93.1% and 66.3%, respectively. Compar-ing results in tissue and plasma samples, hsa-miR-129-1-3p was significantly down-regulated in GC compared to AG (P = 0.0021 and P = 0.024, tissue and plasma, respectively). Moreover, analysis revealed that hsa-miR-215-3p/5p and hsa-miR-934 were significantly deregulated in GC based on Helicobacter pylori (H. pylori) infection status [log2 fold change (FC) = -4.52, P-adjusted = 0.02; log2FC = -4.00, P-adjusted = 0.02; log2FC = 6.09, P-adjusted = 0.02, respectively].
Comprehensive miRNome study provides evidence for gradual deregulation of hsa-miR-196a-5p and hsa-miR-129-1-3p in gastric carcinogenesis and found hsa-miR-215-3p/5p and hsa-miR-934 to be significantly deregulated in H. pylori carrying GC patients.
Core Tip: In this research we aimed to evaluate microRNAs profiles of premalignant and malignant stages of gastric cancer (GC). To date this is the first study analyzing atrophic gastritis (AG) and GC tissue miRNomes in the subjects of European origin using next-generation sequencing approach. We showed that hsa-miR-196a-5p expression in tissue is significant for discrimination between controls and AG or GC, while hsa-miR-129-1-3p is potential candidate for non-invasive GC diagnostic. This study provides novel insights into complex GC pathogenesis cascade and might be highly significant for future studies of new AG or GC associated epigenetic markers or even diagnostic targets.
- Citation: Varkalaite G, Vaitkeviciute E, Inciuraite R, Salteniene V, Juzenas S, Petkevicius V, Gudaityte R, Mickevicius A, Link A, Kupcinskas L, Leja M, Kupcinskas J, Skieceviciene J. Atrophic gastritis and gastric cancer tissue miRNome analysis reveals hsa-miR-129-1 and hsa-miR-196a as potential early diagnostic biomarkers. World J Gastroenterol 2022; 28(6): 653-664
- URL: https://www.wjgnet.com/1007-9327/full/v28/i6/653.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i6.653
Gastric cancer (GC) is the one of the most common malignancy and the fourth leading cause of cancer-related death worldwide[1]. Studies show, that in most cases GC development is a stepwise process: Chronic gastric mucosa inflammation progresses to atrophic gastritis (AG) or intestinal metaplasia (IM), which eventually may become predisposition to GC. This complex cascade involves many factors: Helicobacter pylori
In this study, we aimed to investigate miRNome profile through GC tumorigenesis cascade including precancerous lesions, such as AG. Also, expression of two miRNAs (hsa-miR-129-1 and hsa-miR-196a) was analyzed in plasma samples of the independent cohort of AG and GC patients. Tissue miRNome analysis results revealed distinct miRNA profiles comparing controls (CON), AG, and GC groups. Also, our study findings show that two miRNAs: Hsa-miR-129-1 and hsa-miR-196a may be a relevant biomarker for GC diagnostics.
The study included a total of 125 CON and patients diagnosed with AG and GC, who were divided into the profiling cohort of 60 subjects and validation cohort of 65 subjects. Tissue samples of profiling cohort were collected during the years 2007-2015, while plasma of participants in validation cohort was collected from years 2011-2019 at the Departments of Surgery and Gastroenterology, Hospital of Lithuanian University of Health Sciences (Kaunas, Lithuania). Clinical and phenotypic characteristics of subjects investigated in profiling and validation cohorts are presented in Table 1. H. pylori status was assessed using indirect ELISA to detect serum-specific IgG antigen (Virion/Serion GmbH, Germany). Control group consisted of subjects, who had no signs of atrophy or IM according to Operative Link on Gastritis Assessment (OLGA) staging system (stage 0)[12]. AG group consisted of individuals that had stage I-IV atrophy score in gastric mucosa by OLGA classification. Gastric adenocarcinoma in GC patients was verified by histology and classified according to the American Joint Committee on Cancer TNM Staging Classification and Lauren Classification[13,14]. Adjacent GC (GCaj) samples were biopsy samples obtained from endoscopically healthy appearing gastric mucosa at least 2 cm away from the primary tumor.
| Profiling cohort (n = 60) | Validation cohort (n = 65) | ||||||
| CON (n = 21) | AG (n = 19) | GC (n = 20) | CON (n = 11) | AG (n = 30) | GC (n = 24) | ||
| Age | Mean ± SD | 58.29 ± 15.52 | 69.21 ± 8.78 | 64.95 ± 10.89 | 42.27 ± 12.89 | 68.01 ± 11.81 | 68.33 ± 11.27 |
| Gender (n) | Male | 5 | 3 | 15 | 5 | 9 | 18 |
| Female | 16 | 16 | 5 | 6 | 21 | 6 | |
| Helicobacter pylori infection (n) | Negative | 12 | 10 | 8 | - | 17 | 9 |
| Positive | 9 | 9 | 9 | - | 10 | 4 | |
| Unknown | - | - | 3 | 11 | 3 | 11 | |
| Differentiation grade (n) | G1 | - | - | 4 | - | - | - |
| G2 | - | - | 4 | - | - | 12 | |
| G3 | - | - | 12 | - | - | 12 | |
| Lauren classification (n) | Diffuse | - | - | 10 | - | - | 8 |
| Intestinal | - | - | 10 | - | - | 13 | |
| Mixed | - | - | - | - | - | 2 | |
| Unknown | - | - | - | - | - | 1 | |
| T (n) | T1 | - | - | 6 | - | - | 3 |
| T2 | - | - | 2 | - | - | 5 | |
| T3 | - | - | 8 | - | - | 9 | |
| T4 | - | - | 4 | - | - | 6 | |
| Unknown | - | - | - | - | - | 1 | |
| N (n) | N0 | - | - | 10 | - | - | 6 |
| N1 | - | - | 2 | - | - | 5 | |
| N2 | - | - | 3 | - | - | 4 | |
| N3 | - | - | 5 | - | - | 8 | |
| Unknown | - | - | - | - | - | 1 | |
| M (n) | M0 | - | - | 7 | - | - | 14 |
| M1 | - | - | 2 | - | - | 9 | |
| Unknown | - | - | 11 | - | - | 1 | |
The study was approved by the Kaunas Regional Biomedical Research Ethics Committee (approval No BE-2-10 and BE-2-31) and performed in accordance with the Declaration of Helsinki. All study participants provided written informed consent before enrollment.
Total RNA, including small RNA fraction, was isolated from CON, AG and GC tissues using miRNeasy Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions. Quantification of RNA was performed using Nanodrop2000 spectrophotometer (Thermo Fisher Scientific, United States) and quality of RNA samples was evaluated by Agilent 2100 Bioanalyzer (Agilent Technologies, United States). Circulating nucleic acids, including circulating miRNA fraction, was isolated using QIAamp Circulating Nucleic Acid Kit (Qiagen, Germany) according to manufacturer’s instructions. All isolated samples were stored at -80 °C prior to further analysis.
Small RNA libraries were prepared using Illumina TruSeq Small RNA Sample Preparation Kit (Illumina, United States) according to the manufacturer’s protocol with 1 μg RNA input per sample followed by RNA 3’ adapter ligation, RNA 5’ adapter ligation, cDNA synthesis, polymerase chain reaction (PCR) amplification using unique barcode sequences for each sample and gel size-selection of small RNA library. The yield and quality of sequencing libraries were assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, United States). The small RNA libraries were randomized, pooled 24 samples per lane and sequenced using Illumina HiSeq 2500 (1 × 50 bp single-end reads).
Analysis of raw small RNA-seq data was performed by nf-core/smrnaseq pipeline v.1.0.0 including Nextflow v.20.07.1[15], Java v.11.0.7, and Docker v.19.03.12. In brief, all steps consisted of read quality control using FastQC v.0.11.9, removing 3’ adapter sequences with TrimGalore! v.0.6.5, mapping to mature and hairpin miRNAs (miRBase v.22.1[16]), and GRCh37 human reference genome with Bowtie v.1.3.0[17]. After alignment and trimming sorted BAM files were used for further analysis with edgeR v.3.32.1[18] and mirtop v.0.4.23. MiRNA quality was assessed and summarized using MultiQC v.1.9[19]. Normalized counts were generated using isomiRs package and differential expression analysis was carried out using the DESeq2 Bioconductor package v.1.26.0[20]. The threshold for significant differential expression was Bonferroni[21] adjusted P-value < 0.05 and absolute value of log2 fold change (FC) |log2FC| > 1.
To validate differentially expressed miRNAs in plasma samples, isolated plasma circulating microRNA was reverse transcribed to cDNA using the TaqMan™ MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific, United States). The material was preamplified using the TaqMan PreAmp Master Mix (Applied Biosystems, United States) according to the manufacturer’s protocol. Quantitative real-time PCR (RT-PCR) was performed using the TaqMan MicroRNA Assays: Hsa-miR-129* (Assay ID: 002298), hsa-miR-196a (Assay ID: 241070_mat) on 7500 Fast Real-Time PCR System (Applied Biosystems, United States). All RT-qPCR reactions were run in duplicate in a 20 μL reaction and the relative fold change in miRNA expression was estimated using the 2-ΔΔCt method[22]. Ct values were normalized to the RNU6B (Assay ID: 001093, Thermo Fisher Scientific, United States) endogenous control.
Statistical analysis was performed using RStudio software (R v.3.6.3). Shapiro-Wilk normality test was used to test the normal distribution of data. For normally distributed data, statistical significance was assessed by Student's t-test. If the data did not pass normality tests was performed non-parametric Wilcoxon rank-sum test. A P < 0.05 was considered statistically significant. Area under the receiver operating characteristic curve (AUC-ROC) analysis was performed using pROC R package.
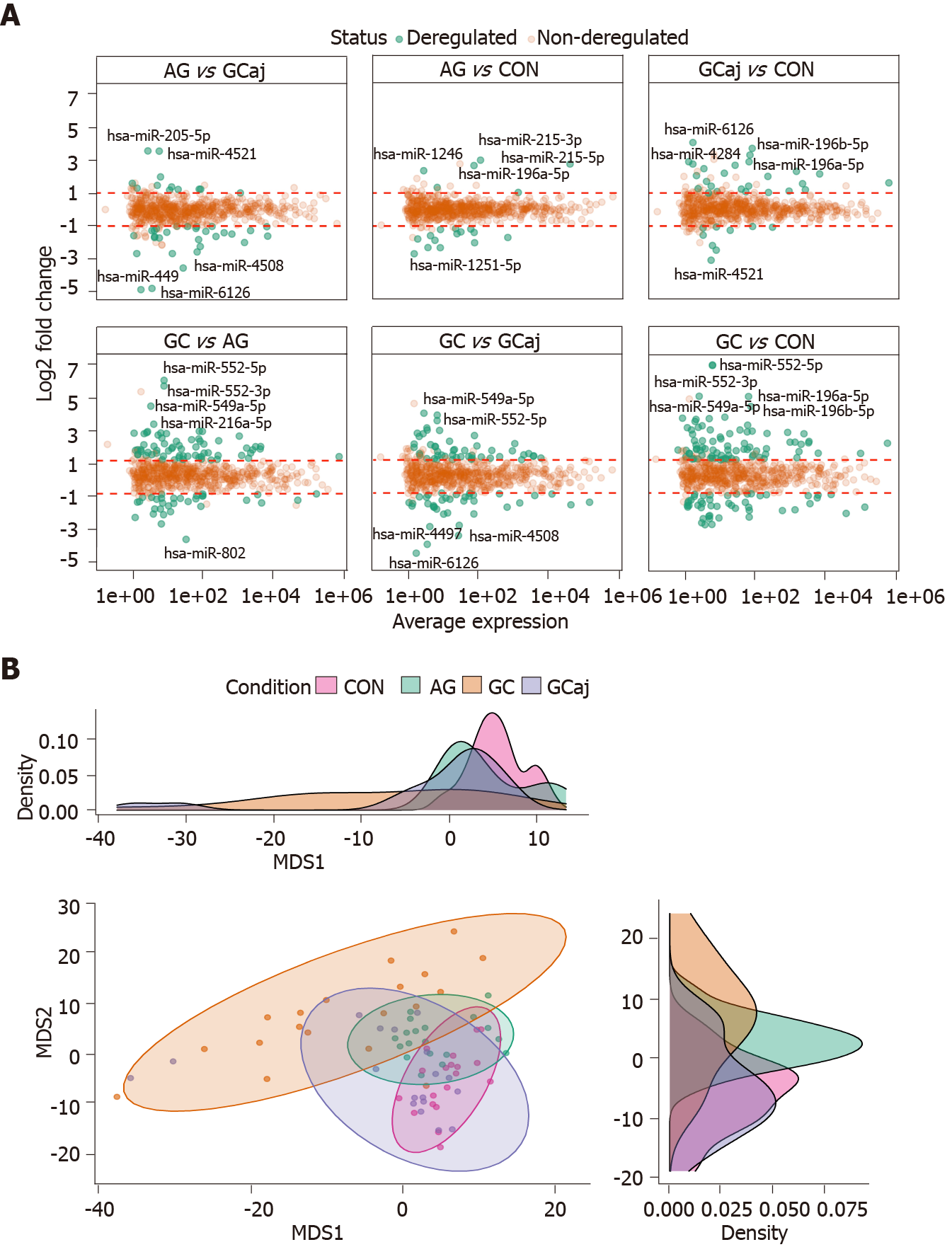
Small RNA sequencing of CON, AG, and paired GC (cancerous and adjacent) tissues in total identified 1037 miRNAs annotated in the miRBase v22.1. Sequencing yielded approx 250 M raw sequencing reads (from 359 K to 16 M reads per sample). After quality control steps 396 low-abundant and non-variable miRNAs and 5 outlying samples were removed resulting in 641 miRNAs and 75 samples which were used for further analysis (Supplementary Figures 1 and 2). The number of deregulated miRNAs corresponded to pathological cascade of GC development. The highest number of deregulated miRNAs were determined when comparing GC and CON groups (129 differentially expressed miRNAs, 82 up-regulated and 47 down-regulated; Supplementary Table 1). Next, 99 differentially expressed miRNAs were identified analyzing GC compared to AG (67 up-regulated and 32 down-regulated; Supplementary Table 2). The lowest number, 20 miRNAs, were found to be deregulated comparing AG and CON (6 up-regulated and 14 down-regulated; Supplementary Table 3). Differential expression results comparing GC vs GCaj, AG vs GCaj, and CON vs GCaj are presented in Supplementary Tables 4, 5 and 6 respectively.
Differential expression results and top five deregulated miRNAs in each case are represented in Figure 1A. Multidimensional scaling analysis of normalized expression values, assessing the similarity structure of miRNomes (Spearman’s correlation distance), revealed 4 clusters, corresponding to the CON, AG, GC cancerous and adjacent tissues (Figure 1B). The AG cluster was intermediate between GC and CON, whereas GCaj was overlapping with AG and CON groups.
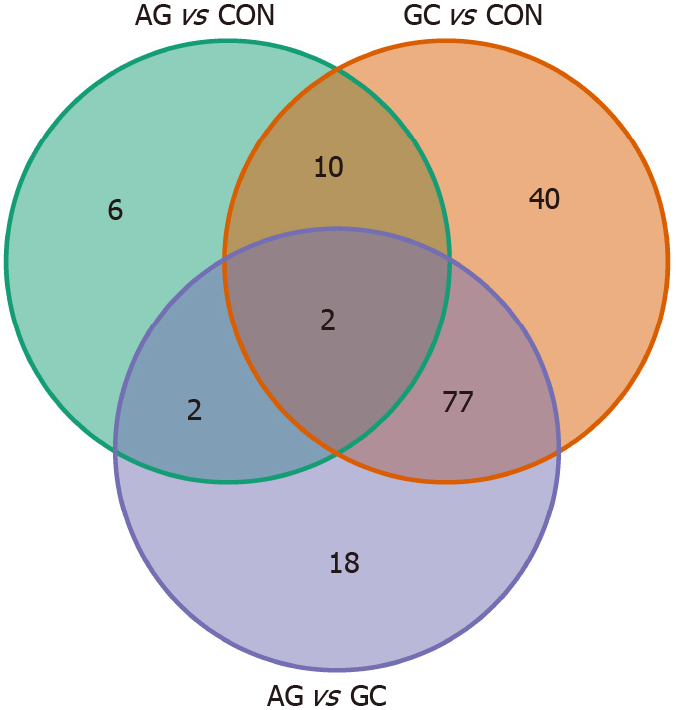
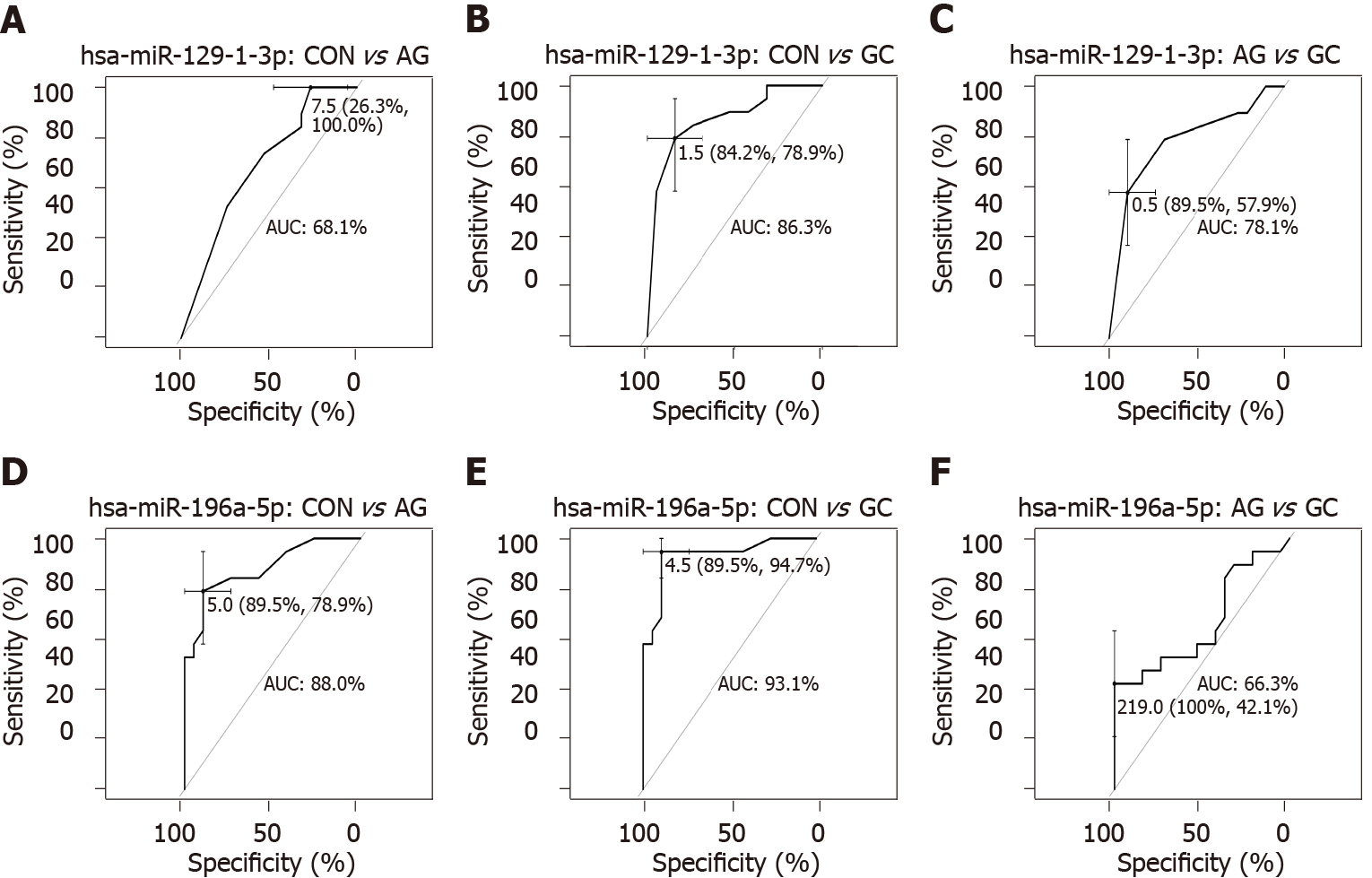
To further study miRNome profiles, altered expression of miRNAs was analyzed in three main comparison groups: AG vs CON, GC vs CON and AG vs GC according to clinical significance. Analyzing uniquely deregulated miRNAs, 40 differentially expressed miRNAs were found when compared GC to CON (25.8% of all deregulated miRNAs), 18 (11.6%) - AG compared to GC, and 6 (3.9%) - AG compared to CON (Figure 2). Most of the deregulated miRNAs (n = 79, 68.7%) were similar between GC vs CON and GC vs AG comparison groups. 12 miRNAs (7.7%) were deregulated in both AG and GC groups when compared to CON. Four miRNAs (2.6%) were similarly deregulated between AG vs CON and AG vs GC groups. Finally, only 2 miRNAs (hsa-miR-129-1-3p and hsa-miR-196a-5p) (1.29%) were identified as deregulated between all comparison groups. AUC-ROC analysis revealed that expression level of hsa-miR-129-1-3p in tissues resulted in AUCs: 68.1%; 86.3%, and 78.1%, CON vs AG, CON vs GC, and AG vs GC, respectively (Figures 3A, 3B and 3C). In addition to this, expression level of hsa-miR-196a-5p could be significant for discrimination of CON vs AG, CON vs GC and AG vs GC and resulted in AUCs: 88.0%, 93.1% and 66.3% (Figures 3D, 3E and 3F).
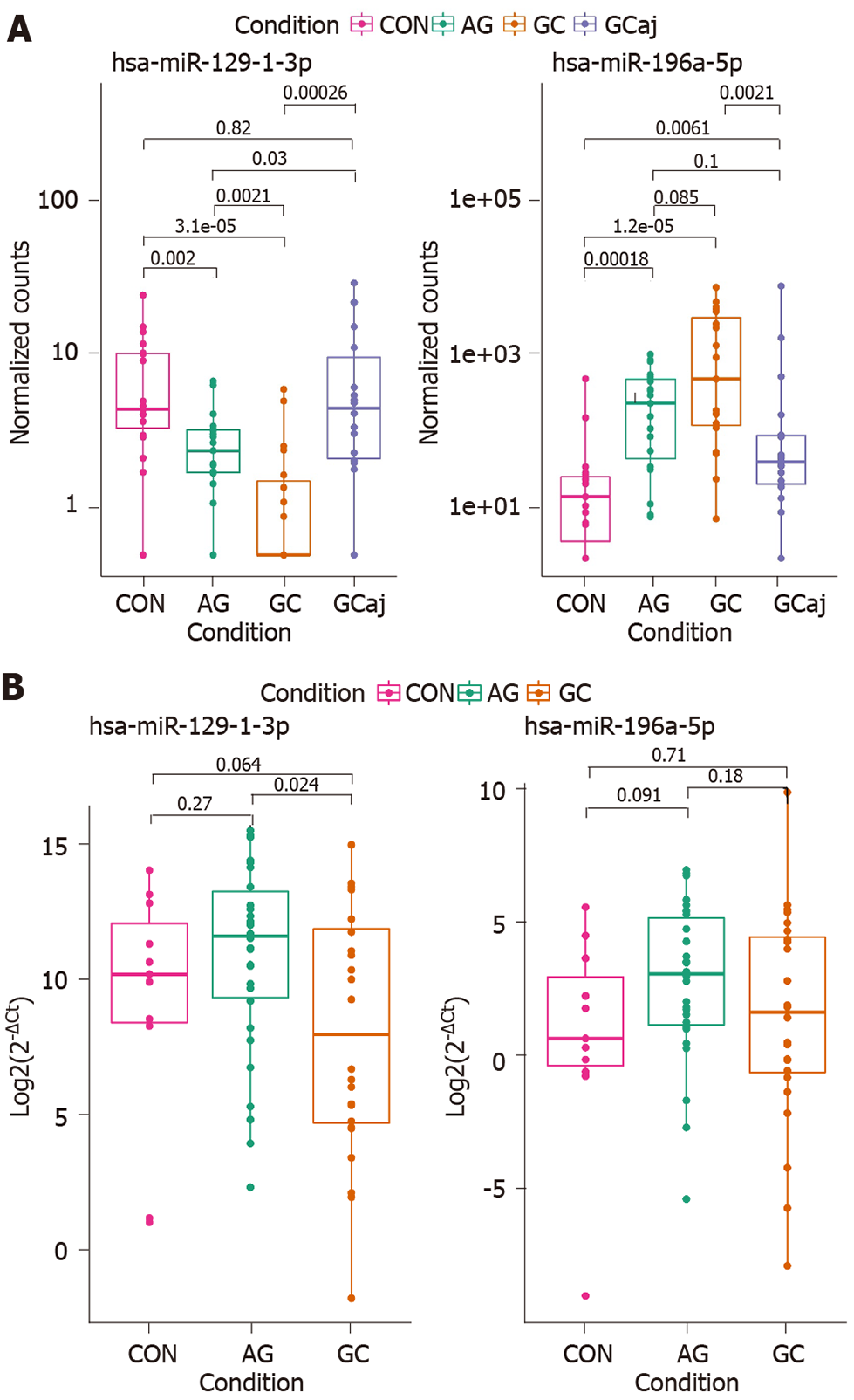
Differential expression analysis of NGS data in tissue samples revealed that hsa-miR-129-1-3p was significantly down-regulated and hsa-miR-196a-5p was up-regulated in AG and GC tissues compared to CON (P = 0.002 and P = 0.00018; P = 1.2 × 10-5 and P = 3.1 × 10-5, respectively). Moreover, hsa-miR-129-1-3p was significantly down-regulated in the case of AG compared to GC (P = 0.0021) and reflected a stepwise process of a pathology (Figure 4A). Therefore, to identify whether the expression changes of these two miRNAs can be detected noninvasively in the body fluids of the patients, hsa-miR-129-1-3p and hsa-miR-196a-5p were selected for RT-qPCR analysis in plasma samples of independent cohort. The analysis showed similar expression patterns in the case of hsa-miR-129-1-3p, which was significantly down-regulated when comparing AG and GC groups (P = 0.024). There were no other significant findings between the groups (Figure 4B).
To investigate role of miRNAs in AG atrophy progression (OLGA classification) and H. pylori-induced GC, differential miRNAs profile analysis in the subgroups of the study was performed. The analysis revealed a minor clustering in AG tissues corresponding to OLGA stages (Supplementary Figures 3A and 3H). H. pylori status in GC tissues (Supplementary Figure 3B). However, no significantly deregulated miRNAs were determined comparing I-II OLGA stages vs III-IV OLGA stages (AG tissue samples). On the other hand, analyzing GC group based on H. pylori infection status
This study represents comprehensive miRNome profiling of premalignant and malignant GC cases by implementing high throughput technologies such as NGS. Although there are several studies reporting profiles of GC tissue miRNAs[23,24], analysis of the association between miRNA expression and AG is very scarce reporting only individual miRNAs[25]. Moreover, based on small RNA-seq findings, two miRNAs were analyzed in subjects’ plasma samples to investigate potential non-invasive markers. To our best knowledge this is the first study analyzing AG and GC tissue miRNomes in the subjects of European origin.
First, our study showed different profiles of deregulated miRNAs between tissue samples of studied groups. In total, 20 differentially expressed miRNAs were identified in AG and 129 - in GC comparing to CON; also 99 deregulated miRNAs - comparing GC and AG groups. MiRNAs such as hsa-miR-3131, hsa-miR-483, hsa-miR-150, hsa-miR-200a-3p, hsa-miR-873-5p were previously reported by the GC profiling studies of Pereira et al[23] and Assumpção et al[24]. Yet, we were able to identify number of novel miRNAs (of which hsa-miR-548ba, hsa-miR-4521, hsa-miR-549a were the most deregulated). There are no data showing the role of these novel miRNAs in inflammatory or tumorous processes of gastric tissue. However, recent studies have shown that hsa-miR-548ba was associated with bladder cancer, hsa-miR-549a with the metastasis of renal cancer, and hsa-miR-4521 with H. pylori infection in esophageal epithelial cells[26-28]. Taking into consideration miRNome of AG, hsa-miR-3591-3p, hsa-miR-122-3p and hsa-miR-122-5p, hsa-miR-451a miRNAs were already reported by Liu et al[29], while the most deregulated miRNAs including hsa-miR-215, hsa-miR-4497, and hsa-miR-1251 were reported for the first time in our study. Previous research showed that hsa-miR-215-5p was deregulated in different lesions of the gastrointes-tinal tract (Barrett’s esophagus, intraepithelial neoplastic lesions, ulcerative colitis)[30-32]. However, hsa-miR-4497 and hsa-miR-452 were not previously associated with AG but were reported to play an important role in GC development[33,34].
Next, we identified hsa-miR-215-3p and hsa-miR-215-5p to be down-regulated while hsa-miR-934 - up-regulated in GC group comparing negative and positive H. pylori infection status. Studies revealed the altered expression of various miRNAs in H. pylori-induced GC tissue samples, including miR-934, miR-146a, miR-375, miR-204[35-37]. Although, hsa-miR-215 deregulation was previously associated with GC[38-40], there is no data showing its link with H. pylori infection.
In addition to this, we showed that two miRNAs (hsa-miR-129-1-3p and hsa-miR-196a-5p) were gradually deregulated comparing all three study groups (CON, AG, and GC) which also corresponds to pathological cascade of GC. In concordance to our results, it has already been shown that hsa-miR-129-1-3p was down-regulated in GC tissues, function as a tumor suppressor in GC and even corresponds to the same expression pattern in gastric juice[41,42]. There is no data regarding the hsa-miR-196a expression in AG tissue, however, investigators have revealed that hsa-miR-196a is overexpressed in GC tissue, plasma, commercial cell lines and promotes cell proliferation[43,44]. ROC-AUC analysis suggests great potential of hsa-miR-196a-5p expression in tissue for discrimination of AG and GC in contrast to CON (AUC = 89.5% and AUC = 89.5%, respectively). Therefore, further studies are needed to confirm this finding.
Finally, selected miRNAs were analyzed in independent cohort of CON, AG, and GC plasma samples by using RT-qPCR. Results showed similar deregulation direction in plasma samples as in the tissue samples. However, significant differences were only determined comparing the expression of hsa-miR-129-1-3p between AG and GC suggesting its potential role in non-invasive diagnostics of malignant cases. No significant expression changes were observed between study groups and hsa-miR-196a-5p. Other studies have shown controversial results: Tsai et al[45] reported that miR-196a/b was up-regulated in both the plasma and tissue of metastatic GC patients, while miRNome profiling study revealed that miR-196a-5p was found to be down-regulated in plasma of patients with precursor lesions of GC compared to non-active gastritis[46].
In our study, using NGS and RT-qPCR techniques we have shown the distinct miRNome profiles of CON, AG, GC, GCaj tissues, and potential of specific miRNAs as non-invasive biomarkers. In addition to this, novel miRNAs not previously reported as AG or GC associated epigenetic markers were identified. We have shown that hsa-miR-196a-5p expression in tissue could be significant for discrimination between CON and AG or GC, confirmed hsa-miR-129-1-3p as non-invasive biomarker in disease progression monitoring, and showed that miRNAs could be a great candidate for future research of new diagnostic approaches.
In conclusion, we showed gradual deregulation of hsa-miR-196a-5p and hsa-miR-129-1-3p in the gastric carcinogenesis pathway and confirmed hsa-miR-129-1-3p as a possible non-invasive biomarker. We also found hsa-miR-215-3p/5p and hsa-miR-934 to be significantly deregulated in GC based on H. pylori infection status. These data provide novel insights into complex GC pathogenesis cascade which could be highly significant for future studies of new diagnostic GC targets.
Gastric cancer (GC) is a complex disease arising from the interaction of environmental (e.g., diet, smoking, etc.) and host-associated factors [e.g., Helicobacter pylori (H. pylori) infection, genetics, etc.]. Due to its silent course, it is also one of the most lethal cancers worldwide as it is usually diagnosed at the advanced stages.
Novel biomarkers that would help to improve GC patients’ diagnosis and prognosis are highly needed. Studies show that microRNAs (miRNAs) play an important role in many cancers and could be a promising biomarker or even therapeutic target.
The objectives of the study were to analyze whole miRNome profiles of control, premalignant and malignant gastric tissues, and select the potential miRNA markers that could have a potential for minimally invasive GC diagnostics.
Total RNA from gastric tissue samples was subjected for small RNA sequencing (smRNA-seq). Plasma total circulating nucleic acids were used for the expression analysis of the most tissue deregulated miRNAs by real-time quantitative polymerase chain reaction. Statistical analysis involved the differential expression and discrimination analyses.
The abundance of altered expression miRNAs corresponded to a pathological cascade of GC development. Hsa-miR-129-1-3p and has-miR-196a-5p were shown to be deregulated in healthy-premalignant-malignant sequence. In addition to this, we showed that down-regulation of hsa-miR-129-1-3p could also be detected non-invasively in GC patients’ plasma samples. Finally, results indicated that hsa-miR-215-3p/5p and hsa-miR-934 were significantly deregulated based on H. pylori infection status for GC patients.
Gastric tissue miRNome study provides extensive profiling of control, premalignant and malignant cases. Based on smRNA-seq results several miRNAs were shown as potential gastric carcinogenesis (hsa-miR-196a-5p and hsa-miR-129-1-3p); and H. Pylori-related (hsa-miR-215-3p/5p and hsa-miR-934) biomarkers.
This study provides novel insights into complex GC pathogenesis cascade and could serve as a reference for future research to support our findings.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Lithuania
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Gao W, Kotelevets SM S-Editor: Wang JJ L-Editor: A P-Editor: Wang JJ
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50630] [Cited by in F6Publishing: 43146] [Article Influence: 14382.0] [Reference Citation Analysis (47)] |
| 2. | McLean MH, El-Omar EM. Genetics of gastric cancer. Nat Rev Gastroenterol Hepatol. 2014;11:664-674. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 250] [Cited by in F6Publishing: 280] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 3. | Tan P, Yeoh KG. Genetics and Molecular Pathogenesis of Gastric Adenocarcinoma. Gastroenterology. 2015;149:1153-1162.e3. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 269] [Cited by in F6Publishing: 311] [Article Influence: 34.6] [Reference Citation Analysis (0)] |
| 4. | Bartel DP. Metazoan MicroRNAs. Cell. 2018;173:20-51. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2329] [Cited by in F6Publishing: 2229] [Article Influence: 371.5] [Reference Citation Analysis (0)] |
| 5. | Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-233. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14460] [Cited by in F6Publishing: 15337] [Article Influence: 1022.5] [Reference Citation Analysis (1)] |
| 6. | Bracken CP, Scott HS, Goodall GJ. A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet. 2016;17:719-732. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 437] [Cited by in F6Publishing: 468] [Article Influence: 58.5] [Reference Citation Analysis (0)] |
| 7. | Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNAs in human cancer. Adv Exp Med Biol. 2013;774:1-20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 463] [Cited by in F6Publishing: 550] [Article Influence: 50.0] [Reference Citation Analysis (0)] |
| 8. | Gyvyte U, Juzenas S, Salteniene V, Kupcinskas J, Poskiene L, Kucinskas L, Jarmalaite S, Stuopelyte K, Steponaitiene R, Hemmrich-Stanisak G, Hübenthal M, Link A, Franke S, Franke A, Pangonyte D, Lesauskaite V, Kupcinskas L, Skieceviciene J. MiRNA profiling of gastrointestinal stromal tumors by next-generation sequencing. Oncotarget. 2017;8:37225-37238. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 9. | Streleckiene G, Inciuraite R, Juzenas S, Salteniene V, Steponaitiene R, Gyvyte U, Kiudelis G, Leja M, Ruzgys P, Satkauskas S, Kupcinskiene E, Franke S, Thon C, Link A, Kupcinskas J, Skieceviciene J. miR-20b and miR-451a Are Involved in Gastric Carcinogenesis through the PI3K/AKT/mTOR Signaling Pathway: Data from Gastric Cancer Patients, Cell Lines and Ins-Gas Mouse Model. Int J Mol Sci. 2020;21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 10. | Link A, Kupcinskas J. MicroRNAs as non-invasive diagnostic biomarkers for gastric cancer: Current insights and future perspectives. World J Gastroenterol. 2018;24:3313-3329. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 73] [Cited by in F6Publishing: 80] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 11. | Juzėnas S, Saltenienė V, Kupcinskas J, Link A, Kiudelis G, Jonaitis L, Jarmalaite S, Kupcinskas L, Malfertheiner P, Skieceviciene J. Analysis of Deregulated microRNAs and Their Target Genes in Gastric Cancer. PLoS One. 2015;10:e0132327. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 12. | Rugge M, Correa P, Di Mario F, El-Omar E, Fiocca R, Geboes K, Genta RM, Graham DY, Hattori T, Malfertheiner P, Nakajima S, Sipponen P, Sung J, Weinstein W, Vieth M. OLGA staging for gastritis: a tutorial. Dig Liver Dis. 2008;40:650-658. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 210] [Cited by in F6Publishing: 197] [Article Influence: 12.3] [Reference Citation Analysis (1)] |
| 13. | Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR, Winchester DP. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more "personalized" approach to cancer staging. CA Cancer J Clin. 2017;67:93-99. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2341] [Cited by in F6Publishing: 3185] [Article Influence: 455.0] [Reference Citation Analysis (2)] |
| 14. | Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31-49. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4011] [Cited by in F6Publishing: 4108] [Article Influence: 146.7] [Reference Citation Analysis (0)] |
| 15. | Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155-D162. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1834] [Cited by in F6Publishing: 2309] [Article Influence: 577.3] [Reference Citation Analysis (0)] |
| 16. | Di Tommaso P, Chatzou M, Floden EW, Barja PP, Palumbo E, Notredame C. Nextflow enables reproducible computational workflows. Nat Biotechnol. 2017;35:316-319. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 859] [Cited by in F6Publishing: 943] [Article Influence: 157.2] [Reference Citation Analysis (0)] |
| 17. | Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15569] [Cited by in F6Publishing: 15270] [Article Influence: 1018.0] [Reference Citation Analysis (0)] |
| 18. | Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047-3048. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2496] [Cited by in F6Publishing: 3205] [Article Influence: 400.6] [Reference Citation Analysis (0)] |
| 19. | Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139-140. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22632] [Cited by in F6Publishing: 24654] [Article Influence: 1643.6] [Reference Citation Analysis (0)] |
| 20. | Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34752] [Cited by in F6Publishing: 43328] [Article Influence: 4814.2] [Reference Citation Analysis (0)] |
| 21. | Ranstam J. Multiple P-values and Bonferroni correction. Osteoarthritis Cartilage. 2016;24:763-764. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 98] [Cited by in F6Publishing: 101] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 22. | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117419] [Cited by in F6Publishing: 121468] [Article Influence: 5281.2] [Reference Citation Analysis (0)] |
| 23. | Pereira A, Moreira F, Vinasco-Sandoval T, Cunha A, Vidal A, Ribeiro-Dos-Santos AM, Pinto P, Magalhães L, Assumpção M, Demachki S, Santos S, Assumpção P, Ribeiro-Dos-Santos Â. miRNome Reveals New Insights Into the Molecular Biology of Field Cancerization in Gastric Cancer. Front Genet. 2019;10:592. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 24. | Assumpção MB, Moreira FC, Hamoy IG, Magalhães L, Vidal A, Pereira A, Burbano R, Khayat A, Silva A, Santos S, Demachki S, Ribeiro-Dos-Santos Â, Assumpção P. High-Throughput miRNA Sequencing Reveals a Field Effect in Gastric Cancer and Suggests an Epigenetic Network Mechanism. Bioinform Biol Insights. 2015;9:111-117. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 25. | Link A, Schirrmeister W, Langner C, Varbanova M, Bornschein J, Wex T, Malfertheiner P. Differential expression of microRNAs in preneoplastic gastric mucosa. Sci Rep. 2015;5:8270. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 40] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 26. | Zhao F, Ge YZ, Zhou LH, Xu LW, Xu Z, Ping WW, Wang M, Zhou CC, Wu R, Jia RP. Identification of hub miRNA biomarkers for bladder cancer by weighted gene coexpression network analysis. Onco Targets Ther. 2017;10:5551-5559. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 27. | Xuan Z, Chen C, Tang W, Ye S, Zheng J, Zhao Y, Shi Z, Zhang L, Sun H, Shao C. TKI-Resistant Renal Cancer Secretes Low-Level Exosomal miR-549a to Induce Vascular Permeability and Angiogenesis to Promote Tumor Metastasis. Front Cell Dev Biol. 2021;9:689947. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 28. | Teng G, Dai Y, Chu Y, Li J, Zhang H, Wu T, Shuai X, Wang W. Helicobacter pylori induces caudal-type homeobox protein 2 and cyclooxygenase 2 expression by modulating microRNAs in esophageal epithelial cells. Cancer Sci. 2018;109:297-307. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 29. | Liu H, Li PW, Yang WQ, Mi H, Pan JL, Huang YC, Hou ZK, Hou QK, Luo Q, Liu FB. Identification of non-invasive biomarkers for chronic atrophic gastritis from serum exosomal microRNAs. BMC Cancer. 2019;19:129. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 30. | Fassan M, Croce CM, Rugge M. miRNAs in precancerous lesions of the gastrointestinal tract. World J Gastroenterol. 2011;17:5231-5239. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 22] [Cited by in F6Publishing: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 31. | Fassan M, Volinia S, Palatini J, Pizzi M, Baffa R, De Bernard M, Battaglia G, Parente P, Croce CM, Zaninotto G, Ancona E, Rugge M. MicroRNA expression profiling in human Barrett's carcinogenesis. Int J Cancer. 2011;129:1661-1670. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 88] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 32. | Wijnhoven BP, Hussey DJ, Watson DI, Tsykin A, Smith CM, Michael MZ; South Australian Oesophageal Research Group. MicroRNA profiling of Barrett's oesophagus and oesophageal adenocarcinoma. Br J Surg. 2010;97:853-861. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 98] [Cited by in F6Publishing: 109] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 33. | Bibi F, Naseer MI, Alvi SA, Yasir M, Jiman-Fatani AA, Sawan A, Abuzenadah AM, Al-Qahtani MH, Azhar EI. microRNA analysis of gastric cancer patients from Saudi Arabian population. BMC Genomics. 2016;17:751. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 34. | Yin C, Zheng X, Xiang H, Li H, Gao M, Meng X, Yang K. Differential expression profile analysis of cisplatinregulated miRNAs in a human gastric cancer cell line. Mol Med Rep. 2019;20:1966-1976. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 35. | Chang H, Kim N, Park JH, Nam RH, Choi YJ, Lee HS, Yoon H, Shin CM, Park YS, Kim JM, Lee DH. Different microRNA expression levels in gastric cancer depending on Helicobacter pylori infection. Gut Liver. 2015;9:188-196. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 54] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 36. | Prinz C, Mese K, Weber D. MicroRNA Changes in Gastric Carcinogenesis: Differential Dysregulation during Helicobacter pylori and EBV Infection. Genes (Basel). 2021;12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 37. | Wang F, Sun G, Zou Y, Zhong F, Ma T, Li X. Protective role of Helicobacter pylori infection in prognosis of gastric cancer: evidence from 2,454 patients with gastric cancer. PLoS One. 2013;8:e62440. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 23] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 38. | Jin Z, Selaru FM, Cheng Y, Kan T, Agarwal R, Mori Y, Olaru AV, Yang J, David S, Hamilton JP, Abraham JM, Harmon J, Duncan M, Montgomery EA, Meltzer SJ. MicroRNA-192 and -215 are upregulated in human gastric cancer in vivo and suppress ALCAM expression in vitro. Oncogene. 2011;30:1577-1585. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 95] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 39. | Deng Y, Huang Z, Xu Y, Jin J, Zhuo W, Zhang C, Zhang X, Shen M, Yan X, Wang L, Wang X, Kang Y, Si J, Zhou T. MiR-215 modulates gastric cancer cell proliferation by targeting RB1. Cancer Lett. 2014;342:27-35. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 74] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 40. | Li N, Zhang QY, Zou JL, Li ZW, Tian TT, Dong B, Liu XJ, Ge S, Zhu Y, Gao J, Shen L. miR-215 promotes malignant progression of gastric cancer by targeting RUNX1. Oncotarget. 2016;7:4817-4828. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 41. | Wang D, Luo L, Guo J. miR-129-1-3p inhibits cell migration by targeting BDKRB2 in gastric cancer. Med Oncol. 2014;31:98. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 42. | Yu X, Luo L, Wu Y, Yu X, Liu Y, Zhao X, Zhang X, Cui L, Ye G, Le Y, Guo J. Gastric juice miR-129 as a potential biomarker for screening gastric cancer. Med Oncol. 2013;30:365. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 43. | Treece AL, Duncan DL, Tang W, Elmore S, Morgan DR, Dominguez RL, Speck O, Meyers MO, Gulley ML. Gastric adenocarcinoma microRNA profiles in fixed tissue and in plasma reveal cancer-associated and Epstein-Barr virus-related expression patterns. Lab Invest. 2016;96:661-671. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 44. | Sun M, Liu XH, Li JH, Yang JS, Zhang EB, Yin DD, Liu ZL, Zhou J, Ding Y, Li SQ, Wang ZX, Cao XF, De W. MiR-196a is upregulated in gastric cancer and promotes cell proliferation by downregulating p27(kip1). Mol Cancer Ther. 2012;11:842-852. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 104] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 45. | Tsai MM, Wang CS, Tsai CY, Huang CG, Lee KF, Huang HW, Lin YH, Chi HC, Kuo LM, Lu PH, Lin KH. Circulating microRNA-196a/b are novel biomarkers associated with metastatic gastric cancer. Eur J Cancer. 2016;64:137-148. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 44] [Article Influence: 5.5] [Reference Citation Analysis (1)] |
| 46. | Lario S, Brunet-Vega A, Quílez ME, Ramírez-Lázaro MJ, Lozano JJ, García-Martínez L, Pericay C, Miquel M, Junquera F, Campo R, Calvet X. Expression profile of circulating microRNAs in the Correa pathway of progression to gastric cancer. United European Gastroenterol J. 2018;6:691-701. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |