Published online Sep 28, 2022. doi: 10.3748/wjg.v28.i36.5338
Peer-review started: July 20, 2022
First decision: August 6, 2022
Revised: August 14, 2022
Accepted: September 6, 2022
Article in press: September 6, 2022
Published online: September 28, 2022
The most important consideration in determining treatment strategies for undifferentiated early gastric cancer (UEGC) is the risk of lymph node metastasis (LNM). Therefore, identifying a potential biomarker that predicts LNM is quite useful in determining treatment.
To develop a machine learning (ML)-based integral procedure to construct the LNM gray-level co-occurrence matrix (GLCM) prediction model.
We retrospectively selected 526 cases of UEGC confirmed through pathological examination after radical gastrectomy without endoscopic treatment in four tertiary hospitals between January 2015 to December 2021. We extracted GLCM-based features from grayscale images and applied ML to the classification of candidate predictive variables. The robustness and clinical utility of each model were evaluated based on the following factors: Receiver operating characteristic curve (ROC), decision curve analysis, and clinical impact curve.
GLCM-based feature extraction significantly correlated with LNM. The top 7 GLCM-based factors included inertia value 0° (IV_0), inertia value 45° (IV_45), inverse gap 0° (IG_0), inverse gap 45° (IG_45), inverse gap full angle (IG_all), Haralick 30° (Haralick_30), Haralick full angle (Haralick_all), and Entropy. The areas under the ROC curve (AUCs) of the random forest classifier (RFC) model, support vector machine, eXtreme gradient boosting, artificial neural network, and decision tree ranged from 0.805 [95% confidence interval (CI): 0.258-1.352] to 0.925 (95%CI: 0.378-1.472) in the training set and from 0.794 (95%CI: 0.237-1.351) to 0.912 (95%CI: 0.355-1.469) in the testing set, respectively. The RFC (training set: AUC: 0.925, 95%CI: 0.378-1.472; testing set: AUC: 0.912, 95%CI: 0.355-1.469) model that incorporates Entropy, Haralick_all, Haralick_30, IG_all, IG_45, IG_0, and IV_45 had the highest predictive accuracy.
The evaluation results indicate that the method of selecting radiological and textural features becomes more effective in the LNM discrimination against UEGC patients. Additionally, the ML-based prediction model developed using the RFC can be used to derive treatment options and identify LNM, which can hence improve clinical outcomes.
Core Tip: Gray-level co-occurrence matrix-based feature extraction can be a robust and promising tool to improve the efficiency in predicting lymph node metastasis of individual undifferentiated early gastric cancer patients. Additionally, machine learning adopts more optimized algorithms and more clear feature extraction. Models developed using random forest classifier have the highest predictive accuracy in terms of Entropy, Haralick full angle, Haralick 30°, inverse gap full angle, inverse gap 45°, inverse gap 0°, and inertia value 45°. Further research is required to develop these models for clinical practice.
- Citation: Wei X, Yan XJ, Guo YY, Zhang J, Wang GR, Fayyaz A, Yu J. Machine learning-based gray-level co-occurrence matrix signature for predicting lymph node metastasis in undifferentiated-type early gastric cancer. World J Gastroenterol 2022; 28(36): 5338-5350
- URL: https://www.wjgnet.com/1007-9327/full/v28/i36/5338.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i36.5338
Gastric cancer (GC) is one of the most common and fatal malignancies worldwide and is an important part of the global cancer burden[1,2]. In GC, undifferentiated early GC (UEGC) differs from differentiated-type GC in terms of clinical features and disease state, and their treatment and prognosis vary[3]. Therefore, UEGC should be identified and diagnosed early.
The incidence of lymphatic vessel invasion and risk of lymph node metastasis (LNM) in UEGC are high in surgical specimens of GC[4,5]. Endoscopic resection (ER), including endoscopic mucosal resection (EMR) and endoscopic submucosal dissection (ESD), has been considered a minimally invasive treatment option for early GC with negligible risk of LNM[6,7]. Nevertheless, indication or curability evaluation has not been conducted for ESD of undifferentiated GC (e.g., poorly differentiated adenocarcinoma, signet ring cell carcinoma, or mucinous adenocarcinoma) due to the potential risk of LNM. Although ER can be used as painless treatment, the LNM incidence after non-curative ER can be as low as 5.1% and as high as 12.2%[8-10]. Additionally, ESD is only applicable to intramucosal cancer with a tumor diameter of ≤ 20 mm and without ulcer lesions; thus, treating lesions that meet the ESD indications through surgery is unnecessary[11,12]. That is, when resection beyond the expanded standard is considered ineffective, the potential risk of LNM cannot be ignored. Hence, additional surgical resection and lymph node dissection should be performed. Unlike differentiated early GC, ER indications of UEGC are limited. Therefore, to address this challenging problem, a precise tool that can predict LNM must be explored.
Previous studies have mainly focused on risk factors for LNM or distant metastasis of differentiated-type early GC[13-15]. However, for UEGC, LNM has different risk factors. Thus, objective and universal evaluation indicators for evaluating its risk are lacking. In this study, we clarified the LNM risk factors of patients with UEGC who underwent surgical resection. Subsequently, we analyzed clinical-pathological factors by introducing gray-level co-occurrence matrix (GLCM) image feature extraction mining to classify LNM risk groups according to the combination of risk factors. This study aims to provide a reference for clinical diagnosis and treatment.
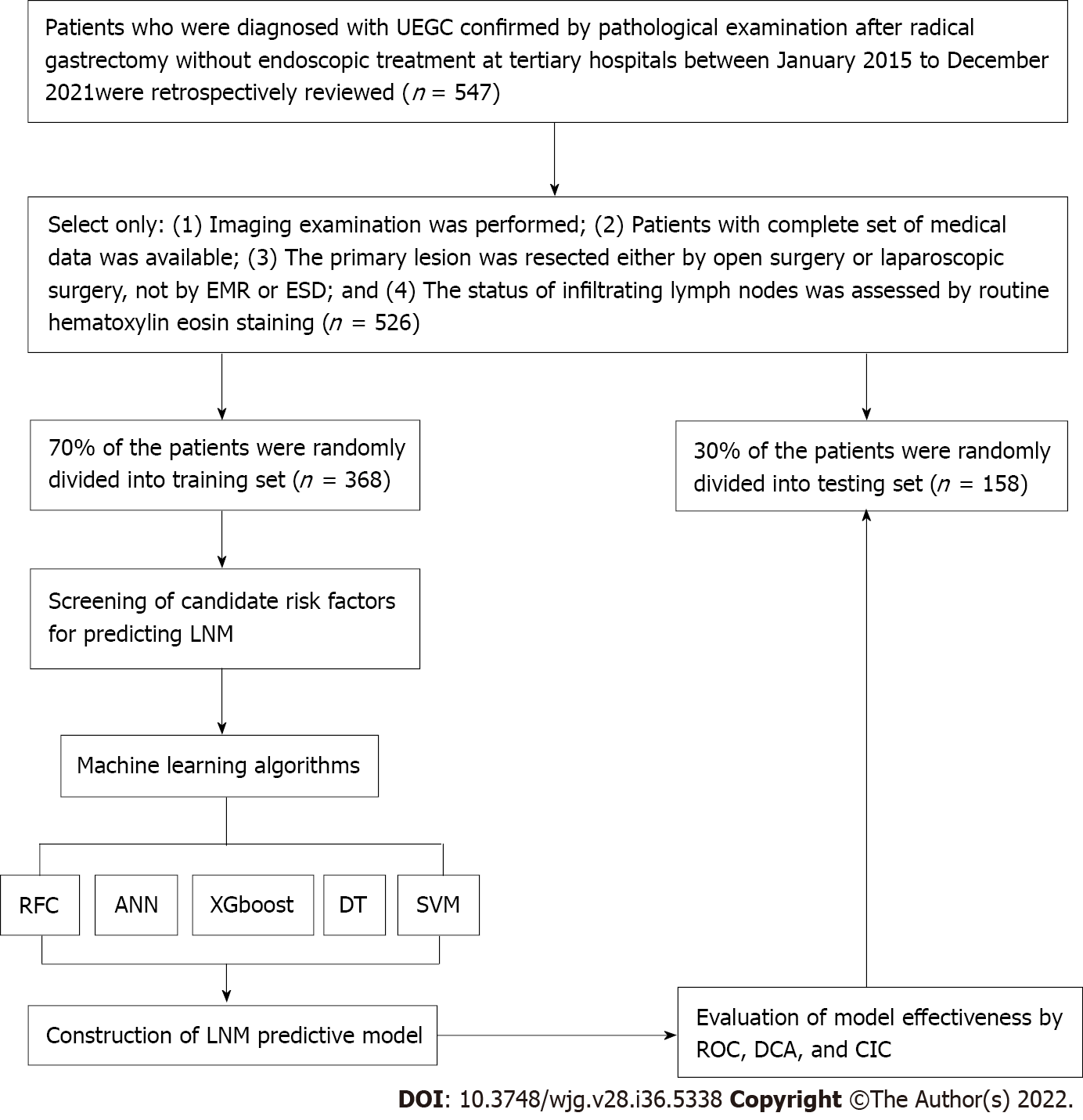
The clinical records of 526 patients who were diagnosed with UEGC were confirmed through pathological examination after radical gastrectomy without endoscopic treatment at four tertiary hospitals. These hospitals are Shaanxi Provincial People’s Hospital, Shaanxi Provincial Tumour Hospital, the First Affiliated Hospital of Xi’an Jiaotong University, and the Second Affiliated Hospital of Xi’an Jiaotong University. The clinical records were between January 2015 to December 2021 and were retrospectively reviewed. The following were the inclusion criteria: (1) Imaging examination was performed; (2) Patients have a complete set of medical data; (3) Primary lesion was resected either via open surgery or laparoscopic surgery and not via EMR or ESD; and (4) The status of infiltrating lymph nodes was assessed through routine hematoxylin-eosin staining. To minimize the confounding effect of unnecessary variables, the following were the exclusion criteria: (1) Sufficient information cannot be extracted or mismatched clinical data of patients; and (2) Patients without complete magnetic resonance imaging (MRI) plain scan or the MRI image quality being unacceptable. This study complies with the provisions of the Helsinki Declaration (revised in 2013) and was approved by the Institutional Review Committee of Shaanxi Provincial People’s Hospital (2021-Y024). Figure 1 presents in detail the patient screening steps and modeling process.
All texture parameter post-processing was conducted on Omni dynamics software (GE pharmaceuticals, Shanghai). Two radiologists who have vast experience in gastrointestinal diagnosis referred to the MRI images to sketch the lesions on the ADC map. First, they manually sketch the entire area with cancer on each layer of the map, avoiding the gas in the intestine, until the whole tumor volume was cut out. Second, the software automatically generates the texture features. In this study, the following are the selected texture parameters of the GLCM: Total frequency, energy value, entropy, inertia value, correlation coefficient, inverse moment, cluster shadow, and cluster prominence.
For variables with missing values (often this missing value is less than 10%), the variable’s mean value should be filled. If ≥ 10% of the given variables are missing, this value is excluded from the variable screening of the final model. Similarly, this study adopted unit feature interpolation for the missing values that meet the interpolation requirements. That is, the missing values can be interpolated using the constant values provided or using the statistical data of each column where these missing values are located (e.g., average value, median value, or the most frequently occurring value)[16,17].
Based on the machine learning (ML) algorithm, the commonly used iterative algorithm models are included: Random forest classifier (RFC), decision tree (DT), support vector machine (SVM), eXtreme gradient boosting (XGBoost), and artificial neural network (ANN). The RFC is an integrated method that forms a cumulative effect by integrating multiple relatively simple evaluators. Random forest is an integrated learning tool based on DT. The SVM is a type of a generalized linear classifier that categorizes data binary through supervised learning. The ANN is a nonlinear equation transformation output algorithm comprising input, hidden, and output layers. Finally, XGBoost is an additive model. In each iteration, only the sub-models in the current step are optimized. In this study, we refer to the guide proposed by Luo et al[18] for the best use of prediction models in biomedical research, that is, the Delphi method, which is used to generate the list of reported items.
For the screening of candidate variables, we mainly rely on the principle of “bag repeatedly put back and extract”, sort according to variables’ weight, and finally obtain the final predictor of the prediction model from the top 10 variables[19]. For the effectiveness evaluation of the prediction model, the receiver operating characteristic (ROC) curve is used to evaluate the accuracy of the model. Meanwhile, the decision curve analysis and clinical impact curve (CIC) were used to evaluate the model’s robustness and differentiation, respectively.
The measurement and counting data in this study are expressed by interquartile spacing (25%, 75%) and percentage (%), respectively. For the comparison between groups, the continuous variables adopt the t-test or Mann-Whitney U test of independent samples (provided that it does not conform to the normal distribution). The counting data adopt the chi-square goodness-of-fit test. Values of Bonferroni corrected probability are used to compare the qualitative data[20]. The prediction model visualization and other data analysis are performed using R software (version 4.0.4, http://www.r-project.org/). For the comparison between groups, P value < 0.05 is considered statistically significant and vice versa.
Table 1 summarizes the baseline characteristics of 526 hospitalized patients with UEGC. For internal validation, the patients were randomly divided into two sets using the caret package: Training set (n = 368, 70%) and validation set (n = 158, 30%). Regarding the LNM rate, the training and validation cohorts were 62 (16.85%) and 29 (18.35%), respectively. In addition to the previously reported clinical-related indicators (e.g., tumor size, infiltration depth, vascular_invasion, and vascular tumor thrombus), significant differences exist between the LNM and non-LNM groups. We found that GLCM-based texture acquisition features also have significant statistical differences between the two groups.
| Variables | Training set | Testing set | ||||||
| Overall (n = 368) | Yes (n = 62) | No (n = 306) | P value | Overall (n = 158) | Yes (n = 29) | No (n = 129) | P value | |
| Age (median, IQR), yr | 51.00 (40.75, 61.00) | 52.50 (40.25, 64.50) | 51.00 (41.00, 60.00) | 0.185 | 47.00 (37.25, 58.75) | 53.00 (39.00, 63.00) | 46.00 (37.00, 57.00) | 0.215 |
| Sex (%) | ||||||||
| Male | 266 (72.3) | 48 (77.4) | 218 (71.2) | 0.404 | 123 (77.8) | 21 (72.4) | 102 (79.1) | 0.594 |
| Female | 102 (27.7) | 14 (22.6) | 88 (28.8) | 35 (22.2) | 8 (27.6) | 27 (20.9) | ||
| Site (%) | ||||||||
| Nearly 1/3 | 81 (22.0) | 10 (16.1) | 71 (23.2) | 0.384 | 37 (23.4) | 9 (31.0) | 28 (21.7) | 0.138 |
| Medium 1/3 | 73 (19.8) | 15 (24.2) | 58 (19.0) | 23 (14.6) | 1 (3.4) | 22 (17.1) | ||
| Far 1/3 | 214 (58.2) | 37 (59.7) | 177 (57.8) | 98 (62.0) | 19 (65.5) | 79 (61.2) | ||
| Ulcer (%) | ||||||||
| Yes | 103 (28.0) | 21 (33.9) | 82 (26.8) | 0.329 | 45 (28.5) | 11 (37.9) | 34 (26.4) | 0.308 |
| No | 265 (72.0) | 41 (66.1) | 224 (73.2) | 113 (71.5) | 18 (62.1) | 95 (73.6) | ||
| Gross_type (%) | ||||||||
| Uplift | 98 (26.6) | 14 (22.6) | 84 (27.5) | 0.587 | 56 (35.4) | 8 (27.6) | 48 (37.2) | 0.227 |
| Flat | 72 (19.6) | 11 (17.7) | 61 (19.9) | 27 (17.1) | 8 (27.6) | 19 (14.7) | ||
| Sunken | 198 (53.8) | 37 (59.7) | 161 (52.6) | 75 (47.5) | 13 (44.8) | 62 (48.1) | ||
| Tumor_size (%) | ||||||||
| ≤ 2 cm | 296 (80.4) | 18 (29.0) | 278 (90.8) | < 0.001 | 120 (75.9) | 6 (20.7) | 114 (88.4) | < 0.001 |
| > 2 cm | 72 (19.6) | 44 (71.0) | 28 (9.2) | 38 (24.1) | 23 (79.3) | 15 (11.6) | ||
| Infiltration_depth (%) | ||||||||
| Mucosal layer | 267 (72.6) | 12 (19.4) | 255 (83.3) | < 0.001 | 113 (71.5) | 9 (31.0) | 104 (80.6) | < 0.001 |
| Submucosa | 101 (27.4) | 50 (80.6) | 51 (16.7) | 45 (28.5) | 20 (69.0) | 25 (19.4) | ||
| Vascular_invasion (%) | ||||||||
| Yes | 124 (33.7) | 42 (67.7) | 82 (26.8) | < 0.001 | 56 (35.4) | 21 (72.4) | 35 (27.1) | < 0.001 |
| No | 244 (66.3) | 20 (32.3) | 224 (73.2) | 102 (64.6) | 8 (27.6) | 94 (72.9) | ||
| VTT (%) | ||||||||
| Yes | 124 (33.7) | 45 (72.6) | 79 (25.8) | < 0.001 | 44 (27.8) | 23 (79.3) | 21 (16.3) | < 0.001 |
| No | 244 (66.3) | 17 (27.4) | 227 (74.2) | 114 (72.2) | 6 (20.7) | 108 (83.7) | ||
| TF (median, IQR) | 3.78 (3.56, 3.99) | 4.13 (3.97, 4.27) | 3.70 (3.51, 3.90) | < 0.001 | 3.79 (3.52, 4.01) | 4.16 (4.00, 4.31) | 3.70 (3.49, 3.93) | < 0.001 |
| EV (median, IQR) | 0.88 (0.64, 1.12) | 0.60 (0.49, 0.68) | 0.98 (0.72, 1.20) | < 0.001 | 0.85 (0.65, 1.09) | 0.64 (0.54, 0.70) | 0.92 (0.72, 1.16) | < 0.001 |
| Entropy (median, IQR) | 8.68 (8.37, 8.98) | 10.51 (10.07, 10.88) | 8.57 (8.33, 8.83) | < 0.001 | 8.79 (8.43, 9.02) | 10.44 (10.16, 10.96) | 8.65 (8.38, 8.89) | < 0.001 |
| IG_all (median, IQR) | 2.16 (1.76, 2.47) | 3.04 (2.64, 3.62) | 2.03 (1.69, 2.30) | < 0.001 | 2.12 (1.75, 2.47) | 2.94 (2.70, 3.54) | 1.97 (1.64, 2.31) | < 0.001 |
| IG_0 (median, IQR) | 2.26 (1.75, 2.66) | 3.34 (2.72, 3.80) | 2.10 (1.69, 2.48) | < 0.001 | 2.41 (1.90, 2.79) | 3.70 (3.16, 4.18) | 2.22 (1.77, 2.62) | < 0.001 |
| IG_45 (median, IQR) | 1.88 (1.54, 2.18) | 2.85 (2.32, 3.26) | 1.78 (1.47, 2.04) | < 0.001 | 1.85 (1.48, 2.18) | 2.73 (2.31, 3.11) | 1.74 (1.40, 2.03) | < 0.001 |
| IG_90 (median, IQR) | 2.34 (1.85, 2.85) | 3.36 (2.89, 3.84) | 2.20 (1.75, 2.63) | < 0.001 | 2.42 (1.94, 2.78) | 3.27 (3.03, 3.61) | 2.25 (1.75, 2.61) | < 0.001 |
| IV_all (median, IQR) | 176.90 (148.98, 207.25) | 134.80 (109.30, 163.02) | 182.00 (156.00, 210.75) | < 0.001 | 175.50 (143.25, 200.75) | 133.50 (105.80, 155.70) | 183.00 (154.00, 206.00) | < 0.001 |
| IV_all_SD (median, IQR) | 4584.00 (3148.00, 6602.50) | 2166.50 (1340.50, 3535.00) | 5025.00 (3747.00, 7011.75) | < 0.001 | 4940.50 (2987.25, 6682.00) | 2849.00 (1841.00, 3428.00) | 5618.00 (3813.00, 6897.00) | < 0.001 |
| IV_0 (median, IQR) | 149.85 (122.75, 186.75) | 96.40 (78.95, 125.82) | 163.20 (134.00, 195.65) | < 0.001 | 146.70 (112.78, 185.57) | 74.10 (65.60, 90.60) | 158.40 (131.40, 196.20) | < 0.001 |
| IV_45 (median, IQR) | 239.55 (201.40, 284.75) | 164.40 (123.83, 188.62) | 254.30 (220.67, 290.60) | < 0.001 | 226.25 (201.25, 266.67) | 157.40 (133.90, 193.80) | 243.90 (214.30, 273.50) | < 0.001 |
| IV_90 (median, IQR) | 129.00 (103.00, 154.00) | 101.00 (77.75, 119.00) | 134.00 (109.25, 159.00) | < 0.001 | 124.50 (109.00, 150.75) | 105.00 (77.00, 118.00) | 133.00 (117.00, 156.00) | < 0.001 |
| Haralick_all (median, IQR) | 0.10 (0.09, 0.10) | 0.12 (0.11, 0.13) | 0.09 (0.09, 0.10) | < 0.001 | 0.10 (0.09, 0.10) | 0.12 (0.12, 0.14) | 0.09 (0.09, 0.10) | < 0.001 |
| Haralick_30 (median, IQR) | 0.10 (0.09, 0.11) | 0.14 (0.12, 0.15) | 0.10 (0.09, 0.11) | < 0.001 | 0.10 (0.09, 0.11) | 0.14 (0.13, 0.15) | 0.10 (0.09, 0.11) | < 0.001 |
| Haralick_45 (median, IQR) | 0.09 (0.08, 0.10) | 0.11 (0.10, 0.12) | 0.09 (0.08, 0.10) | < 0.001 | 0.09 (0.08, 0.10) | 0.11 (0.10, 0.13) | 0.09 (0.08, 0.10) | < 0.001 |
| Haralick_90 (median, IQR) | 0.11 (0.10, 0.13) | 0.14 (0.12, 0.16) | 0.11 (0.09, 0.12) | < 0.001 | 0.12 (0.10, 0.13) | 0.15 (0.12, 0.16) | 0.11 (0.09, 0.13) | < 0.001 |
| CSV (median, IQR) | 106.00 (102.00, 111.00) | 108.00 (105.00, 111.00) | 106.00 (101.00, 111.00) | 0.001 | 107.00 (102.25, 111.00) | 109.00 (105.00, 113.00) | 107.00 (102.00, 111.00) | 0.007 |
| CP (median, IQR) | 65.50 (60.00, 70.00) | 68.00 (66.00, 71.00) | 64.00 (59.00, 70.00) | < 0.001 | 64.00 (60.00, 68.00) | 67.00 (64.00, 68.00) | 63.00 (59.00, 68.00) | 0.002 |
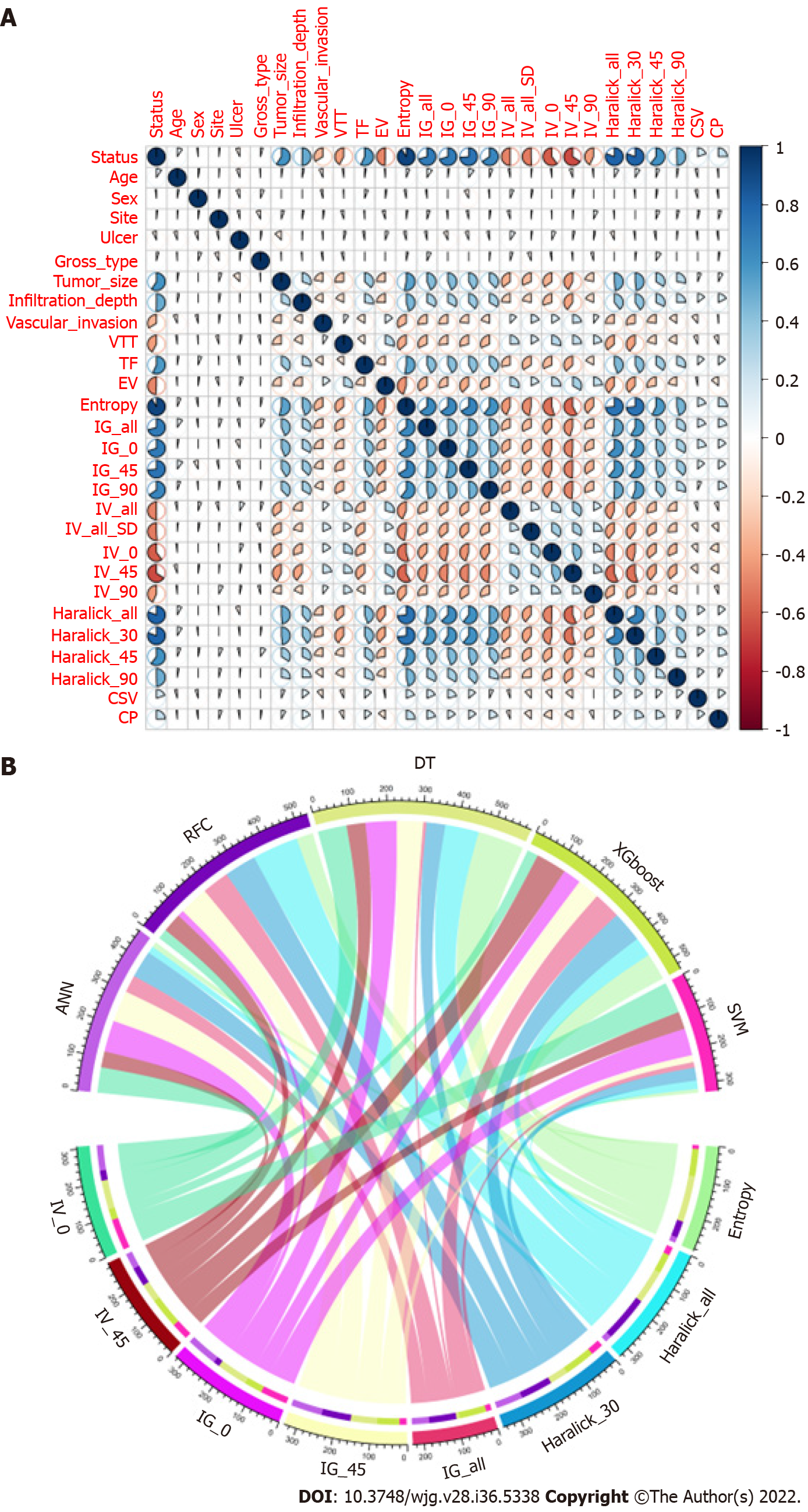
We conducted a correlation analysis on the variables with significant differences based on the statistical difference analysis of baseline data. As shown in Figure 2A, the correlation matrix (based on Pearson correlation analysis) indicates that the characteristic variables in the GLCM and LNM had a strong correlation degree (r > 0.6). For example, Entropy, Haralick full angle (Haralick_all), Haralick 30° (Haralick_30), Inverse gap full angle (IG_all), Inverse gap 45° (IG_45), Inverse gap 0° (IG_0), etc. were highly correlated with LNM. This suggests that these potential candidate variables can be used as LNM predictors and for the construction of subsequent models. Interestingly, in the subsequent models developed based on ML algorithms, we found that Entropy, Haralick_all, Haralick_30, IG_all, IG_45, IG_0, and Inertia value 45° (IV_45) occupied high weights as the top 7 GLCM-based factors (Figure 2B). Specifically, Entropy has the largest weight among these factors.
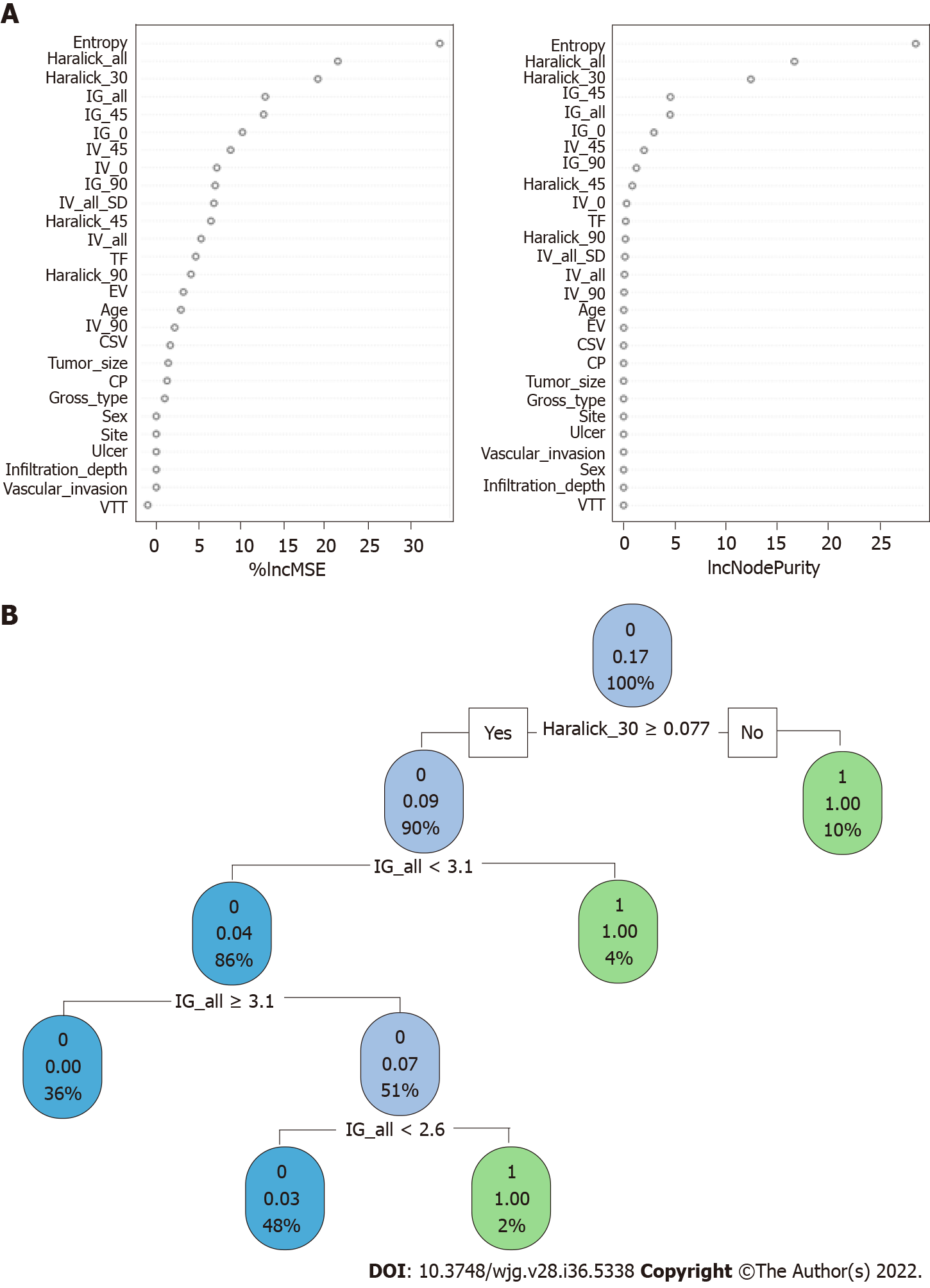
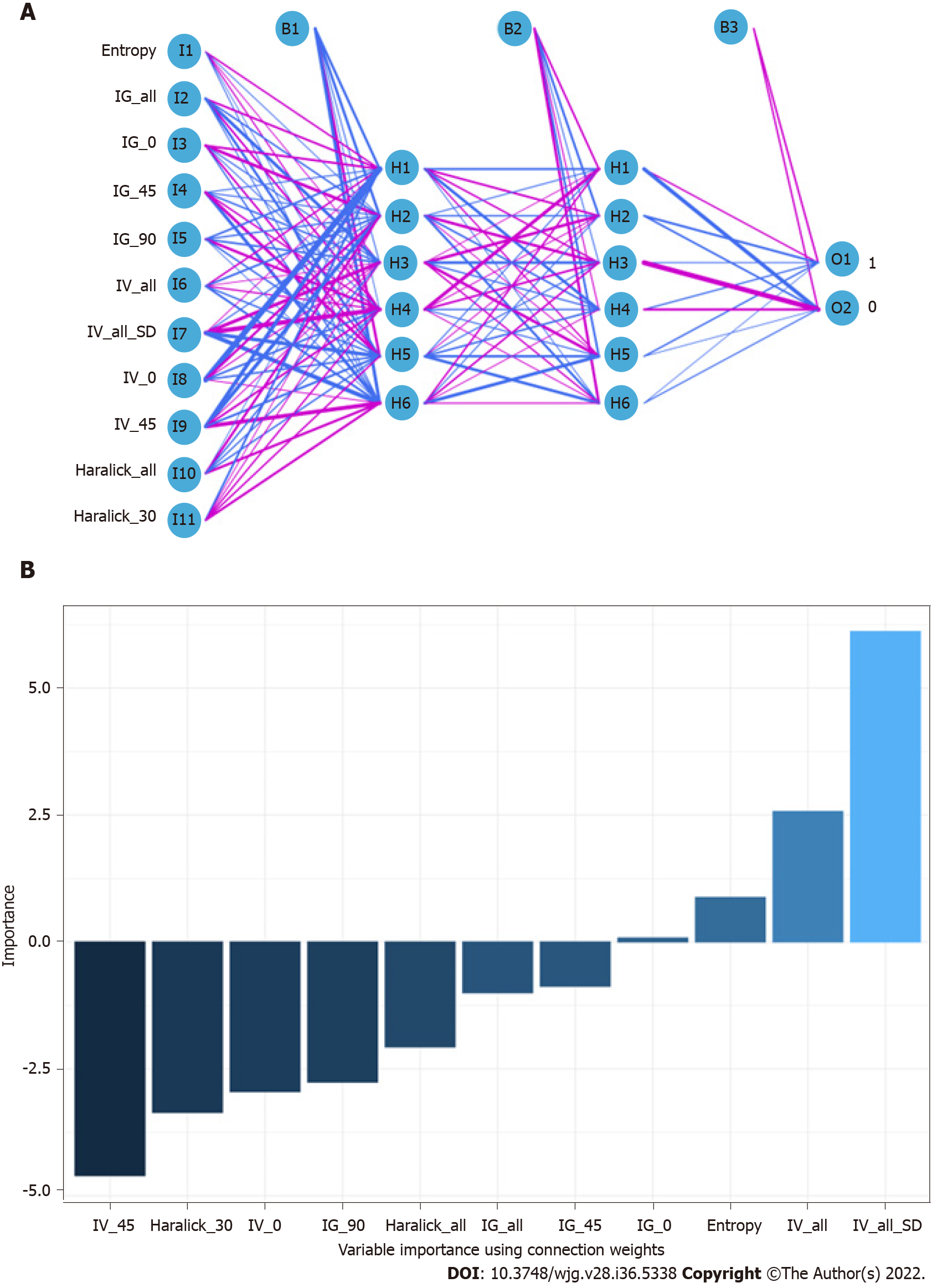
When constructing the RFC model [training set: Areas under the ROC curve (AUC): 0.925, 95% confidence interval (CI): 0.378-1.472; testing set: AUC: 0.912, 95%CI: 0.355-1.469], we repeatedly randomly selected N samples from the original training set N to generate the new training set DT and then generate M DTs to form a random forest according to the above steps. As shown in Figure 3A and Supplementary Table 1, the smallest Gini index after splitting was selected, including that for Entropy, Haralick_all, Haralick_30, IG_all, IG_45, IG_0, and IV_45. Similarly, Haralick_30 and IG_all served as important weight at DT branches (training set: AUC: 0.856, 95%CI: 0.309-1.403; testing set: AUC: 0.813, 95%CI: 0.256-1.370) (Figure 3B). In the ANN model (Figure 4), the accuracy of the prediction model developed using the prediction variables in the GLCM can also reach 0.887 (95%CI: 0.340-1.434) and 0.837 (95%CI: 0.280-1.394) in the training and verification sets, respectively. Although this accuracy is slightly inferior to that of the RFC model, it is better than those of other prediction models (i.e., DT, XGBoost, and SVM). Table 2, Supplementary Table 1, and Figure 5 summarize the predictive performance of ML-based models. In general, the prediction model constructed by using any ML algorithm was better than the logistic regression algorithm in predicting LNM, further confirming the superiority of ML algorithm, especially the robustness of the RFC.
| Model | Training set | Testing set | Variables1 | ||
| AUC mean | AUC 95%CI | AUC mean | AUC 95%CI | ||
| RFC | 0.925 | 0.378-1.472 | 0.912 | 0.355-1.469 | Entropy, Haralick_all, Haralick_30, IG_all, IG_45, IG_0, IV_45 |
| ANN | 0.887 | 0.340-1.434 | 0.837 | 0.280-1.394 | Entropy, IG_all, IG_0, IG_45, IG_90, IV_all, IV_all_SD, IV_0, IV_45, Haralick_all, Haralick_30 |
| DT | 0.856 | 0.309-1.403 | 0.813 | 0.256-1.370 | Entropy, Haralick_all, Haralick_30, IG_all, IG_45, IG_0, IV_45 |
| XGboost | 0.814 | 0.267-1.361 | 0.807 | 0.250-1.364 | Entropy, Haralick_all, Haralick_30, IG_all, IG_45, IG_0, IV_45, IG_90 |
| SVM | 0.805 | 0.258-1.352 | 0.794 | 0.237-1.351 | Entropy, Haralick_all, Haralick_30, IG_all, IG_45, IG_0, IV_45 |
| GLM | 0.796 | 0.229-1.362 | 0.799 | 0.233-1.365 | Entropy, Haralick_all, Haralick_30, IG_all, IG_45, IG_0, IV_45 |
| Radiologist | 0.789 | 0.242-1.336 | 0.801 | 0.254-1.348 | - |
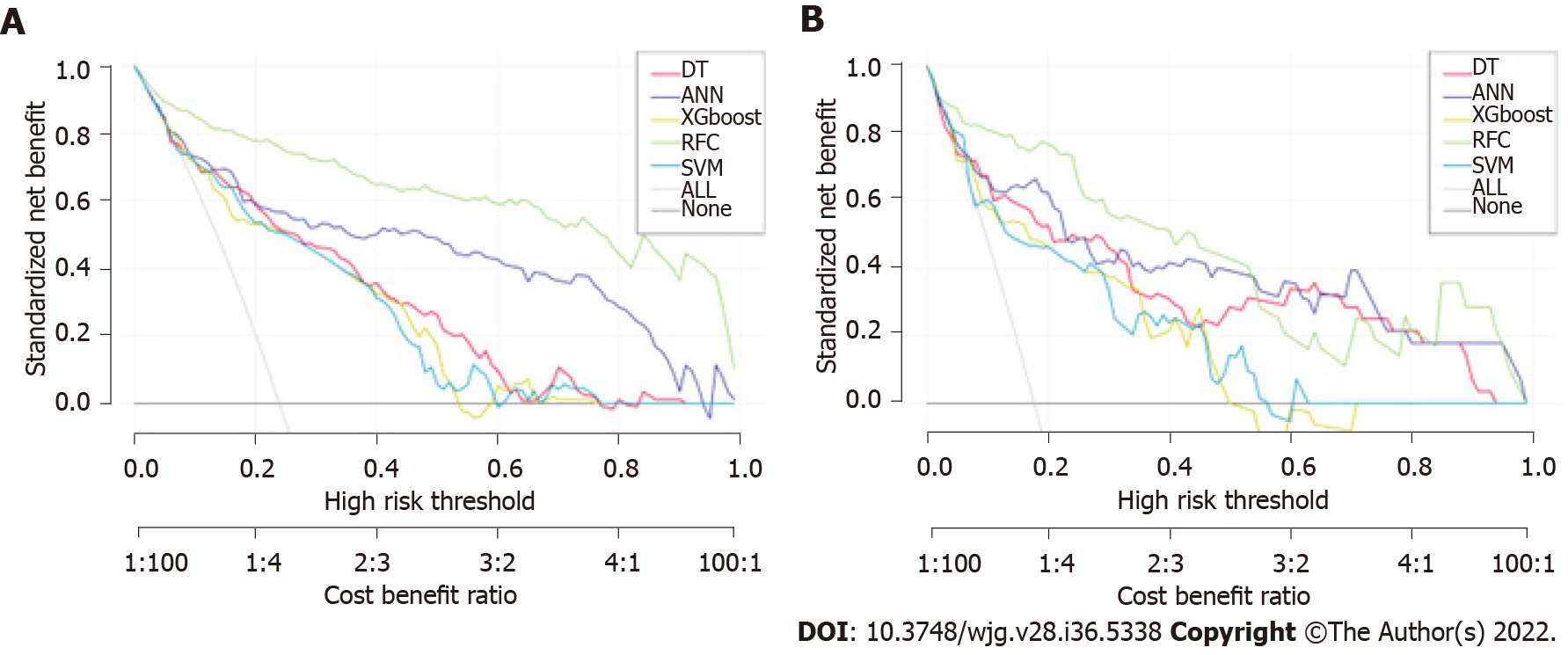
The prediction efficiency of the RFC model was the best in the process of precise stratification of LNM patients. To further evaluate the “stratification effect” of the RFC, results of CIC analysis indicate that high-risk LNM was accurately distinguished using the RFC model, and “cross-linking” did not occur in the stratification process. The results of this model for the validation and training sets were consistent (Supplementary Table 2), implying that the robustness and LNM discrimination of the RFC model were satisfactory.
The standard treatment for early GC is surgery. However, recently, ER has become the standard local treatment for some patients with early GC without LNM[21]. For a long time, it has been used to treat differentiated-type early GC limited to the mucosa, with a diameter of < 2 cm[22,23]. Recent studies have shown that ER indications have been expanded in many studies, even including UEGC and ≤ 2 cm diameter, without ulcer or lymphatic vessel invasion[24]. However, whether UEGC can accept the standard treatment of ER remains a subject of debate. That is, additional surgery should be performed if curability is considered questionable. Given this situation, the risk factors of LNM or distant metastasis and mortality after non-curative ER of UEGC should be investigated. Previous studies have also shown that patients with two or more risk factors (e.g., ulcer, submucosal invasion, and positive vertical margin) benefit greatly from surgical resection after ER that cannot be cured by UEGC[14,25]. However, due to the heterogeneity of clinical characteristics, risk stratification based on these predictions provides a simple prediction, which is challenging to apply in clinical practice.
The potential application of the GLCM in the prediction of LNM of UEGC has not been systematically explored thus far. In this study, GLCM-based features were extracted from underlying grayscale images collected through MRI. We developed an LNM risk prediction model for patients with UEGC using an ML-based algorithm. The following are the two important findings of our study. First, the accurate risk stratification of UEGC patients who should undergo additional surgery depends on the added value of the GLCM. Second, a new ML-based prediction model was used to identify patients and whether they have LNM. According to previous studies[26], texture analysis can quantify the spatial differences of pixels and the subtle differences reflected in gray values, which is consistent with the conclusion of this study. To some extent, we used GLCM features to gather spatial information and reduced the overfitting effect by replacing the softmax layer with the ML-based algorithm.
In this study, we created five types of ML-based models (i.e., RFC, ANN, DT, XGBoost, and SVM), which used GLCM features to predict LNM. Interestingly, there were differences in the prediction efficiency obtained by ML-based models of different algorithms. For example, the RFC model had the highest predictive accuracy, which was achieved by incorporating Entropy, Haralick_all, Haralick_30, IG_all, IG_45, IG_0, and IV_45. Meanwhile, the ANN, DT, XGBoost, and SVM exhibited an inferior performance compared with the RFC. This suggests that the accuracy of the RFC in predicting LNM is superior to that of the ML model. A previous study indicated that a random forest algorithm is more efficient in processing classification problems, which is consistent with the results of this study[27]. Meanwhile, DT is not as good as the RFC in terms of fitting, and the low prediction ability of the ANN model indicates that an “overfitting” phenomenon may occur. In general, different ML models show consistent accuracy, indicating that the prediction performance of ML can be improved through data processing.
Our results confirm a GLCM-based LNM classification, which has an ideal predictive effect on the diagnosis and treatment of patients with UEGC. However, the following problems were inevitably encountered in this study. First, because this study involved a retrospective analysis, the case inclusion criteria may have a certain bias on the results, which remains to be confirmed by a large sample of prospective studies in the future. Second, there were relatively few selected cases in this study, and only some parameters of the GLCM were extracted. Thus, the results of its prediction model should be verified by external data. Third, when data from multi-center and large sample studies are available in the future, it is crucial to predict the presence or absence of LNM. Additionally, the GLCM is an important imaging sequence of UEGC, and hence we will further perform other image texture analyses in subsequent research.
GLCM-based feature extraction could, in general, serve as a robust and promising tool to improve predictive efficiency for LNM in individual UEGC patients. ML adopts the algorithm of “classification and pruning” and clearer feature extraction, leading to better data fitting than the conventional prediction model. The model constructed using the RFC had the highest predictive accuracy, with the following being the most important predictors: Entropy, Haralick_all, Haralick_30, IG_all, IG_45, IG_0, and IV_45. In the future, we are still required to validate and optimize these prediction models using datasets of various scenarios to better apply them to clinical practice.
Gray-level co-occurrence matrix (GLCM) based feature extraction could serve as a robust and promising tool to improve the predictive efficiency for lymph node metastasis (LNM) of individual undifferentiated early gastric cancer (UEGC) patients. Additionally, machine learning (ML) adopts more optimized algorithms and more clear feature extraction. Models built using random forest classifier (RFC) have the highest predictive accuracy in Entropy, Haralick full angle (Haralick_all), Haralick 30° (Haralick_30), Inverse gap full angle (IG_all), Inverse gap 45° (IG_45), Inverse gap 0° (IG_0), and Inertia value 45° (IV_45). Further research is needed to develop these models for clinical practice.
The evaluation results indicate that the method of selecting radiological and textural features becomes more effective in the discrimination of LNM from UEGC patients. In addition, an ML-based prediction model developed using RFC can be used to derive treatment options and identify LNM that can improve clinical outcomes.
GLCM based feature extraction significantly correlated with LNM. The top 7 GLCM based factors included Inertia value 0°, IV_45, IG_0, IG_45, IG_all, Haralick_30, Haralick_all, and Entropy. The areas under the receiver operating characteristic (ROC) curve (AUCs) of the RFC model, support vector machine (SVM), eXtreme gradient boosting (XGBoost), artificial neural network (ANN), and decision tree (DT) ranged from 0.805 [95% confidence interval (CI): 0.258-1.352] to 0.925 (95%CI: 0.378-1.472) in the training set and from 0.794 (95%CI: 0.237-1.351) to 0.912 (95%CI: 0.355-1.469) in the testing set, respectively. The RFC (training set: AUC: 0.925, 95%CI: 0.378-1.472; testing set: AUC: 0.912, 95%CI: 0.355-1.469) model incorporating Entropy, Haralick_all, Haralick_30, IG_all, IG_45, IG_0, and IV_45 had the highest predictive accuracy.
We retrospectively selected 526 cases of UEGC confirmed by pathological examination after radical gastrectomy without endoscopic treatment in four tertiary hospitals between January 2015 to December 2021. GLCM-based features were extracted from grayscale images and ML was applied to the classification of candidate predictive variables. In order to evaluate robustness and clinical utility of each model, the following were made: ROC, decision curve analysis, and clinical impact curve.
Identifying a potential biomarker that predicts LNM is proven to be very useful in determining treatment.
To develop a ML-based integral procedure to construct the LNM gray level co-occurrence matrix (GLCM) prediction model.
The risk of LNM is the most important consideration in determining treatment strategies for UEGC. Therefore, identifying a potential biomarker that predicts LNM is proven to be very useful in determining treatment.
The authors thank all study participants for consenting to the use of their medical records.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A, A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: He D, China; Pantelis AG, Greece; Toyoshima O, Japan S-Editor: Wang JJ L-Editor: A P-Editor: Wang JJ
| 1. | Puliga E, Corso S, Pietrantonio F, Giordano S. Microsatellite instability in Gastric Cancer: Between lights and shadows. Cancer Treat Rev. 2021;95:102175. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 76] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 2. | Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23:700-713. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 900] [Cited by in F6Publishing: 1163] [Article Influence: 116.3] [Reference Citation Analysis (0)] |
| 3. | Lee A, Chung H. Endoscopic Resection of Undifferentiated-type Early Gastric Cancer. J Gastric Cancer. 2020;20:345-354. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 4. | Horiuchi Y, Fujisaki J, Yamamoto N, Ishizuka N, Ishiyama A, Yoshio T, Hirasawa T, Yamamoto Y, Nagahama M, Takahashi H, Tsuchida T. Undifferentiated-type predominant mixed-type early gastric cancer is a significant risk factor for requiring additional surgeries after endoscopic submucosal dissection. Sci Rep. 2020;10:6748. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 5. | Ono H, Kondo H, Gotoda T, Shirao K, Yamaguchi H, Saito D, Hosokawa K, Shimoda T, Yoshida S. Endoscopic mucosal resection for treatment of early gastric cancer. Gut. 2001;48:225-229. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1134] [Cited by in F6Publishing: 1104] [Article Influence: 48.0] [Reference Citation Analysis (4)] |
| 6. | Lee JH, Kim JJ. Endoscopic mucosal resection of early gastric cancer: Experiences in Korea. World J Gastroenterol. 2007;13:3657-3661. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 15] [Cited by in F6Publishing: 24] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 7. | Ohkuwa M, Hosokawa K, Boku N, Ohtu A, Tajiri H, Yoshida S. New endoscopic treatment for intramucosal gastric tumors using an insulated-tip diathermic knife. Endoscopy. 2001;33:221-226. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 295] [Cited by in F6Publishing: 314] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 8. | Zhao X, Cai A, Xi H, Chen L, Peng Z, Li P, Liu N, Cui J, Li H. Predictive Factors for Lymph Node Metastasis in Undifferentiated Early Gastric Cancer: a Systematic Review and Meta-analysis. J Gastrointest Surg. 2017;21:700-711. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 9. | Kato M, Nishida T, Yamamoto K, Hayashi S, Kitamura S, Yabuta T, Yoshio T, Nakamura T, Komori M, Kawai N, Nishihara A, Nakanishi F, Nakahara M, Ogiyama H, Kinoshita K, Yamada T, Iijima H, Tsujii M, Takehara T. Scheduled endoscopic surveillance controls secondary cancer after curative endoscopic resection for early gastric cancer: a multicentre retrospective cohort study by Osaka University ESD study group. Gut. 2013;62:1425-1432. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 158] [Cited by in F6Publishing: 181] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 10. | Hahn KY, Park JC, Kim EH, Shin S, Park CH, Chung H, Shin SK, Lee SK, Lee YC. Incidence and impact of scheduled endoscopic surveillance on recurrence after curative endoscopic resection for early gastric cancer. Gastrointest Endosc. 2016;84:628-638.e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 11. | Nasu J, Doi T, Endo H, Nishina T, Hirasaki S, Hyodo I. Characteristics of metachronous multiple early gastric cancers after endoscopic mucosal resection. Endoscopy. 2005;37:990-993. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 115] [Cited by in F6Publishing: 124] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 12. | Nakajima T, Oda I, Gotoda T, Hamanaka H, Eguchi T, Yokoi C, Saito D. Metachronous gastric cancers after endoscopic resection: how effective is annual endoscopic surveillance? Gastric Cancer. 2006;9:93-98. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 157] [Cited by in F6Publishing: 160] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 13. | Kawata N, Kakushima N, Takizawa K, Tanaka M, Makuuchi R, Tokunaga M, Tanizawa Y, Bando E, Kawamura T, Sugino T, Kusafuka K, Shimoda T, Nakajima T, Terashima M, Ono H. Risk factors for lymph node metastasis and long-term outcomes of patients with early gastric cancer after non-curative endoscopic submucosal dissection. Surg Endosc. 2017;31:1607-1616. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 14. | Suzuki H, Oda I, Abe S, Sekiguchi M, Nonaka S, Yoshinaga S, Saito Y, Fukagawa T, Katai H. Clinical outcomes of early gastric cancer patients after noncurative endoscopic submucosal dissection in a large consecutive patient series. Gastric Cancer. 2017;20:679-689. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 67] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 15. | Hirasawa T, Gotoda T, Miyata S, Kato Y, Shimoda T, Taniguchi H, Fujisaki J, Sano T, Yamaguchi T. Incidence of lymph node metastasis and the feasibility of endoscopic resection for undifferentiated-type early gastric cancer. Gastric Cancer. 2009;12:148-152. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 344] [Cited by in F6Publishing: 368] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 16. | Donders AR, van der Heijden GJ, Stijnen T, Moons KG. Review: a gentle introduction to imputation of missing values. J Clin Epidemiol. 2006;59:1087-1091. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1422] [Cited by in F6Publishing: 1362] [Article Influence: 75.7] [Reference Citation Analysis (0)] |
| 17. | Carpenter JR, Smuk M. Missing data: A statistical framework for practice. Biom J. 2021;63:915-947. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 54] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 18. | Luo W, Phung D, Tran T, Gupta S, Rana S, Karmakar C, Shilton A, Yearwood J, Dimitrova N, Ho TB, Venkatesh S, Berk M. Guidelines for Developing and Reporting Machine Learning Predictive Models in Biomedical Research: A Multidisciplinary View. J Med Internet Res. 2016;18:e323. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 366] [Cited by in F6Publishing: 490] [Article Influence: 61.3] [Reference Citation Analysis (0)] |
| 19. | Fan J, Lv J. A Selective Overview of Variable Selection in High Dimensional Feature Space. Stat Sin. 2010;20:101-148. [PubMed] [Cited in This Article: ] |
| 20. | Armstrong RA. When to use the Bonferroni correction. Ophthalmic Physiol Opt. 2014;34:502-508. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1208] [Cited by in F6Publishing: 1269] [Article Influence: 126.9] [Reference Citation Analysis (0)] |
| 21. | Goto O, Fujishiro M, Kodashima S, Ono S, Omata M. Outcomes of endoscopic submucosal dissection for early gastric cancer with special reference to validation for curability criteria. Endoscopy. 2009;41:118-122. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 130] [Cited by in F6Publishing: 149] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 22. | Gotoda T, Yanagisawa A, Sasako M, Ono H, Nakanishi Y, Shimoda T, Kato Y. Incidence of lymph node metastasis from early gastric cancer: estimation with a large number of cases at two large centers. Gastric Cancer. 2000;3:219-225. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1308] [Cited by in F6Publishing: 1270] [Article Influence: 52.9] [Reference Citation Analysis (0)] |
| 23. | Kojima T, Parra-Blanco A, Takahashi H, Fujita R. Outcome of endoscopic mucosal resection for early gastric cancer: review of the Japanese literature. Gastrointest Endosc. 1998;48:550-4; discussion 554. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 201] [Cited by in F6Publishing: 211] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 24. | Soetikno R, Kaltenbach T, Yeh R, Gotoda T. Endoscopic mucosal resection for early cancers of the upper gastrointestinal tract. J Clin Oncol. 2005;23:4490-4498. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 408] [Cited by in F6Publishing: 443] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 25. | Kim ER, Lee H, Min BH, Lee JH, Rhee PL, Kim JJ, Kim KM, Kim S. Effect of rescue surgery after non-curative endoscopic resection of early gastric cancer. Br J Surg. 2015;102:1394-1401. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 58] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 26. | Naik A, Edla DR, Dharavath R. Prediction of Malignancy in Lung Nodules Using Combination of Deep, Fractal, and Gray-Level Co-Occurrence Matrix Features. Big Data. 2021;9:480-498. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 27. | Li J, Tian Y, Zhu Y, Zhou T, Li J, Ding K. A multicenter random forest model for effective prognosis prediction in collaborative clinical research network. Artif Intell Med. 2020;103:101814. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |