Published online Nov 14, 2020. doi: 10.3748/wjg.v26.i42.6556
Peer-review started: August 29, 2020
First decision: September 12, 2020
Revised: September 24, 2020
Accepted: October 20, 2020
Article in press: October 20, 2020
Published online: November 14, 2020
In the last two decades, the vision of a unique carcinogenesis model for colorectal carcinoma (CRC) has completely changed. In addition to the adenoma to carcinoma transition, colorectal carcinogenesis can also occur via the serrated pathway. Small non-coding RNA, known as microRNAs (miRNAs), were also shown to be involved in progression towards malignancy. Furthermore, increased expression of certain miRNAs in premalignant sessile serrated lesions (SSLs) was found, emphasizing their role in the serrated pathway progression towards colon cancer. Since miRNAs function as post-transcriptional gene regulators, they have enormous potential to be used as useful biomarkers for CRC and screening in patients with SSLs particularly. In this review, we have summarized the most relevant information about the specific role of miRNAs and their relevant signaling pathways among different serrated lesions and polyps as well as in serrated adenocarcinoma. Additional focus is put on the correlation between gut immunity and miRNA expression in the serrated pathway, which remains unstudied.
Core Tip: In addition to the adenoma to carcinoma transition, colorectal carcinogenesis can also occur via the serrated pathway. In most serrated polyps, the pathway is believed to include the acquisition of a mutation in a gene that regulates mitogen-activated protein kinase (MAPK) pathway, disruptions to the Wnt signaling pathway and widespread methylation of CpG islands. Moreover, there are less data about different microRNAs (miRNAs) expression profiling in serrated adenomas with different grades of dysplasia. In contrast to the conventional colorectal carcinogenesis, the pivotal role of miRNAs and their relevant signaling pathways in the serrated pathway of carcinogenesis is still to be elucidated because of an insufficient number of studies conducted to clarify separate steps in the process.
- Citation: Peruhova M, Peshevska-Sekulovska M, Krastev B, Panayotova G, Georgieva V, Konakchieva R, Nikolaev G, Velikova TV. What could microRNA expression tell us more about colorectal serrated pathway carcinogenesis? World J Gastroenterol 2020; 26(42): 6556-6571
- URL: https://www.wjgnet.com/1007-9327/full/v26/i42/6556.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i42.6556
Colorectal cancer (CRC) is the most prevalent cancer in Western countries and the second cause of cancer-related death[1]. Obesity, sedentary lifestyle, tobacco and alcohol consumption are considered the driving factor behind the growth of CRC[2]. In the last two decades the vision of a unique carcinogenesis model for CRC has completely changed. The most prevalent genetic events accompanying CRC development are mutations that de-regulate the Wnt signaling cascade. In particular, inactivating mutations in the tumor suppressor adenomatous polyposis coli (APC) are considered the earliest genetic lesions sufficient to initiate tumorigenesis[3].
In addition to the adenoma to carcinoma sequence, colorectal carcinogenesis can also occur via the serrated pathway. After the identification of serrated carcinomas by Jass et al[4] in 1992, the underlying genetic and epigenetic alterations have been described. In most serrated polyps, the pathway is believed to be the acquisition of a mutation in a gene that regulates mitogen-activated protein kinase (MAPK) pathway, disruptions to the Wnt signaling pathway and widespread methylation of CpG islands[5,6].
A class of small non-coding RNAs, designated as microRNAs (miRNAs), are involved in progression towards malignancy. miRNAs act as tumor suppressors or oncogenes depending on the characteristics of their downstream targets[7]. They function as post-transcriptional gene regulators and have been increasingly recognized as useful biomarkers for CRC[8].
A plethora of studies have documented aberrant miRNA levels in CRC, but only a few of them relate to serrated pathway carcinogenesis[9]. There is even less data about different miRNA expression profiling in serrated adenomas with different grades of dysplasia[10]. In contrast to the conventional colorectal carcinogenesis, the pivotal role of miRNAs in the serrated pathway is still to be elucidated because of the insufficient number of studies conducted to clarify separate steps in serrated carcinogenesis[11].
Many of the published reviews in the English literature about the serrated pathway have been focused on histological, endoscopic, and molecular features[12,13]. However, there are a few data about post-transcriptional gene regulation, in particular, the expression of miRNAs in the serrated pathway in CRC. We aimed to interrogate the role of miRNAs in relevant signaling pathways in serrated carcinogenesis.
Emerging new approaches revealed increased expression of certain miRNAs in premalignant sessile serrated lesions (SSLs), emphasizing their role in the serrated pathway progression towards colon cancer[14]. This could make miRNAs potential biomarkers for screening in patients with SSLs[15,16].
In this review, we summarized the most relevant information about the specific role of miRNAs among different serrated lesions and polyps as well as in serrated adenocarcinoma (SAC). Additionally, the review is the first that looks at the correlation between gut immunity and miRNA expression in the serrated pathway.
Based on the literature, the percentage prevalence of serrated pathway is highly variable, ranging from 15% up to 30% of all CRCs[17-20].
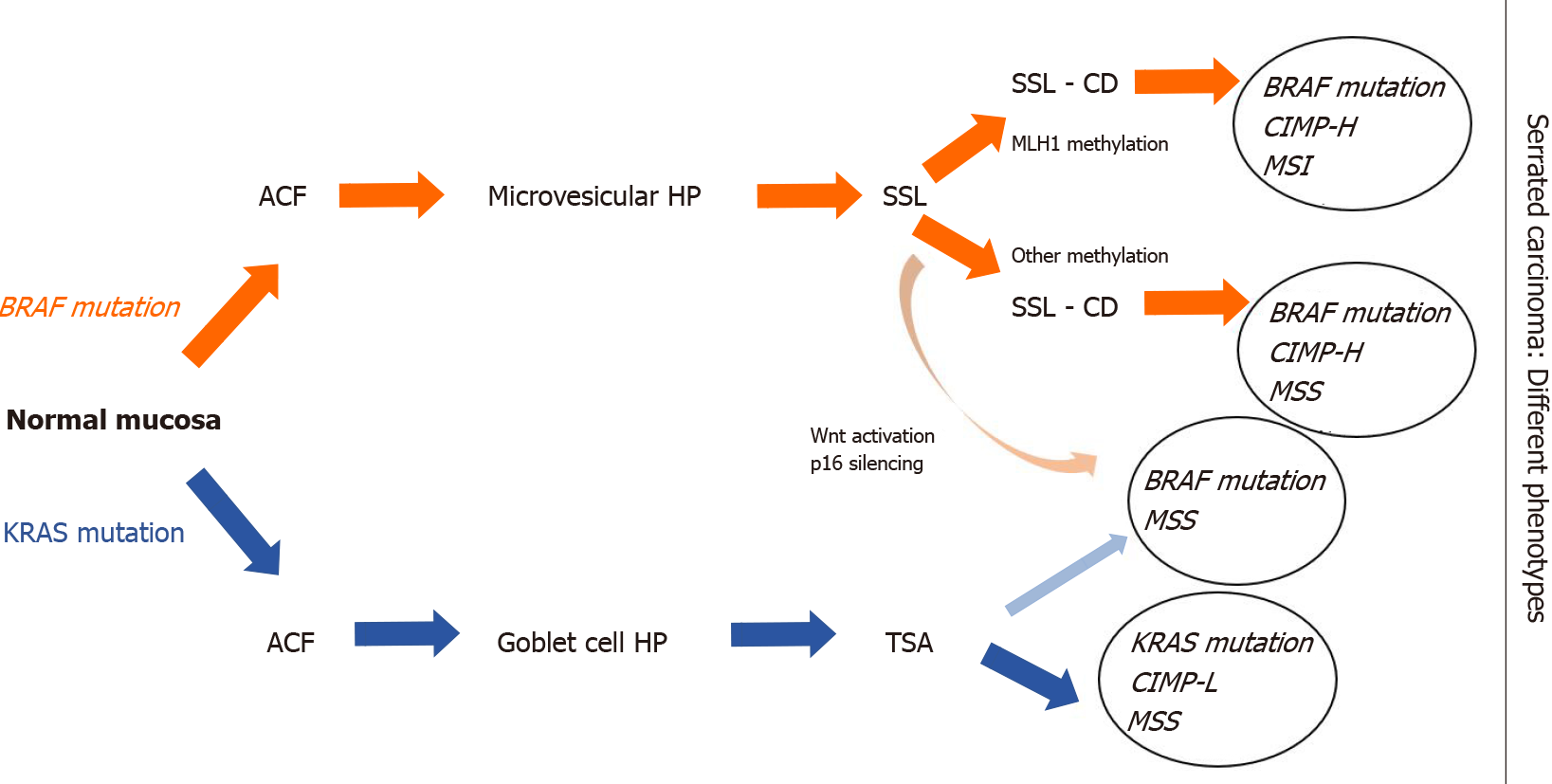
According to the 5th edition of WHO classification of colorectal serrated lesions and polyps, they are classified into three histopathological subtypes: Hyperplastic polyps (HPs), SSLs, and traditional serrated adenomas (TSAs)[21] (Figure 1). TSAs are extremely rare < 1% of all colorectal polyps, while HPs are the most common, comprising approximately 75% of all serrated polyps. SSLs (previously known as sessile serrated adenomas or sessile serrated polyps) cause nearly 25% of serrated polyps[22].
HPs are usually small, rarely cause symptoms, and have minimal malignant potential. However, it was established that HPs could progress to SSLs or TSAs for a period of 7.5 years[23]. In this context, HPs may predispose to cancer because of their ability to transform into serrated lesions[24]. These lesions could be found anywhere in the colon, but they are mostly placed in the distal colon (70%-80%)[25]. It was established that HPs, with right-side localization, are more likely to have malignant potential[26-28].
Clinical characteristics, such as size, location, and endoscopic appearance, can support the identification of SSLs but are not sufficient for their identification. Approximately 10% of SSLs could lead to sporadic CRCs via the serrated polyp-carcinoma sequence[29].
In most series, TSAs account for < 1% of all colorectal polyps, represent about 1%-2% of the serrated lesions and are located predominantly in the left colon[30-32].
SAC is characterized by mainly right-sided location of the colon, specific molecular features and female predominance. Percentage prevalence of SAC is about 7.5%-8.7% of all CRCs and according to the literature it has worse prognosis than conventional CRC[6,33].
Toyota et al[34] introduced the CpG island methylator phenotype (CIMP) in 1999. Methylation is an epigenetic process where a methyl group (CH3) is added to the cytosine nucleotide at a CpG dinucleotide group. The process of methylation of gene promoters is a physiological mechanism by which gene expression is regulated without altering the DNA sequence[35,36].
Transcriptional silencing of essential tumor suppressor genes, caused by aberrant DNA methylation, could promote neoplastic growth. This aberrant methylator has been called the CIMP and is thought to be important in the serrated pathway in CRC[37].
Using eight markers, Ogino et al[38] classified CIMP in CRC into three subgroups, CIMP-low (CIMP-L), CIMP-high (CIMP-H), and CIMP-negative, according to the numbers of methylated promoters.
With the growing impact of translational research and molecular pathology, the CRC pathogenesis became more elucidated based on the association of CIMP and key mutations in KRAS, BRAF, PIK3CA, TP53, and APC. Furthermore, microsatellite instability (MSI), caused by dysfunction of DNA mismatch repair (MMR) genes, is considered another critical pathway in carcinogenesis[39].
The MSI mechanism in CRC was first described in relation to Lynch syndrome, where germline mutations take place in specific MMR genes such as MLH1, MSH2, MSH6, and PMS2[40]. Germline deletions at 3’ end of the EPCAM gene which lead to decreased MSH2 expression were also demonstrated as a recurrent cause of Lynch syndrome[41]. Furthermore, functional relevance of MSH3 mutations for the development and inheritance of CRC were reported, but their role in the serrated pathway needs further analysis and more cohort studies[42,43]. Evidence has shown that mutations in MSI are vital points in the developing malignancy in 3%-15% of all CRC[42,43]. About 80% of MSI CRCs are characterized by the hypermethylation of MLH1, while 20% of MSI CRCs by mutations in MMR genes[44]. MSI status could be subclassified into MSI-high (MSI-H), MSI-low (MSI-L) and microsatellite stable (MSS) according to the number of mutations in microsatellite sequences[45].
Alteration of MMR genes due to epigenetic silencing by sporadic, acquired hypermethylation of the MLH1 gene promoter leads to the serrated pathway in CRC[44].
Serrated colorectal malignancies are characterized by CIMP-H, MLH1 promoter hypermethylation, and MSI and BRAF mutations[46].
Serrated colorectal lesions rarely bеаr truncating APC mutations, but the most frequent genetic alterations involve BRAF mutations, whereas KRAS mutations are less common[47]. Both KRAS and BRAF belong to the MAPK signaling pathway, mediating cell proliferation, apoptosis and differentiation[48].
BRAF gene encodes a protein called B-Raf, which plays a pivotal role in regulating the MAPK/ERKs signaling pathway[49]. Recent findings in molecular biology demonstrated that mutations in BRAF are found in about 10% of CRC patients[50]. BRAF-mutated CRCs are associated with the female gender, often right-sided, mucinous histology, and advanced stage[51]. BRAF mutations are considered as early events in CIMP cancers by inhibition of normal apoptosis in colonic mucosa[52]. Many recent studies classified two different molecular phenotypes of CRC based on BRAF mutation status: BRAF V600E- and non-V600-mutated CRC[53]. А correlation between serrated carcinogenesis and BRAF V600E mutation was established, which induce CIMP-H status and methylation of MLH1 promoter[54]. In contrast to the conventional adenomas, the earliest event in serrated precursor lesions are BRAF mutations and hypermethylation, which leads to transformation of aberrant crypt foci (ACF) to microvesicular HP and then to SSLs. Methylation and loss of key tumor suppressor genes such as p16 and MLH1 are the key points in SSLs’ progression to SAC[55]. Interesting information about the BRAF mutated/MSS SACs was reported by Bond et al[55]. They found out that hypermethylation events occurred in BRAF mutated SACs more often than in conventional pathway (respectively 60% and 3%)[55]. BRAF V600E-mutated CRCs are with dismal prognosis and resistance to standard systemic chemotherapy[56,57].
Another significant driver in the serrated pathway is KRAS mutations[58]. Opposite to the traditional model of Vogelstein, where aberrant activation of Wnt pathway has been observed, high frequency of KRAS mutations was established in TSAs. In contrast to SSLs, TSA lesions showed MGMT hypermethylation, but not MLH1 promoter hypermethylation. Based on this evidence, a non-MLH1 mutating SSL could progress to a TSA and ultimately develop into a BRAF-mutated MSS tumor (Figure 2)[59,60].
miRNAs were discovered in Caenorhabditis elegans by Lee et al[61] in 1993 while studying the gene lin-14. However, the scientific community became aware of the importance of miRNAs seven years later when they were recognized as a specific class of biological regulators. miRNAs are small, single-stranded, non-coding RNAs (18-24 nucleotides) that can post-transcriptionally regulate the expression of various oncogenes and tumor suppressor genes[62]. Also, they play an essential role in cancer development, proliferation, regression, and metastasis. Even though their role in cancer progression is yet to be elucidated, several studies reported the influence of specific miRNA alterations in premalignant and malignant lesions[63-66]. miRNA expression profiling gives us the opportunity to understand and identify differences between benign and malignant lesions of the colon mucosa, as well as to stratify benign lesions according to their malignant potential[67].
In this scenario, several studies showed a unique miRNA signature in different types of colonic polyps, as well as in the progression of serrated lesions.
Tsikitis et al[68] profiled miRNA patterns in screen-detected polyps in relation to histologic features and cancer-related risk. miRNA expression analysis was carried out on biopsy specimens from 109 patients. The specimens were obtained from normal mucosa (NM), HPs, tubular adenomas (TAs), tubulovillous adenomas, or high-grade dysplasia (TVHGs), SSLs, and TSAs. They have not found a significant difference in the expression of miRNA between TSAs and SSLs. miRNAs expression pattern was similar in TSAs and HGTVs, whilst there were several differentially expressed miRNAs between HPNMs and TSAs. Additionally, they performed pairwise comparisons of non-serrated tissues and serrated lesions. miRNAs-222 and miRNA-214 were significantly downregulated by 2.35- and 1.51-fold respectively in serrated polyps, whereas miRNA-335 was significantly overexpressed by two-fold in non-serrated tissues. Tsikitis et al[68] drew the conclusion that the downregulation of miRNA-125b and miRNA-320a in the serrated pathway may be used as independent predictors of progression with a concordance index of 84.7%.
Opposite to the serrated pathway, in the conventional adenoma-carcinoma sequence, many studies showed a high expression of miRNA-125b in advanced tumor size. Another correlation was found between the overexpression of miRNA-125b, which leads to repression of the endogenous level of p53 protein in human CRC cells. Cancer progression and poor outcomes were associated with overexpression of miRNA-125b in the conventional colorectal pathway[69].
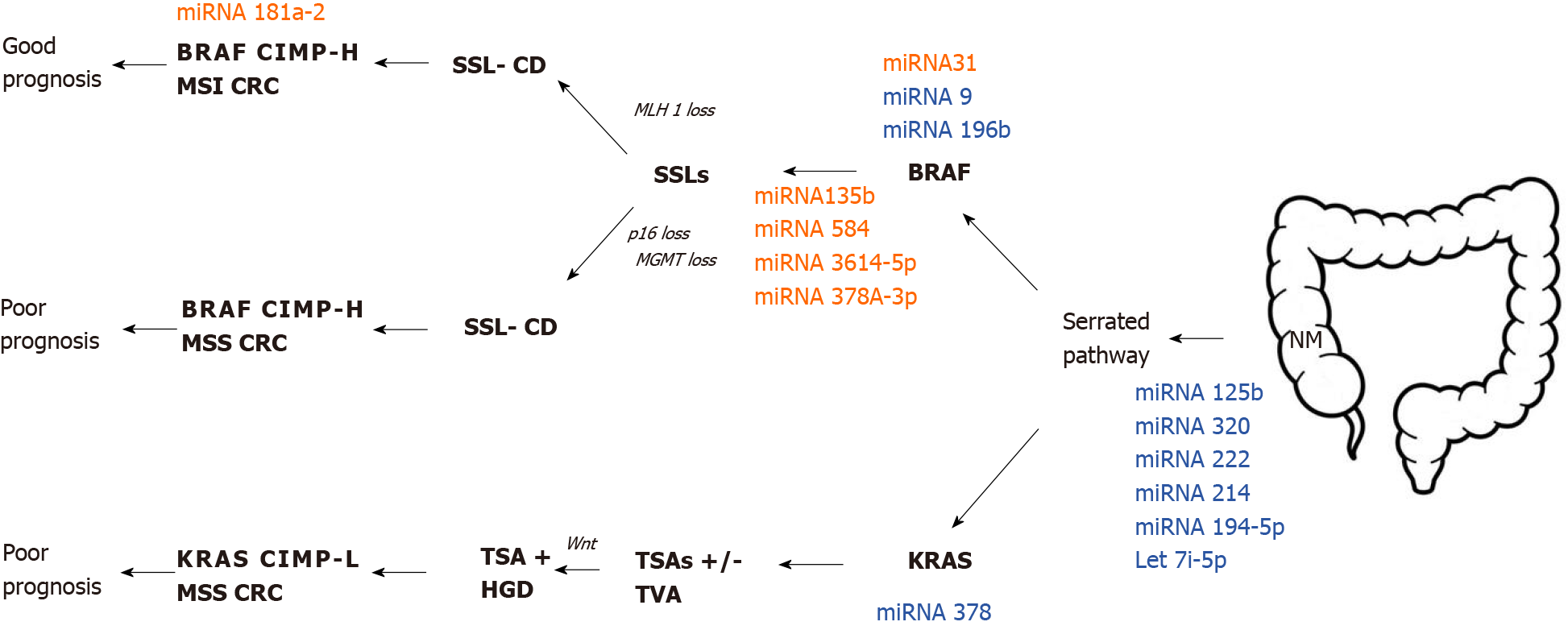
However, many studies showed that miRNA-31 plays a pivotal role in serrated carcinogenesis. In this scenario, miRNA-31 is located at 9p21.3 and is frequently overexpressed in sessile serrated adenomas. Aoki et al[70] analyzed in their case report miRNA-31 expression using quantitative reverse transcription-PCR in patients with early invasive CRC with HP component. Their results showed higher miRNA-31 expression in the carcinoma component compared to HP component. They revealed that progression of HP (or SSLs) to SAC is likely to be associated with overexpression of miRNA-31.
To shed light on the role of miRNA31 on the serrated pathway, Kanth et al[11] conducted a study of 108 colon biopsies with distinct histology types. Different expression was established in 23 miRNAs between NM and serrated lesions. Additionally, six miRNAs showed a different expression pattern between SSLs and HPs, as miRNA-31-5p has been the most significantly modulated.
Nosho et al[71] based on miRNA array analysis, identified that miRNA-31 was the most upregulated in BRAF (V600E) mutation, compared to BRAF-wild type CRCs. Moreover, they performed transfection of the miRNA-31 inhibitor and consequently showed that miRNA-31 might regulate BRAF activation in CRCs. Therefore, miRNA-31 could be used as a diagnostic biomarker as well as a feasible therapeutic target in the future. Finally, they proved that high miRNA-31 expression was associated with shorter prognosis in patients with CRC.
Higher miRNA-31 expression was associated with cell proliferation and survival in development in CRC, as well as tumor invasion and poor prognosis[72-75]. Kubota et al[76] pointed out that miRNA-31 could be a potential prognostic biomarker in their study of patients with stage IV of CRC. They also found out a correlation between miRNA-31 overexpression and poor tumor differentiation, as well as advanced disease stages.
Recent studies showed the presence of miRNA-31 in the serum of patients with metastatic CRC, who were treated with anti-EGFR therapy. Igarashi et al[77] found out a correlation between high miRNA-31-5p expression and shorter PFS in CRC patients treated with anti-EGFR therapeutics. Their theory suggested that miRNA-31-5p could be a useful prognostic biomarker for anti-EGFR therapy.
Even though the underlying mechanisms of the role of miRNA-31-5p in CRC remain unknown. It has been postulated that miRNA-31 can directly bind to the 3' untranslated region (3' UTR) of SATB2, which takes part in regulation of transcription and chromatin remodeling. Overexpression of miRNA-31-5p could induce epithelial-mesenchymal transition, tumorigenesis, and progression in CRC[78].
Furthermore, another correlation between the expression of miRNA-31 and CRC-associated fibroblast (CAFs) was established, but not in vivo experimental models. Yang et al[79] elucidated that miRNA-31 inhibits autophagy in CAFs and alters colorectal proliferation and invasion of CRC cells. Thus, more studies must be conducted in this direction because of the lack of in vivo experimental models.
In many studies, it has been reported that overexpression of miRNA-135-B has been associated with APC dysfunction in CRC, leading to the promotion of tumor-proliferation, progression, and invasion[63,80]. It was established that miRNA-135-B had been associated with the serrated pathway and colorectal carcinogenesis.
Only few studies indicate that specific miRNA profiles can be used to distinguish neoplastic from benign lesions in colon mucosa[6]. A study by Kanth et al[11] was the first that showed the overexpression of specific miRNAs in serrated polyps or serrated carcinoma. In summary, they provided a comprehensive analysis of miRNA gene expression in SSLs, by identifying miRNA-135B, miRNA-378A, miRNA-548, miRNA-9, and miRNA-196B. miRNA-378A-3p was significantly downregulated in SSLs compared to normal colon mucosa. They suggested that these miRNAs are good predictors in SSLs to carcinoma transformation. Additionally, they discovered that miRNA-9 and miRNA-196b were also de-regulated in SSL compared to HP. These miRNAs showed different expression patterns in BRAF mutated-MSI tumors. Interestingly, reduced expression of miRNA-196B has been detected in the plasma of patients with CIMP-positive SSLs or MSI colon cancers[11].
MiRNA-21 is one of the most eminent miRNAs involved in the genesis and progression of CRC. Evidence implied that miRNA-21 negatively regulates tumor suppressor phosphatase and tensin homolog (PTEN) gene, which played an essential role in cell proliferation and invasion in CRC[81-84]. An interesting study by Ghareib et al[85] established that miRNA-21 in serum could be feasible, non-invasive biomarker with high sensitivity and specificity (95.8% and 91.7%) for early detection and prognosis in patients with CRC.
In addition, Chen et al[86] report a correlation between tissue and serum miRNA-21 overexpression and poor prognosis in patients with CRC. It is more significant in colon cancers, compared to rectal.
Another interesting study by Yau et al[87] presents the potential role of fecal-based miRNA-21 and miRNA-92a as non-invasive biomarkers for CRC screening. They reported higher expression of miRNA-21 and miRNA-92a in patients with advanced distal CRC compared to the proximal localization, without significant value in the detection of early CRC.
miRNA-21 down-regulates tumor suppressor PDCD4, thus stimulating cancer cell invasion and intravasation. Moreover, the high level of miRNA-21 was associated with metastasis and resistance to chemotherapy of 5-FU in CRC. Thus, it makes miRNA-21 a potential non-invasive biomarker for diagnostic and prognosis for CRC[88].
Recently, several studies have reported the correlation between expression of miRNA-21 and serrated pathway in CRC. A study by Schmitz et al[89] demonstrated different expression of miRNA-21 among NM, HPs, and SSLs. They found overexpression of miRNA-21 in SSLs, whereas normal colon mucosa and HPs exhibited no differences. Opposite to them, Kanth et al[11] proved that there was no statistically significant expression of miRNA-21 in SSLs.
Future investigations are necessary to find out the correlation between expression levels of miRNA-21 and genetic and epigenetic alterations of SSLs.
miRNA-181 plays a pivotal role in regulation at the post-transcriptional level in many different types of cancer. More specifically, the expression of miRNA-181a and miRNA-181b are strongly associated with the mutation status of the tumor suppressor gene p53 in colorectal carcinogenesis[90]. The underlying mechanism of how miRNA-181a influences conventional colorectal carcinogenesis could be based on up-regulation miRNA-181a through the activation of the Wnt/β-catenin pathway[91].
Little is known about the expression of miRNA-181a in the serrated pathway. A comprehensive analysis of miRNA profile in SACs and MSI-H CRC has been carried out by Kondelova et al[10] Interesting information about the molecular features of miRNA expression in SACs and MSI-H CRC has been elucidated. Microarray assay showed that 223 miRNAs were differently expressed, as 75 of them were downregulated in SACs compared to MSI-H CRC. On the other hand, 148 miRNAs were upregulated in the same comparison group. Notably, only miRNA-181a-2 showed significant overexpression in MSI-H CRC compared to SACs. It has been established that miRNA-181a-2 has an inverse correlation with nicotinamide phosphoribosyl transferase, which is a transcription factor playing a significant role in organogenesis and stem cell development[92].
In conclusion, their analysis showed that miRNA-181a-2 plays a role in development in different subtypes of CRC from the serrated pathological pathway. Additionally, the up-regulation of miRNA181a-2 was associated with MSI-H status. This study may be a foundation for further researches aiming to elucidate the function of miRNA-181a-2 in CRC[10].
Slattery et al[15] have carried out promising research about different miRNA expression between NM and different types of polyps. They made a comprehensive analysis of miRNA expression among adenomatous polyp (AD), SSLs, and HPs. This study identified 19 differently expressed miRNAs between AD and HP such as let-7i-5p, miRNA-1229-5p, miRNA-1234-5p, miRNA-1249, miRNA-1268B, miRNA-1275, miRNA-194-5p, miRNA-215, miRNA-2392, miRNA-30b-5p, miRNA-331-3p, miRNA-3653, miRNA-3960, miRNA-4281, miRNA-4689, mRNA-4739, miRNA-518a-5p, miRNA-6510-5p and miRNA-939-5p. They concluded that the expression of the above-mentioned miRNAs in HP and SSLs are down-regulated and are related to MSI and CIMP. On the other hand, ADs have upregulated miRNA expression and are associated with TP53 and KRAS-mutations. Additionally, their study aimed to identify different miRNA expression and molecular pathways in colorectal carcinogenesis through genomic landscaping of colon polyps[15]. An overview of putative miRNA profile expression in the serrated colorectal pathway is presented in Figure 3.
Human gut microbiota comprises approximately 39 trillion microorganisms that colonize the adult gut system[93]. It plays a significant role in maintaining homeostasis of the intestinal immune system, which represents a natural barrier to pathogen infection[94] but also maintain oral tolerance in the gut. Gut homeostasis can be disturbed by environmental factors such as lifestyle, diets, infections, and antibiotics, leading to dysbiosis. Many recent studies have demonstrated the association between gut dysbiosis and colorectal carcinogenesis[95]. Evidence suggest that Fusobacterium nucleatum (F. nucleatum) has overabundance in gut microbiota in dysbiosis[96]. This finding is in agreement with the fact that F. nucleatum is involved in mucosal inflammation and contributes to the progression of CRC[97,98]. There are plenty of studies that investigate interactions between F. nucleatum and conventional adenoma to carcinoma sequences[99-101]. Ito et al[102] focused on F. nucleatum and serrated carcinoma pathway. In particular, they investigated the putative correlation between F. nucleatum and miRNA-31 expression. However, the results of the study did not indicate a significant association between miRNA-31 and F. nucleatum. Nevertheless, Yu et al[103] showed that invasive F. nucleatum might play a role in developing proximal colon carcinogenesis through the serrated neoplasia process, which may play a less significant role in the traditional adenomas-carcinoma sequence. Bacterial biofilms may not support F. nucleatum infiltrate tumor tissues.
Longitudinal studies of immune infiltrate in resected CRC tumors have shown the role of the immune response in the pathophysiology of CRC. miRNAs, as non-coding RNAs, are capable of controlling several post-transcription target genes and performing essential roles in cell proliferation, differentiation, and apoptosis, including the immune cells[104]. In other words, miRNAs are necessary for maintaining the functioning of the immune system. However, abnormal expression of miRNAs is often found in various forms of tumors that contributes to immune deficiencies or immune evasion. Li et al[104] focused on the possible functions of miRNAs in CRC immune response control and the use of specific miRNA targets for CRC therapy. It is assumed that miRNAs possess an immunomodulatory role and can potentially be a part of the anti-cancer target pipeline. However, there may be some drawbacks and threats of using miRNAs as immunotherapeutics.
As discussed above, different miRNA profile variations from the transition of NM to adenoma and CRC identified some miRNA as contributors to those transformations. Moreover, serum miRNAs may be used as markers to track certain changes accompanying carcinogenesis[105]. miRNA profiles obtained in standard colorectal mucosa differ from those in adenomas and CRC. Oncogenes such as c-Met and KRAS, together with the miRNAs could also have pro- or anti-CRC effects, including influencing the immune system. More interestingly, some miRNAs increased their expression in developing CRC, whereas others reduced their expression, such as miRNA-30b[106]. Furthermore, evidence indicates that miRNAs not only participate in colorectal carcinogenesis, but can be used as biomarkers for diagnosing, managing, and follow up the patients.
It is well-known that one of the mechanisms for cancer invasion is to establish complex pathways for disarming the immune system and evading immune surveillance. Nakanishi et al[106] demonstrated that in human serrated tumors, the expression of atypical protein kinases C (PKC) is decreased. Simultaneous inactivation of the encoding genes in the intestinal epithelium of the mouse culminated in random serrated tumorigenesis with a highly reactive and immunosuppressive stroma leading to advanced cancer development. Whereas epithelial PKC deficiency resulted in the death of immunogenic cells and the infiltration of CD8+ T cells that repressed tumor initiation, IFN, and CD8+ T cell responses were impaired by PKC loss, resulting in tumorigenesis[106].
Some tumors may stimulate the immune cells in the tumor stroma to produce a variety of inhibiting cytokines such as transforming growth factor (TGF-β) and IL-10, which suppress the recruitment and activation of antitumor T lymphocytes[107]. Furthermore, IL-6 suppresses the ability of dendritic cells to present antigens by activating the signal transducer and transcription activator 3 (STAT3) and lessens CD4+ T cell-mediated immune response[108]. Thus, an immunotherapy that utilizes monoclonal antibodies that antagonize immunosuppressive cytokines or inactivate immunosuppressive cells may enhance tolerance to cancer and prevent tumor growth[16]. Our team also documented that IL-6 upregulation is crucial for developing both IBD and CRC well before the upregulation of other Th17/Treg associated genes (TGFb1, IL-10, IL-23, and FoxP3 transcription factor) that are critical primarily for the development of CRC[109]. An additional study revealed that intratumoral IL-17-mediated signaling might inhibit immunotherapy responses[110].
In line with this, synergistic therapeutic efficacy was demonstrated by combined therapy with TGF-β receptor inhibitor and anti-PD-L1 checkpoint blockade. A study of human samples confirmed the importance of atypical PKCs during the immunosurveillance defects in human serrated CRC. These results give insight into how this poor-prognosis subtype of CRC to be diagnosed and treated[106].
Since miRNAs modify the differentiation, activation, and distribution of the various immune cells and the intricate cytokine network, miRNAs play an essential role in both innate and adaptive immune responses. miRNAs are closely involved in processes such as control of innate and adaptive immunity activation, regulation of inflammation and cytokine network, trafficking and cytokine crosstalk between the tumor and its microenvironment, miRNAs are promising targets for immunotherapy of different gastroenterological cancers[111]. Thus, miRNAs exert regulatory and protective functions in the digestive system and antitumor defense against gastroenterological cancers development.
In line with this, KRAS-IRF2 (interferon regulatory factor 2) axis also impacts the immune system towards immune suppression[112]. The clinical significance of this observation is the immunotherapy resistance in CRC. However, the biological functions and mechanisms of oncogenic KRAS in resistance to immune checkpoint blockade therapy are not fully understood.
Additionally, although various studies have examined the immune environment of CRCs with MSI, only one analysis assessed the immune microenvironment of serrated precursor lesions, including sessile serrated adenoma with dysplasia (SSA-D)[113]. Rau et al[113] studied the density of intraepithelial lymphocytes (IELs) in various serrated polyps and SSAs-D. The investigators observed that the amount of IELs was substantially higher in SSA-D than in SSAs, which displayed significantly higher numbers of IELs relative to HPs and typical adenomas. In their research, Acosta-Gonzalez et al[114] examined the immune properties of the serrated carcinogenesis system and its association with morphological stepwise dysplasia-carcinoma development and MSI status. They confirmed the higher density of IELs in lesions of MSI-H tumors. Additionally, other studies have shown that the total number of frameshift mutations in MSI CRCs correlates with lymphocyte infiltrating tumor density, specifically CD8+ lymphocyte density[115].
Nevertheless, the serrated pathway has two outcomes that differ in their clinical and prognostic characteristics as well as in their methylome profile and histological and molecular characteristics: (1) SSLs; or (2) Sporadic CRC showing MSI-H[42]. The latter subtype of CRC is correlated with deep immune invasion and has a better prognosis than the former[116].
The latest approaches in transcriptomics used to classify human CRC have shown that mesenchymal and/or desmoplastic involvement, together with an immunosuppressive microenvironment, are essential determinants of the worst prognosis of CRC. Importantly, these aggressive CRCs harbor the traits of serrated tumors, suggesting that how aggressive the CRC becomes is determined by initiation by this alternate mechanism. Moreover, molecular markers and profiles of gene expression have indicated that at least two CRC subgroups exist within the serrated pathway: (1) An inflammatory subtype with features of stromal/mesenchymal high immune infiltration (referred to “mesenchymal serrated” CRCs); and (2) MSI (“classical serrated”). BRAF mutation characterized with immune suppression in the tumor environment[117].
However, the tumor stroma's possible activation and the type of immune response associated with the CRC tumor stroma are not yet well understood. SAC may be infiltrated by CD45+ cells that express PD-L1 and decrease CD8+ T cells, which determines that there are multiple immune mechanisms to avoid the immune response[106]. Nevertheless, to create more efficient therapies, understanding the pathogenesis, including the tumor environment on the immunological settings, for both forms of serrated CRC is essential. Although emerging data show that immunotherapy is a promising choice for patients with multiple cancer forms still, there is a substantial clinical gap between the identification of serrated precursor lesions and the effective therapies for treating them.
With the growing influence of translational research and molecular pathology, the serrated pathway carcinogenesis became more elucidated based on the association of CIMP and key mutations in BRAF, KRAS, PIK3CA, TP53, and APC. Furthermore, MSI caused by dysfunction of DNA MMR genes, is considered as another critical pathway in carcinogenesis.
In this review we summarized the most relevant information that have been published in the literature so far about miRNA expression in serrated pathway. Furthermore, we intended to answer the question could miRNA expression tell us more about colorectal serrated pathway carcinogenesis. The answer may come from several studies that have been published related to this issue. The data showed a unique miRNA signature in different types of colonic polyps, as well as in the progression of serrated lesions. Besides, those miRNAs play an important role in serrated carcinogenesis, proliferation, regression, and metastasis. Existing evidence support that miRNAs expression profiling, including miRNA-125b, miRNA-222, miRNA-214, miRNA-335 miRNA-31 miRNA-135-B miRNA-21 miRNA-181a-2, etc., allows us to understand and identify differences between benign and malignant lesions of the colon mucosa, as well as to stratify benign lesions according to their malignant potential.
Moreover, serum miRNAs may be used as markers to track specific changes accompanying serrated carcinogenesis. This assertion is based on the fact that there is a significant difference of miRNA expression between serrated and conventional pathway in colorectal carcinogenesis.
The immunopathology of CRC attracted growing attention since an association between gut dysbiosis and colorectal carcinogenesis was suggested by recent authors. miRNAs are putative regulators of several post-transcription target genes and are thought to play essential role in differentiation and proliferation of immune cells. It is assumed that, different miRNA profile pattern may contribute to alterations in gut immunity and dysbiosis, leading to transition events of NM to adenoma.
The specific miRNA expression in serrated pathway, could be useful tool to find appropriate diagnostic, prognostic and treatment response markers in clinical practice. Thus, in order to understand the real significance of miRNAs in this clinical setting, further studies must be conducted.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Bulgaria
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: De Palma FDE S-Editor: Huang P L-Editor: A P-Editor: Liu JH
| 1. | Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-E386. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20108] [Cited by in F6Publishing: 19762] [Article Influence: 2195.8] [Reference Citation Analysis (17)] |
| 2. | Rawla P, Sunkara T, Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz Gastroenterol. 2019;14:89-103. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 444] [Cited by in F6Publishing: 801] [Article Influence: 160.2] [Reference Citation Analysis (0)] |
| 3. | Farooqi AA, de la Roche M, Djamgoz MBA, Siddik ZH. Overview of the oncogenic signaling pathways in colorectal cancer: Mechanistic insights. Semin Cancer Biol. 2019;58:65-79. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 79] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 4. | Jass JR, Smith M. Sialic acid and epithelial differentiation in colorectal polyps and cancer--a morphological, mucin and lectin histochemical study. Pathology. 1992;24:233-242. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 76] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 5. | O'Brien MJ, Yang S, Clebanoff JL, Mulcahy E, Farraye FA, Amorosino M, Swan N. Hyperplastic (serrated) polyps of the colorectum: relationship of CpG island methylator phenotype and K-ras mutation to location and histologic subtype. Am J Surg Pathol. 2004;28:423-434. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 143] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 6. | Mäkinen MJ. Colorectal serrated adenocarcinoma. Histopathology. 2007;50:131-150. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 214] [Cited by in F6Publishing: 196] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 7. | Bartley AN, Yao H, Barkoh BA, Ivan C, Mishra BM, Rashid A, Calin GA, Luthra R, Hamilton SR. Complex patterns of altered MicroRNA expression during the adenoma-adenocarcinoma sequence for microsatellite-stable colorectal cancer. Clin Cancer Res. 2011;17:7283-7293. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 63] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 8. | Schee K, Fodstad Ø, Flatmark K. MicroRNAs as biomarkers in colorectal cancer. Am J Pathol. 2010;177:1592-1599. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 9. | Wong K, Xie G. Updates on the Molecular Genetics of Colorectal Cancer. Colorec Cancer. 2017;3:1. [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 10. | Kondelova A, Alburquerque-González B, Vychytilova-Faltejskova P, García-Solano J, Prochazka V, Kala Z, Pérez F, Slaby O, Conesa-Zamora P. miR-181a-2* expression is different amongst carcinomas from the colorectal serrated route. Mutagenesis. 2020;35:233-241. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 11. | Kanth P, Hazel MW, Boucher KM, Yang Z, Wang L, Bronner MP, Boylan KE, Burt RW, Westover M, Neklason DW, Delker DA. Small RNA sequencing of sessile serrated polyps identifies microRNA profile associated with colon cancer. Genes Chromosomes Cancer. 2019;58:23-33. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 12. | Satorres C, García-Campos M, Bustamante-Balén M. Molecular Features of the Serrated Pathway to Colorectal Cancer: Current Knowledge and Future Directions. Gut Liver. 2020;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 13. | Zhang XT, Zhang QW, Liu F, Lin XL, Chen JN, Li XB. Endoscopic features of sessile serrated adenoma/polyps under narrowband imaging: A retrospective study. J Dig Dis. 2019;20:135-142. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 14. | Okugawa Y, Grady WM, Goel A. Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology 2015; 149: 1204-1225. e12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 424] [Cited by in F6Publishing: 484] [Article Influence: 53.8] [Reference Citation Analysis (1)] |
| 15. | Slattery ML, Herrick JS, Wolff RK, Mullany LE, Stevens JR, Samowitz W. The miRNA landscape of colorectal polyps. Genes Chromosomes Cancer. 2017;56:347-353. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 16. | Ito M, Mitsuhashi K, Igarashi H, Nosho K, Naito T, Yoshii S, Takahashi H, Fujita M, Sukawa Y, Yamamoto E, Takahashi T, Adachi Y, Nojima M, Sasaki Y, Tokino T, Baba Y, Maruyama R, Suzuki H, Imai K, Yamamoto H, Shinomura Y. MicroRNA-31 expression in relation to BRAF mutation, CpG island methylation and colorectal continuum in serrated lesions. Int J Cancer. 2014;135:2507-2515. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 17. | Leggett B, Whitehall V. Role of the serrated pathway in colorectal cancer pathogenesis. Gastroenterology. 2010;138:2088-2100. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 677] [Cited by in F6Publishing: 678] [Article Influence: 48.4] [Reference Citation Analysis (1)] |
| 18. | Snover DC. Update on the serrated pathway to colorectal carcinoma. Hum Pathol. 2011;42:1-10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 439] [Cited by in F6Publishing: 451] [Article Influence: 32.2] [Reference Citation Analysis (0)] |
| 19. | O'Brien MJ, Zhao Q, Yang S. Colorectal serrated pathway cancers and precursors. Histopathology. 2015;66:49-65. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 76] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 20. | Bettington M, Walker N, Clouston A, Brown I, Leggett B, Whitehall V. The serrated pathway to colorectal carcinoma: current concepts and challenges. Histopathology. 2013;62:367-386. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 309] [Cited by in F6Publishing: 323] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 21. | Nagtegaal ID, Odze RD, Klimstra D, Paradis V, Rugge M, Schirmacher P, Washington KM, Carneiro F, Cree IA; WHO Classification of Tumours Editorial Board. The 2019 WHO classification of tumours of the digestive system. Histopathology. 2020;76:182-188. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1833] [Cited by in F6Publishing: 1673] [Article Influence: 418.3] [Reference Citation Analysis (2)] |
| 22. | Fan C, Younis A, Bookhout CE, Crockett SD. Management of Serrated Polyps of the Colon. Curr Treat Options Gastroenterol. 2018;16:182-202. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 23. | Yang S, Farraye FA, Mack C, Posnik O, O'Brien MJ. BRAF and KRAS Mutations in hyperplastic polyps and serrated adenomas of the colorectum: relationship to histology and CpG island methylation status. Am J Surg Pathol. 2004;28:1452-1459. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 230] [Cited by in F6Publishing: 237] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 24. | Hawkins NJ, Bariol C, Ward RL. The serrated neoplasia pathway. Pathology. 2002;34:548-555. [PubMed] [Cited in This Article: ] |
| 25. | Cao H, He N, Song S, Xu M, Piao M, Yan F, Wang B. Is surveillance colonoscopy necessary for patients with sporadic gastric hyperplastic polyps? PLoS One. 2015;10:e0122996. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 26. | Goldstein NS, Bhanot P, Odish E, Hunter S. Hyperplastic-like colon polyps that preceded microsatellite-unstable adenocarcinomas. Am J Clin Pathol. 2003;119:778-796. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 54] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 27. | Longacre TA, Fenoglio-Preiser CM. Mixed hyperplastic adenomatous polyps/serrated adenomas. A distinct form of colorectal neoplasia. Am J Surg Pathol. 1990;14:524-537. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 448] [Cited by in F6Publishing: 412] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 28. | Torlakovic E, Skovlund E, Snover DC, Torlakovic G, Nesland JM. Morphologic reappraisal of serrated colorectal polyps. Am J Surg Pathol. 2003;27:65-81. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 446] [Cited by in F6Publishing: 480] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 29. | Rosenberg DW, Yang S, Pleau DC, Greenspan EJ, Stevens RG, Rajan TV, Heinen CD, Levine J, Zhou Y, O'Brien MJ. Mutations in BRAF and KRAS differentially distinguish serrated versus non-serrated hyperplastic aberrant crypt foci in humans. Cancer Res. 2007;67:3551-3554. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 145] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 30. | Carr NJ, Mahajan H, Tan KL, Hawkins NJ, Ward RL. Serrated and non-serrated polyps of the colorectum: their prevalence in an unselected case series and correlation of BRAF mutation analysis with the diagnosis of sessile serrated adenoma. J Clin Pathol. 2009;62:516-518. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 154] [Cited by in F6Publishing: 158] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 31. | Spring KJ, Zhao ZZ, Karamatic R, Walsh MD, Whitehall VL, Pike T, Simms LA, Young J, James M, Montgomery GW, Appleyard M, Hewett D, Togashi K, Jass JR, Leggett BA. High prevalence of sessile serrated adenomas with BRAF mutations: a prospective study of patients undergoing colonoscopy. Gastroenterology. 2006;131:1400-1407. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 411] [Cited by in F6Publishing: 397] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 32. | McCarthy AJ, Serra S, Chetty R. Traditional serrated adenoma: an overview of pathology and emphasis on molecular pathogenesis. BMJ Open Gastroenterol. 2019;6:e000317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 33. | García-Solano J, Pérez-Guillermo M, Conesa-Zamora P, Acosta-Ortega J, Trujillo-Santos J, Cerezuela-Fuentes P, Mäkinen MJ. Clinicopathologic study of 85 colorectal serrated adenocarcinomas: further insights into the full recognition of a new subset of colorectal carcinoma. Hum Pathol. 2010;41:1359-1368. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 76] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 34. | Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999;96:8681-8686. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1834] [Cited by in F6Publishing: 1803] [Article Influence: 72.1] [Reference Citation Analysis (0)] |
| 35. | Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042-2054. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2489] [Cited by in F6Publishing: 2366] [Article Influence: 112.7] [Reference Citation Analysis (0)] |
| 36. | Nagasaka T, Koi M, Kloor M, Gebert J, Vilkin A, Nishida N, Shin SK, Sasamoto H, Tanaka N, Matsubara N, Boland CR, Goel A. Mutations in both KRAS and BRAF may contribute to the methylator phenotype in colon cancer. Gastroenterology 2008; 134: 1950-1960, 1960. e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 98] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 37. | Fernando WC, Miranda MS, Worthley DL, Togashi K, Watters DJ, Leggett BA, Spring KJ. The CIMP Phenotype in BRAF Mutant Serrated Polyps from a Prospective Colonoscopy Patient Cohort. Gastroenterol Res Pract. 2014;2014:374926. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 38. | Ogino S, Kawasaki T, Kirkner GJ, Kraft P, Loda M, Fuchs CS. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 2007;9:305-314. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 266] [Cited by in F6Publishing: 288] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 39. | Advani SM, Advani P, DeSantis SM, Brown D, VonVille HM, Lam M, Loree JM, Mehrvarz Sarshekeh A, Bressler J, Lopez DS, Daniel CR, Swartz MD, Kopetz S. Clinical, Pathological, and Molecular Characteristics of CpG Island Methylator Phenotype in Colorectal Cancer: A Systematic Review and Meta-analysis. Transl Oncol. 2018;11:1188-1201. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 40. | Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643-649. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2881] [Cited by in F6Publishing: 2762] [Article Influence: 106.2] [Reference Citation Analysis (0)] |
| 41. | Kuiper RP, Vissers LE, Venkatachalam R, Bodmer D, Hoenselaar E, Goossens M, Haufe A, Kamping E, Niessen RC, Hogervorst FB, Gille JJ, Redeker B, Tops CM, van Gijn ME, van den Ouweland AM, Rahner N, Steinke V, Kahl P, Holinski-Feder E, Morak M, Kloor M, Stemmler S, Betz B, Hutter P, Bunyan DJ, Syngal S, Culver JO, Graham T, Chan TL, Nagtegaal ID, van Krieken JH, Schackert HK, Hoogerbrugge N, van Kessel AG, Ligtenberg MJ. Recurrence and variability of germline EPCAM deletions in Lynch syndrome. Hum Mutat. 2011;32:407-414. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 110] [Cited by in F6Publishing: 118] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 42. | De Palma FDE, D'Argenio V, Pol J, Kroemer G, Maiuri MC, Salvatore F. The Molecular Hallmarks of the Serrated Pathway in Colorectal Cancer. Cancers (Basel). 2019;11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 88] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 43. | Hashimoto T, Yamashita S, Yoshida H, Taniguchi H, Ushijima T, Yamada T, Saito Y, Ochiai A, Sekine S, Hiraoka N. WNT Pathway Gene Mutations Are Associated With the Presence of Dysplasia in Colorectal Sessile Serrated Adenoma/Polyps. Am J Surg Pathol. 2017;41:1188-1197. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 44. | Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology 2010; 138: 2073-2087. e3. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1290] [Cited by in F6Publishing: 1357] [Article Influence: 96.9] [Reference Citation Analysis (0)] |
| 45. | Murphy KM, Zhang S, Geiger T, Hafez MJ, Bacher J, Berg KD, Eshleman JR. Comparison of the microsatellite instability analysis system and the Bethesda panel for the determination of microsatellite instability in colorectal cancers. J Mol Diagn. 2006;8:305-311. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 191] [Cited by in F6Publishing: 204] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 46. | Kim JH, Bae JM, Cho NY, Kang GH. Distinct features between MLH1-methylated and unmethylated colorectal carcinomas with the CpG island methylator phenotype: implications in the serrated neoplasia pathway. Oncotarget. 2016;7:14095-14111. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 47. | Travaglino A, D'Armiento FP, Cassese G, Campanino MR, Borrelli G, Pignatiello S, Luglio G, Maione F, De Palma GD, D'Armiento M. Clinicopathological factors associated with BRAF-V600E mutation in colorectal serrated adenomas. Histopathology. 2019;75:160-173. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 48. | Sebolt-Leopold JS, Herrera R. Targeting the mitogen-activated protein kinase cascade to treat cancer. Nat Rev Cancer. 2004;4:937-947. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 894] [Cited by in F6Publishing: 918] [Article Influence: 45.9] [Reference Citation Analysis (0)] |
| 49. | Mármol I, Sánchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int J Mol Sci. 2017;18. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 520] [Cited by in F6Publishing: 704] [Article Influence: 100.6] [Reference Citation Analysis (1)] |
| 50. | Tejpar S, Bertagnolli M, Bosman F, Lenz HJ, Garraway L, Waldman F, Warren R, Bild A, Collins-Brennan D, Hahn H, Harkin DP, Kennedy R, Ilyas M, Morreau H, Proutski V, Swanton C, Tomlinson I, Delorenzi M, Fiocca R, Van Cutsem E, Roth A. Prognostic and predictive biomarkers in resected colon cancer: current status and future perspectives for integrating genomics into biomarker discovery. Oncologist. 2010;15:390-404. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 121] [Cited by in F6Publishing: 129] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 51. | Caputo F, Santini C, Bardasi C, Cerma K, Casadei-Gardini A, Spallanzani A, Andrikou K, Cascinu S, Gelsomino F. BRAF-Mutated Colorectal Cancer: Clinical and Molecular Insights. Int J Mol Sci. 2019;20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 77] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 52. | Hughes LA, Khalid-de Bakker CA, Smits KM, van den Brandt PA, Jonkers D, Ahuja N, Herman JG, Weijenberg MP, van Engeland M. The CpG island methylator phenotype in colorectal cancer: progress and problems. Biochim Biophys Acta. 2012;1825:77-85. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 69] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 53. | Jones JC, Renfro LA, Al-Shamsi HO, Schrock AB, Rankin A, Zhang BY, Kasi PM, Voss JS, Leal AD, Sun J, Ross J, Ali SM, Hubbard JM, Kipp BR, McWilliams RR, Kopetz S, Wolff RA, Grothey A. Non-V600 BRAF Mutations Define a Clinically Distinct Molecular Subtype of Metastatic Colorectal Cancer. J Clin Oncol. 2017;35:2624-2630. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 180] [Cited by in F6Publishing: 226] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 54. | Kambara T, Simms LA, Whitehall VL, Spring KJ, Wynter CV, Walsh MD, Barker MA, Arnold S, McGivern A, Matsubara N, Tanaka N, Higuchi T, Young J, Jass JR, Leggett BA. BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut. 2004;53:1137-1144. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 536] [Cited by in F6Publishing: 571] [Article Influence: 28.6] [Reference Citation Analysis (1)] |
| 55. | Bond CE, Whitehall VLJ. How the BRAF V600E Mutation Defines a Distinct Subgroup of Colorectal Cancer: Molecular and Clinical Implications. Gastroenterol Res Pract. 2018;2018:9250757. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 56. | Sinicrope FA, Shi Q, Smyrk TC, Thibodeau SN, Dienstmann R, Guinney J, Bot BM, Tejpar S, Delorenzi M, Goldberg RM, Mahoney M, Sargent DJ, Alberts SR. Molecular markers identify subtypes of stage III colon cancer associated with patient outcomes. Gastroenterology. 2015;148:88-99. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 227] [Cited by in F6Publishing: 239] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 57. | O'Brien MJ, Yang S, Mack C, Xu H, Huang CS, Mulcahy E, Amorosino M, Farraye FA. Comparison of microsatellite instability, CpG island methylation phenotype, BRAF and KRAS status in serrated polyps and traditional adenomas indicates separate pathways to distinct colorectal carcinoma end points. Am J Surg Pathol. 2006;30:1491-1501. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 368] [Cited by in F6Publishing: 399] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 58. | Yamane L, Scapulatempo-Neto C, Reis RM, Guimarães DP. Serrated pathway in colorectal carcinogenesis. World J Gastroenterol. 2014;20:2634-2640. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 68] [Cited by in F6Publishing: 66] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 59. | Bettington ML, Walker NI, Rosty C, Brown IS, Clouston AD, McKeone DM, Pearson SA, Klein K, Leggett BA, Whitehall VL. A clinicopathological and molecular analysis of 200 traditional serrated adenomas. Mod Pathol. 2015;28:414-427. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 120] [Cited by in F6Publishing: 123] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 60. | Bettington M, Walker N, Rosty C, Brown I, Clouston A, McKeone D, Pearson SA, Leggett B, Whitehall V. Clinicopathological and molecular features of sessile serrated adenomas with dysplasia or carcinoma. Gut. 2017;66:97-106. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 129] [Cited by in F6Publishing: 133] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 61. | Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993; 75:843-854. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8672] [Cited by in F6Publishing: 8408] [Article Influence: 271.2] [Reference Citation Analysis (0)] |
| 62. | To KK, Tong CW, Wu M, Cho WC. MicroRNAs in the prognosis and therapy of colorectal cancer: From bench to bedside. World J Gastroenterol. 2018;24:2949-2973. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 116] [Cited by in F6Publishing: 138] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 63. | Nagel R, le Sage C, Diosdado B, van der Waal M, Oude Vrielink JA, Bolijn A, Meijer GA, Agami R. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008;68:5795-5802. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 341] [Cited by in F6Publishing: 374] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 64. | Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu CG, Calin GA, Croce CM, Harris CC. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425-436. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 966] [Cited by in F6Publishing: 1166] [Article Influence: 72.9] [Reference Citation Analysis (0)] |
| 65. | Balaguer F, Link A, Lozano JJ, Cuatrecasas M, Nagasaka T, Boland CR, Goel A. Epigenetic silencing of miR-137 is an early event in colorectal carcinogenesis. Cancer Res. 2010;70:6609-6618. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 226] [Cited by in F6Publishing: 242] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 66. | Nagy ZB, Wichmann B, Kalmár A, Galamb O, Barták BK, Spisák S, Tulassay Z, Molnár B. Colorectal adenoma and carcinoma specific miRNA profiles in biopsy and their expression in plasma specimens. Clin Epigenetics. 2017;9:22. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 67. | Oberg AL, French AJ, Sarver AL, Subramanian S, Morlan BW, Riska SM, Borralho PM, Cunningham JM, Boardman LA, Wang L, Smyrk TC, Asmann Y, Steer CJ, Thibodeau SN. miRNA expression in colon polyps provides evidence for a multihit model of colon cancer. PLoS One. 2011;6:e20465. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 110] [Cited by in F6Publishing: 123] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 68. | Tsikitis VL, Potter A, Mori M, Buckmeier JA, Preece CR, Harrington CA, Bartley AN, Bhattacharyya AK, Hamilton SR, Lance MP, Thompson PA. MicroRNA Signatures of Colonic Polyps on Screening and Histology. Cancer Prev Res (Phila). 2016;9:942-949. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 69. | Nishida N, Yokobori T, Mimori K, Sudo T, Tanaka F, Shibata K, Ishii H, Doki Y, Kuwano H, Mori M. MicroRNA miR-125b is a prognostic marker in human colorectal cancer. Int J Oncol. 2011;38:1437-1443. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 65] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 70. | Aoki H, Nosho K, Igarashi H, Ito M, Mitsuhashi K, Naito T, Yamamoto E, Tanuma T, Nomura M, Maguchi H, Shinohara T, Suzuki H, Yamamoto H, Shinomura Y. MicroRNA-31 expression in colorectal serrated pathway progression. World J Gastroenterol. 2014;20:12346-12349. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 6] [Cited by in F6Publishing: 6] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 71. | Nosho K, Igarashi H, Nojima M, Ito M, Maruyama R, Yoshii S, Naito T, Sukawa Y, Mikami M, Sumioka W, Yamamoto E, Kurokawa S, Adachi Y, Takahashi H, Okuda H, Kusumi T, Hosokawa M, Fujita M, Hasegawa T, Okita K, Hirata K, Suzuki H, Yamamoto H, Shinomura Y. Association of microRNA-31 with BRAF mutation, colorectal cancer survival and serrated pathway. Carcinogenesis. 2014;35:776-783. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 83] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 72. | Yu T, Ma P, Wu D, Shu Y, Gao W. Functions and mechanisms of microRNA-31 in human cancers. Biomed Pharmacother. 2018;108:1162-1169. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 73. | Strubberg AM, Madison BB. MicroRNAs in the etiology of colorectal cancer: pathways and clinical implications. Dis Model Mech. 2017;10:197-214. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 98] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 74. | Cekaite L, Rantala JK, Bruun J, Guriby M, Agesen TH, Danielsen SA, Lind GE, Nesbakken A, Kallioniemi O, Lothe RA, Skotheim RI. MiR-9, -31, and -182 deregulation promote proliferation and tumor cell survival in colon cancer. Neoplasia. 2012;14:868-879. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 117] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 75. | Cottonham CL, Kaneko S, Xu L. miR-21 and miR-31 converge on TIAM1 to regulate migration and invasion of colon carcinoma cells. J Biol Chem. 2010;285:35293-35302. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 194] [Cited by in F6Publishing: 212] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 76. | Kubota N, Taniguchi F, Nyuya A, Umeda Y, Mori Y, Fujiwara T, Tanioka H, Tsuruta A, Yamaguchi Y, Nagasaka T. Upregulation of microRNA-31 is associated with poor prognosis in patients with advanced colorectal cancer. Oncol Lett. 2020;19:2685-2694. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 77. | Igarashi H, Kurihara H, Mitsuhashi K, Ito M, Okuda H, Kanno S, Naito T, Yoshii S, Takahashi H, Kusumi T, Hasegawa T, Sukawa Y, Adachi Y, Okita K, Hirata K, Imamura Y, Baba Y, Imai K, Suzuki H, Yamamoto H, Nosho K, Shinomura Y. Association of MicroRNA-31-5p with Clinical Efficacy of Anti-EGFR Therapy in Patients with Metastatic Colorectal Cancer. Ann Surg Oncol. 2015;22:2640-2648. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 78. | Yang MH, Yu J, Chen N, Wang XY, Liu XY, Wang S, Ding YQ. Elevated microRNA-31 expression regulates colorectal cancer progression by repressing its target gene SATB2. PLoS One. 2013;8:e85353. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 76] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 79. | Yang X, Xu X, Zhu J, Zhang S, Wu Y, Wu Y, Zhao K, Xing C, Cao J, Zhu H, Li M, Ye Z, Peng W. miR-31 affects colorectal cancer cells by inhibiting autophagy in cancer-associated fibroblasts. Oncotarget. 2016;7:79617-79628. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 53] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 80. | Valeri N, Braconi C, Gasparini P, Murgia C, Lampis A, Paulus-Hock V, Hart JR, Ueno L, Grivennikov SI, Lovat F, Paone A, Cascione L, Sumani KM, Veronese A, Fabbri M, Carasi S, Alder H, Lanza G, Gafa' R, Moyer MP, Ridgway RA, Cordero J, Nuovo GJ, Frankel WL, Rugge M, Fassan M, Groden J, Vogt PK, Karin M, Sansom OJ, Croce CM. MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell. 2014;25:469-483. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 231] [Cited by in F6Publishing: 235] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 81. | Schee K, Boye K, Abrahamsen TW, Fodstad Ø, Flatmark K. Clinical relevance of microRNA miR-21, miR-31, miR-92a, miR-101, miR-106a and miR-145 in colorectal cancer. BMC Cancer. 2012;12:505. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 113] [Cited by in F6Publishing: 132] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 82. | Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128-2136. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1350] [Cited by in F6Publishing: 1417] [Article Influence: 83.4] [Reference Citation Analysis (0)] |
| 83. | Xiong B, Cheng Y, Ma L, Zhang C. MiR-21 regulates biological behavior through the PTEN/PI-3 K/Akt signaling pathway in human colorectal cancer cells. Int J Oncol. 2013;42:219-228. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 146] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 84. | Wu Y, Song Y, Xiong Y, Wang X, Xu K, Han B, Bai Y, Li L, Zhang Y, Zhou L. MicroRNA-21 (Mir-21) Promotes Cell Growth and Invasion by Repressing Tumor Suppressor PTEN in Colorectal Cancer. Cell Physiol Biochem. 2017;43:945-958. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 120] [Cited by in F6Publishing: 126] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 85. | Ghareib AF, Mohamed RH, Abd El-Fatah AR, Saadawy SF. Assessment of Serum MicroRNA-21 Gene Expression for Diagnosis and Prognosis of Colorectal Cancer. J Gastrointest Cancer. 2020;51:818-823. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 86. | Chen Z, Liu H, Jin W, Ding Z, Zheng S, Yu Y. Tissue microRNA-21 expression predicted recurrence and poor survival in patients with colorectal cancer - a meta-analysis. Onco Targets Ther. 2016;9:2615-2624. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 87. | Yau TO, Tang CM, Harriss EK, Dickins B, Polytarchou C. Faecal microRNAs as a non-invasive tool in the diagnosis of colonic adenomas and colorectal cancer: A meta-analysis. Sci Rep. 2019;9:9491. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 88. | Ding L, Lan Z, Xiong X, Ao H, Feng Y, Gu H, Yu M, Cui Q. The Dual Role of MicroRNAs in Colorectal Cancer Progression. Int J Mol Sci. 2018;19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 79] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 89. | Schmitz KJ, Hey S, Schinwald A, Wohlschlaeger J, Baba HA, Worm K, Schmid KW. Differential expression of microRNA 181b and microRNA 21 in hyperplastic polyps and sessile serrated adenomas of the colon. Virchows Arch. 2009;455:49-54. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 27] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 90. | Xi Y, Formentini A, Chien M, Weir DB, Russo JJ, Ju J, Kornmann M, Ju J. Prognostic Values of microRNAs in Colorectal Cancer. Biomark Insights. 2006;2:113-121. [PubMed] [Cited in This Article: ] |
| 91. | Han P, Li JW, Zhang BM, Lv JC, Li YM, Gu XY, Yu ZW, Jia YH, Bai XF, Li L, Liu YL, Cui BB. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol Cancer. 2017;16:9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 286] [Cited by in F6Publishing: 371] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 92. | Khan JA, Forouhar F, Tao X, Tong L. Nicotinamide adenine dinucleotide metabolism as an attractive target for drug discovery. Expert Opin Ther Targets. 2007;11:695-705. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 128] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 93. | Sender R, Fuchs S, Milo R. Are We Really Vastly Outnumbered? Cell. 2016;164:337-340. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1022] [Cited by in F6Publishing: 1111] [Article Influence: 138.9] [Reference Citation Analysis (0)] |
| 94. | Gagnière J, Raisch J, Veziant J, Barnich N, Bonnet R, Buc E, Bringer MA, Pezet D, Bonnet M. Gut microbiota imbalance and colorectal cancer. World J Gastroenterol. 2016;22:501-518. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 491] [Cited by in F6Publishing: 475] [Article Influence: 59.4] [Reference Citation Analysis (0)] |
| 95. | Zitvogel L, Ayyoub M, Routy B, Kroemer G. Microbiome and Anticancer Immunosurveillance. Cell. 2016;165:276-287. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 257] [Cited by in F6Publishing: 299] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 96. | Holt RA, Cochrane K. Tumor Potentiating Mechanisms of Fusobacterium nucleatum, A Multifaceted Microbe. Gastroenterology. 2017;152:694-696. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 97. | Shang FM, Liu HL. Fusobacterium nucleatum and colorectal cancer: A review. World J Gastrointest Oncol. 2018;10:71-81. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 177] [Cited by in F6Publishing: 154] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 98. | Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, Neuberg D, Huang K, Guevara F, Nelson T, Chipashvili O, Hagan T, Walker M, Ramachandran A, Diosdado B, Serna G, Mulet N, Landolfi S, Ramon Y Cajal S, Fasani R, Aguirre AJ, Ng K, Élez E, Ogino S, Tabernero J, Fuchs CS, Hahn WC, Nuciforo P, Meyerson M. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358:1443-1448. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 649] [Cited by in F6Publishing: 819] [Article Influence: 117.0] [Reference Citation Analysis (0)] |
| 99. | Jahani-Sherafat S, Alebouyeh M, Moghim S, Ahmadi Amoli H, Ghasemian-Safaei H. Role of gut microbiota in the pathogenesis of colorectal cancer; a review article. Gastroenterol Hepatol Bed Bench. 2018;11:101-109. [PubMed] [Cited in This Article: ] |
| 100. | Peters BA, Dominianni C, Shapiro JA, Church TR, Wu J, Miller G, Yuen E, Freiman H, Lustbader I, Salik J, Friedlander C, Hayes RB, Ahn J. The gut microbiota in conventional and serrated precursors of colorectal cancer. Microbiome. 2016;4:69. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 136] [Cited by in F6Publishing: 149] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 101. | Yoon H, Kim N, Park JH, Kim YS, Lee J, Kim HW, Choi YJ, Shin CM, Park YS, Lee DH, Jung HC. Comparisons of Gut Microbiota Among Healthy Control, Patients With Conventional Adenoma, Sessile Serrated Adenoma, and Colorectal Cancer. J Cancer Prev. 2017;22:108-114. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 102. | Ito M, Kanno S, Nosho K, Sukawa Y, Mitsuhashi K, Kurihara H, Igarashi H, Takahashi T, Tachibana M, Takahashi H, Yoshii S, Takenouchi T, Hasegawa T, Okita K, Hirata K, Maruyama R, Suzuki H, Imai K, Yamamoto H, Shinomura Y. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int J Cancer. 2015;137:1258-1268. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 171] [Cited by in F6Publishing: 205] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 103. | Yu J, Chen Y, Fu X, Zhou X, Peng Y, Shi L, Chen T, Wu Y. Invasive Fusobacterium nucleatum may play a role in the carcinogenesis of proximal colon cancer through the serrated neoplasia pathway. Int J Cancer. 2016;139:1318-1326. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 93] [Cited by in F6Publishing: 106] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 104. | Li X, Nie J, Mei Q, Han WD. MicroRNAs: Novel immunotherapeutic targets in colorectal carcinoma. World J Gastroenterol. 2016;22:5317-5331. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 45] [Cited by in F6Publishing: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 105. | Liu G, Li B. Role of miRNA in transformation from normal tissue to colorectal adenoma and cancer. J Cancer Res Ther. 2019;15:278-285. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 16] [Reference Citation Analysis (0)] |
| 106. | Nakanishi Y, Duran A, L'Hermitte A, Shelton PM, Nakanishi N, Reina-Campos M, Huang J, Soldevila F, Baaten BJG, Tauriello DVF, Castilla EA, Bhangoo MS, Bao F, Sigal D, Diaz-Meco MT, Moscat J. Simultaneous Loss of Both Atypical Protein Kinase C Genes in the Intestinal Epithelium Drives Serrated Intestinal Cancer by Impairing Immunosurveillance. Immunity 2018; 49: 1132-1147. e7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 107. | Takeuchi Y, Nishikawa H. Roles of regulatory T cells in cancer immunity. Int Immunol. 2016;28:401-409. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 287] [Cited by in F6Publishing: 351] [Article Influence: 43.9] [Reference Citation Analysis (1)] |
| 108. | Kitamura H, Ohno Y, Toyoshima Y, Ohtake J, Homma S, Kawamura H, Takahashi N, Taketomi A. Interleukin-6/STAT3 signaling as a promising target to improve the efficacy of cancer immunotherapy. Cancer Sci. 2017;108:1947-1952. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 179] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 109. | Velikova TV, Miteva L, Stanilov N, Spassova Z, Stanilova SA. Interleukin-6 compared to the other Th17/Treg related cytokines in inflammatory bowel disease and colorectal cancer. World J Gastroenterol. 2020;26:1912-1925. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 27] [Cited by in F6Publishing: 34] [Article Influence: 8.5] [Reference Citation Analysis (1)] |
| 110. | Llosa NJ, Luber B, Tam AJ, Smith KN, Siegel N, Awan AH, Fan H, Oke T, Zhang J, Domingue J, Engle EL, Roberts CA, Bartlett BR, Aulakh LK, Thompson ED, Taube JM, Durham JN, Sears CL, Le DT, Diaz LA, Pardoll DM, Wang H, Anders RA, Housseau F. Intratumoral Adaptive Immunosuppression and Type 17 Immunity in Mismatch Repair Proficient Colorectal Tumors. Clin Cancer Res. 2019;25:5250-5259. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 111. | Yang Y, Alderman C, Sehlaoui A, Xiao Y, Wang W. MicroRNAs as Immunotherapy Targets for Treating Gastroenterological Cancers. Can J Gastroenterol Hepatol. 2018;2018:9740357. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 112. | Liao W, Overman MJ, Boutin AT, Shang X, Zhao D, Dey P, Li J, Wang G, Lan Z, Li J, Tang M, Jiang S, Ma X, Chen P, Katkhuda R, Korphaisarn K, Chakravarti D, Chang A, Spring DJ, Chang Q, Zhang J, Maru DM, Maeda DY, Zebala JA, Kopetz S, Wang YA, DePinho RA. KRAS-IRF2 Axis Drives Immune Suppression and Immune Therapy Resistance in Colorectal Cancer. Cancer Cell 2019; 35: 559-572. e7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 259] [Cited by in F6Publishing: 322] [Article Influence: 64.4] [Reference Citation Analysis (0)] |
| 113. | Rau TT, Atreya R, Aust D, Baretton G, Eck M, Erlenbach-Wünsch K, Hartmann A, Lugli A, Stöhr R, Vieth M, Wirsing AM, Zlobec I, Katzenberger T. Inflammatory response in serrated precursor lesions of the colon classified according to WHO entities, clinical parameters and phenotype-genotype correlation. J Pathol Clin Res. 2016;2:113-124. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 114. | Acosta-Gonzalez G, Ouseph M, Lombardo K, Lu S, Glickman J, Resnick MB. Immune environment in serrated lesions of the colon: intraepithelial lymphocyte density, PD-1, and PD-L1 expression correlate with serrated neoplasia pathway progression. Hum Pathol. 2019;83:115-123. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 115. | Maby P, Tougeron D, Hamieh M, Mlecnik B, Kora H, Bindea G, Angell HK, Fredriksen T, Elie N, Fauquembergue E, Drouet A, Leprince J, Benichou J, Mauillon J, Le Pessot F, Sesboüé R, Tuech JJ, Sabourin JC, Michel P, Frébourg T, Galon J, Latouche JB. Correlation between Density of CD8+ T-cell Infiltrate in Microsatellite Unstable Colorectal Cancers and Frameshift Mutations: A Rationale for Personalized Immunotherapy. Cancer Res. 2015;75:3446-3455. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 156] [Cited by in F6Publishing: 189] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 116. | García-Solano J, Turpin MC, Torres-Moreno D, Huertas-López F, Tuomisto A, Mäkinen MJ, Conesa A, Conesa-Zamora P. Two histologically colorectal carcinomas subsets from the serrated pathway show different methylome signatures and diagnostic biomarkers. Clin Epigenetics. 2018;10:141. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 117. | Nakanishi Y, Diaz-Meco MT, Moscat J. Serrated Colorectal Cancer: The Road Less Travelled? Trends Cancer. 2019;5:742-754. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |