Published online Jul 21, 2006. doi: 10.3748/wjg.v12.i27.4352
Revised: November 29, 2005
Accepted: January 9, 2006
Published online: July 21, 2006
AIM: To investigate the influences of enteral, parenteral nutrition and probiotics delivered by gut on intestinal microecology, epithelial tight junctions, immune and barrier function of rats with abdominal infection.
METHODS: Rat abdominal infection models established with cecal ligation and perforation method, were divided into three groups: parenteral nutrition (PN group, n = 7), PN+enteral nutrition (EN group, n = 7) and PN + EN + probiotics (probiotics group, n = 7) via the needle jejunostomy and neck vein for five days. The total nutritional supplement of the three groups was isonitrogenic and isocaloric. Probiotics was delivered by jejunostomy 10 mL/d (1 × 108 cfu/mL). The rats were killed on the sixth day. The feces in the cecum were cultured for anaerobic bacterial growth and analyzed with bacterial group DNA fingerprint profile with random amplified polymorphic DNA. The transmembrane binding proteins (occludin) and IgA level in plasma cells of intestine epithelium in colon and terminal ileum were measured by an immunohistochemistry method. The ultrastructure of intestinal epithelial tight junctions in colon and small intestine was observed by electron-microscopy. Vena cava blood and the homogenated tissue of liver, lung and mesenteric lymph nodes were cultured to determine the bacterial translocations, and endotoxin in the blood from portal vein was detected.
RESULTS: (1) The amount of bacteria of gut species in EN group and probiotic group was higher than that in PN group. The DNA-profiles in EN group and probiotic group were similar to that of normal rats. The number of DNA-profiles in probiotics group was much more than that in PN group and EN group. Moreover, there were strange stripes in PN group. (2) The expression of occludin and IgA in the small and large intestine in EN group (2.309 ± 0.336, 15.440 ± 2.383) and probiotic group (2.938 ± 0.515, 16.230 ± 3.183) was improved as compared with PN group (1.207 ± 0.587, P < 0.05, 11.189 ± 2.108, P < 0.01). The expression of occludin in probiotic group (intestine: 2.93 ± 0.515; cecum: 3.40 ± 0.617) was higher than that in EN group (intestine: 2.309 ± 0.336; cecum: 2.076 ± 0.670; P < 0.05). The expression of IgA, especially in EN group (intestine: 15.440 ± 2.383) and probiotic EN group (large intestine: 12.516 ± 1.542) significantly increased as compared with PN group (intestine: 11.189 ± 2.108; cecum: 10.160 ± 1.643; P < 0.01). The intestinal epithelial tight junctions and microvilli of the probiotic group were more intact than those in the PN group. (3) The bacterial translocations in blood, liver, lung and mesenteric lymph nodes, and the levels of endotoxin were significantly reduced in probiotic (0.082 ± 0.029) and EN (0.125 ± 0.040) groups as compared with PN group (0.403 ± 0.181, P < 0.05).
CONCLUSION: Application of EN combined with probiotics could improve the expression of transmembrane binding proteins (occludin) and IgA, correct the intestinal flora disturbance, maintain gut barrier functions and tight junctions, and reduce the occurrence of gut bacterial translocation.
- Citation: Shen TY, Qin HL, Gao ZG, Fan XB, Hang XM, Jiang YQ. Influences of enteral nutrition combined with probiotics on gut microflora and barrier function of rats with abdominal infection. World J Gastroenterol 2006; 12(27): 4352-4358
- URL: https://www.wjgnet.com/1007-9327/full/v12/i27/4352.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i27.4352
Since Deitch[1] put forward the concept of bacterial translocation in the 1980s, it has been commonly accepted that bacteria and endotoxin translocations are always associated with the destruction of gut barrier, which could also cause systemic infection reactive syndrome (SIRS), pyemia, shock, and multi-organs dysfunction syndrome (MODS), and increase the death rate of patients with severe illness. Hence, the mechanism of bacterial translocation and the protective and therapeutic agents, as well as the relation between gut original infection and bacteria translocation and their influence on intestinal function have attracted much clinical attention. Though development of new antibiotics has made much progress, the death rate in MODS is still quite high. Under such circumstances, some scholars proposed that the therapy of infection should be directed at its “source’’ not its “target”[2]. And methods such as taking the unabsorbered antibiotics orally, implementing enteral nutrition at early stage, and decontaminating the intestine were used to help recover the physiology state of intestinal tract, to reduce the possibility of gut microflora disorder and bacterial translocation and maintain normal gut function.
In recent years, researchers have tried to improve the gut microbial and bacterial translocation by probiotics[3,4]. However, the thorough and detailed mechanisms are lacking. The present study aimed to determine the effect of probiotics on the ultrastructure of tight junctions, transmembrane protein-occludin expression, enterocyte IgA levels, gut mucus form, gut microflora and bacterial translocation by parenteral nutrition (PN), enteral nutrition (EN) and enteral nutrition combined with probiotics supplied to rats with abdominal infection. It will provide experimental evidence for using probiotics in clinical therapy of abdominal infections.
Fifty-eight SD rats weighing 250-320 g (Fudan University Medical Animal Center, Shanghai) were anesthetized with intraperitoneal injections of 2% saline-ketamin injection liquid (30 mg/g weight). The rats were fixed in a supine position and prepared in a sterile manner for catheterization and operation. A silastic catheter (0.6 mm in inner diameter, 1.0 mm in outer diameter) was inserted through the external jugular vein into the superior vena cava. The catheter was tunneled subcutaneously to the midscapular region and guarded by the flexible spring, and then hooked up to an infusion pump (B. Braun). The abdominal infection model was established by cecum perforation and ligation (CPL)[5], here one third of the cecum distal was ligated and perforated with holes through which a piece of rubber patch was placed and the pinhead hole (#12) fixed. A silastic catheter (1.2 mm in diameter) was placed by jejunostomy 30-40 cm away from the cecum, and the catheter was tunneled abdominally subcutaneously to the back. The rats were maintained in individual metabolic cages[6]. The silastic catheters of jejunum and neck vein were traversed together through the spring and linked to the infusion pump on the circumrotate equipment[7]. The PN and EN nutrient solution was infused at a constant infusion rate by 2 pumps for 5 d during the experimental period.
After establishment of model, all rats received intravenous 0.9% saline solution at 2.0 mL/h for 24 h. Rats were divided into three groups, PN group (n = 7): supported by PN; PN + EN group (EN group, n = 7): supported by PN + EN(peptide), EN was dripped through jejunum tube; PN + EN + probiotic group (probiotic group, n = 7): supported by PN + EN(peptide), the probiotics was infused by jejunostomy tube at about 10 mL/d. The 3 groups were isonitrogenic and isocaloric (Table 1) during 1-5 d. In the PN group, 100% calorie and nitrogen were supplied by the PN solution at the infusion speed of 3.3 mL/h and 80 mL/d. In EN groups, 80% calorie and nitrogen were supplied by the PN solution at an infusion speed of 2.6 mL/h, and 64 mL/d, and the other 20% were supported by EN (Pepti-2000) at 16 mL/d, which were diluted 2:1 to 24 mL with saline solution and the infusion speed maintained at 1 mL/h. In the probiotic group, the probiotics (supported by Shanghai Jiaotong University Limited Company) were added at 10 mL/d. The daily dose of amino acids was 2.5 g of nitrogen per kilogram, the amount of non-protein calories was 348 kJ/d and the total nitrogen was 504 mg/d. The rats were killed and samples taken in the morning of the 6th d.
| Prescription | Dose |
| 50% Glucose (mL) | 33 |
| 8.5% Novamin (mL) | 45 |
| 20% Intralipid (mL) | 17 |
| Soluvita (mL) | 1 |
| Addamel (mL) | 1 |
| 10% KCl (mL) | 1 |
| 10% NaCl (mL) | 1 |
| RI (U) | 4 |
| Heparin (U) | 40 |
| Non-protein calorie (kJ) | 348 |
| Total nitrogen (mg) | 504 |
Probiotics containing live lactobacillus plantarum (activity 1 × 108 cfu/mL) was infused at about 10 mL/d by jejunostomy tube twice per day through 1-5 d.
The fecal sample (0.1 g) in cecum was placed in 9 mL Ringer dilution solution(which contained cysteine) and incubated anaerobically at 36 ± 1°C supplemented with brucella blood agar (2 d, total anaerobic CFU), bacteroides bile esculin agar (2 d), egg yolk agar (after equal volumes of 95% ethanol were added to the dilutions and the preparations stood for 30 min to select for clostridial spores), and Rogosa SL agar (Difco) (2 d for lactobacilli and 4 d for bifidobacteria, after Lactobacillus colonies were marked at 2 d). The dilutions were removed from the anaerobic glove box and inoculated (100 μL inocula) into plates which contained the following media: supplementary brucella blood agar (2 d, total aerobic CFU), MacConkey agar (Difco) (1 d, enterobacteria), bile esculin azide agar (Difco) (1 d, Cl Perfrigens) and were incubated aerobically at 37°C. To analyze the total Lactobacillus population, 10 colonies were picked randomly from a dilution agar plate containing about 100 colonies. The bacterial colonies were counted and identified by Microscan Autoscan-4 Machine (Dade Behringcom).
The fecal samples in cecum from each subject were also examined by PCR-denaturing gradient gel electrophoresis (DGGE) profiles. To extract bacterial DNA, 1 mL of fecal homogenate in pH 7.0 phosphate buffer (the buffer used for the azoreductase assay) was centrifuged at 14 600 g for 5 min (5°C). DNA was extracted from the resulting pellet with a Fast DNA kit (BIO 101, Vista, CA) by using CLS-TC (a cell lysis solution used for animal tissues and bacteria). The V2-V3 region of the 16S rDNA gene (positions from 339 to 539 in the Escherichia coli gene) of bacteria in the fecal samples was amplified by using primers bacteria ITS PS2 (5’-TG(C/T) ACA CACCGC CCG T-3’), PL2 (5’-GGG T (G/C/T) CCC CAT TC(A/G)G-3’). PCR was performed with 0.2-mL tubes by using a PCR Express thermal cycler (Hybaid, Teddington, UK). Each reaction mixture (50 μL) contained reaction buffer (10 mmol/L [final concentration] Tris-HCl, 2.5 mmol/L [final concentration] MgCl2, 50 mmol/L [final concentration] KCl [pH 8.3]), each deoxynucleoside triphosphate at a concentration of 200 μmol/L, 20 pmoL of each primer, 1 μL of fecal DNA, and 2.5 U of Taq DNA polymerase (Boehringer, Mannheim, Germany). The following amplification program was used: 94°C for 3 min, followed by 30 cycles consisting of 94°C for 30 s, 56°C for 30 s, and 68°C for 60 s, and finally 7 min at 68°C. DGGE was performed by using a DCode universal mutation detection system (Bio-Rad, Richmond, CA) and gels that were 16 cm × 16 cm × 1 mm; 6% polyacrylamide gels were prepared and electrophoresed with 1 × TAE buffer prepared from 50 × TAE buffer (2 mol/L Tris base, 1 mol/L glacial acetic acid, 50 mmol/L EDTA). The denaturing gradient was formed by using two 6% acrylamide (acrylamide/bisacrylamide ratio, 37.5:1) stock solutions (Bio-Rad). The gels contained a 22%-55% gradient of urea and formamide that increased in the direction of electrophoresis. A 100% denaturing solution contained 400 mL/L formamide and 7.0 mol/L urea. Electrophoresis was performed at 130 V (constant voltage) and 60°C for about 4.5 h. Electrophoresis was stopped when a xylene cyanol dye marker reached the bottom of a gel. The gels were stained with an ethidium bromide solution (5 μg/mL) for 20 min, washed with deionized water, and reviewed by UV transillumination[8,9].
The distal ileum and colon tissue were taken to determine the occludin and IgA expression by indirect-immune histochemical fluorescence.
Occludin: The tissue was embedded into the frozen acetone at -20°C.Then it was sliced, and routinely dehydrated. To remove the endogenous peroxidase enzyme activity, the slides were treated with 0.1%-1% H2O2 at 37°C for 5-10 min. Then they were washed with PBS twice, and incubated at room temperature overnight with primary antibody 1:300 (goat anti-rat occludin antibody, Santa Cruz). And then they were washed with PBS 3 times and incubated with the second antibody 1:200 (anti-goat IgG, Santa Cruz) for 30 min, then washed again with PBS 3 times and the formula AB enzyme reagent was added. Then they were washed with PBS again and incubated with peroxidase enzyme for 30 s-10 min. After that, the slides were washed by H2O2, dehydrated and mounted.
IgA: The tissue was embedded into paraffin and sliced, then routinely dewaxed. The slides were washed with PBS 3 times, incubated with 3.3% H2O2 at room temperature for 10-15 min to block the endogenous peroxidase enzyme, then washed with PBS again, and albumin interdiction solution was added for 5 min to reduce the nonspecific background staining. Then slides were washed with PBS, and the first antibody 1:500 (mouse anti-rat IgA antibody, Sigma) was added at 4°C refrigerator overnight. After that slides were washed with PBS again, and second antibody 1:100(anti-mouse antibody, DAKO) labeled with biotin was added. Slides were then washed with PBS again, and visualised with DBA enzyme-substrate illustration system, then dyed again and developed.
The data were analyzed by HPIAS1000 high definition color image manipulation system. Under the same expanding multiple (400), the occludin density was determined per field of vision by light-densimeter (at least five fields per slide).
The gut tissues about 0.5 cm in length were taken from distal ileum and colon 5 cm away from the valva ileocaecalis. One fixation procedure was used for conventional thin-section electron microscopy. The fixation procedure included incubation with OsO4 alone (1% or 2% in phosphate buffer) at 0°C for 30 min. After fixation, the ileum and cecum were washed extensively in Veronal acetate buffer (90 mmol/L, pH 6.0), stained by incubation at 0°C for 60 min in uranyl-magnesium acetate (0.5%) in the same buffer, washed again, dehydrated, and embedded. Thin sections were doubly stained with uranyl acetate and lead nitrate and the change of tight junction and microvilli were observed (2 × 104) using a Philip EM 400 electron microscope.
Blood from vena cava (1 mL), mesenteric lymphatic tissue (5 g), liver and lung tissue, were collected, homogenized and placed into a tube containing cardio-cerebral leachate,incubated at 35°C. After 18-24 h samples were stained with methylene blue-eosin method, and continued to be cultured at 35°C. 18-24 h later, the number of bacterium was counted by Microscan Autoscan-4 machine.
Blood from the portal vein (1-2 mL) was taken into heparin lithium anticoagulant centrifuged at 500 r/min for 5 min. The supernatants were mixed with the limuluslysate reagent to examine the endotoxin level.
Analysis of data was performed by χ2 and Dunnette t test with statistical program SPSS10.0, the experimental data were expressed as mean ± SD of the samples between 3 groups. The level of significance was set at P < 0.05 or P < 0.01.
The CPL model was established successfully in 58 SD rats; 37 rats died, with the total death rate during 6 d of 63.8%. The death rate in PN group (78.7%) was higher than that in probiotic group (41.6%, χ2 = 5.658, P = 0.0174 < 0.05, Table 2) and EN group (46.1%, χ2 = 4.691, P = 0.030 < 0.05, Table 3). Interloop abscess (70%), liver abscess (35%) and lung abscess (20%) were observed in the surviving rats, and the positive rate of the blood culture reached 37%.
| Ingredients | Contents |
| Protein(g) | 19.9 |
| Protein hydrolysate (g) | 19.9 |
| Nitrogen (g) | 2.9 |
| Fat (g) | 4.9 |
| Vegetable oil (g) | 2.45 |
| MCT (g) | 2.45 |
| Linoleic acid | 1.3 |
| Carbohydrates (g) | 93.1 |
| Glucose (g) | 1.8 |
| Maltose (g) | 5.5 |
| Amylose (g) | 84.4 |
| Lactose (g) | < 1.3 |
| Organic acid (g) | 0.1 |
| Minerals (g) | 2.6 |
| Vitamins (g) | 0.4 |
| Nonprotein energy (kJ) | 2090 |
| Protein (En%) | 16 |
| Fat (En%) | 9 |
| Carbohydrates (En%) | 75 |
| Osmotic pressure (mOsm/L) | 410 |
| PH | 6.0 |
In the cecum, the amount of all genus (Enterobacillus, t = -0.276, P = 0.018 < 0.05, Bifidobacteria, t = -2.749, P = 0.017 < 0.05, Lactobacillus, t = -8.033, P = 0.002 < 0.01, Cl Perfrigens, t = -2.317, P = 0.031 < 0.05) of the EN group were more than that of the PN group. In the probiotic group the amount of Enterobacilus (t = -2.30, P = 0.390 < 0.05), lactobacillus (t = -9.935, P = 0.001 < 0.01) and bifidobacteria (t = -5.644, P = 0.002 < 0.01) increased, and the amount of Cl Perfrigens (t = -3.144, P = 0.384 < 0.05) decreased as compared with PN group.
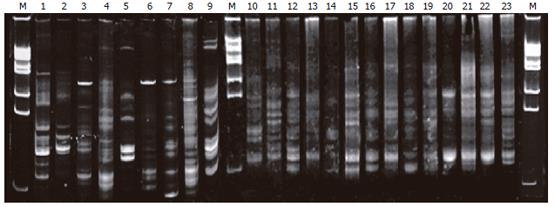
The profiles of DNA sequence in PN group on electro-phoresis (lanes 3-9) were less than that in the normal group (lanes 1-2), EN group (lanes 10-16), and probiotic group (lanes 17-23); lanes 3, 6, 7 presented with a new 16S rDNA sequence in the profile (labeled in Figure 1). The sequence profiles of expression in probiotics and EN groups were similar to that in the normal group (lanes 9-10).
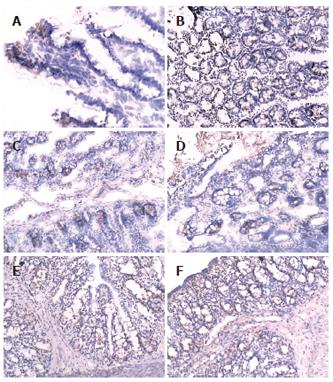
There was more occludin expression in the probiotic group and EN group on the surface, intracellular and inter-cell spaces of the gut epithelial cells as compared with PN group (Figures 2A-F). The occludin positive expression area per measured-window in EN group (intestine, t = -4.955, P = 0.034 < 0.05, cecum, t = -5.407, P = 0.019 < 0.05, Table 4) and probiotics group were higher than that in PN group (intestine, t = -5.426, P = 0.002 < 0.01, cecum, t = -6.112, P = 0.006 < 0.01), and that in the probiotic group was higher than that in EN group(intestine, t = -2.645, P = 0.023 < 0.05, cecum, t = -2.307, P = 0.042 < 0.05).
The expression of IgA in EN group (intestine, t = -4.995, P = 0.003 < 0.01, cecum, t = -2.850, P = 0.014 < 0.05) and probiotic group (intestine, t = -3.575, P = 0.024 < 0.05, cecum, t = -4.946, P = 0.004 < 0.01) was higher than that in PN group, especially the small intestine of the EN group and the cecum of the probiotic group.
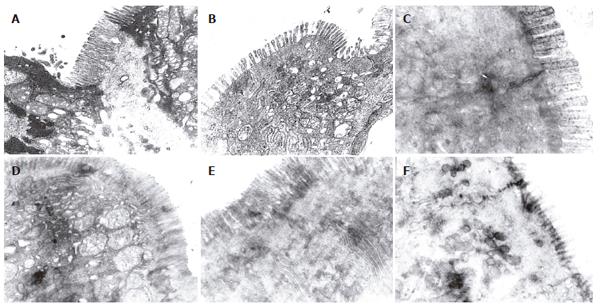
There were more integrated tight junctions, less mito-chondrion endoplasm and microvilli brushed in EN group and probiotics group as compared with PN group (Figures 3A-F). The readability of the tight junction in probiotics group was clearer than that in PN group and EN group.
Bacterium translocation rate (BTR) in PN group (60.7%) was higher than that in probiotic group (28.6%, χ2 = 5.853, P = 0.016 < 0.05, Table 5) and EN group (32.1%, χ2 = 4.595, P = 0.032 < 0.05, Table 5), whereas there was no marked difference of the BTR between the probiotics group and EN group (χ2 = 0.0845, P = 0.7713 > 0.05)
The endotoxin level of the PN group was higher than that in EN group (t = 3.954, P = 0.002 < 0.01 ) and in probiotic group (t = 4.615, P = 0.001 < 0.01).
PN has achieved significant effect as an important therapeutic method for patients with severe illness. However, long-term utility of PN may induce injury of the gut barrier and bacteria translocation. On the other hand, many therapeutic approaches including enteral nutrition and glutamine have been used to improve gut barrier function and positive results have been reported. Nevertheless, they all have certain shortfalls and defects, especially regarding their effect on the gut microbiota[6]. With stress impairments such as wounding, surgery, and infection, not only is the gut barrier disrupted, but normal gut microbiota is also destroyed due to the change in the normal physiology of the gut and the use of antibiotics. The conditional pathogen can predominantly reproduce, disperse and translocate to extra intestinal organs and disseminate throughout the body through blood circulation. As the probiotics are autochthonous habitual bacteria, extra supplement with them could inhibit the potential pathogen’s activity by adhering and colonizing to the intestinal epithelial cell, depressing the pathogenicity of the pathogens, and restoring the normal physiology of the gut. Utility of microorganism regulators such as lactobacillus, bifydobacteria, enterococci and the toxinless, harmless and safe aerophilic bacillus could improve the microflora, reduce the bacterial translocation and protect the gut barrier, and satisfactory results have been reported[7]. Considering the specific effect of the probiotics, Bengmark[10] proposed a new concept of econutrition in 1996, suggesting that. Adding the probiotics to the traditional enteral nutrition, can reduce the excess growth of the pathogen by its antagonist action, improve the environment of the gut by enhancing gut microflora’s fermenting activity, thus maintaining the intestinal function and the gut microbiology, improving the nutritional state and immune capability against disease, reducing the infection rate of the patients with critical illness”.
In the current study, a model of rats with abdominal infection was established to observe the influences of different nutritional pathway and probiotics on the gut ecology and barrier function by CLP method. In traditional fecal anaerobic cultivation, gut microflora disorder was observed and conditional pathogenic bacteria (Cl Perfrigens) increased markedly in PN group. Microflora partly recovered in EN group because of the effect of the EN itself. Both the amount and the species were more than that of the PN group, but the number of conditional pathogen was increased also. However, while the amount of lactobacillus and bifydobacteria increased by adding the probiotic, the Cl Perfrigens were inhibited effectively. Meanwhile the bacteria of normal rats were logarithm mean of the major bacteria: Enterobacillus (5.732), lactobacillus (6.903), bifydobacteria (8.941), Cl Perfrigens (5.132). The amount of gut flora in the probiotic group was closer to that of normal rats, suggesting the relevant intestinal pathogens could be controlled using probiotics. In this study, it was difficult to identify the type or subtype of bacterium and only some local bacteria were determined simultaneously by quantitative analysis because of limited experimental condition. Therefore, the common method of anaerobic cultivation could not reflect the whole profiles of gut preponderant bacterial colonies. Fortunately, molecular biology technology (DNA footprint profile of the microbial gene group) has been used in the analysis of the microbiota in recent years, which could not only quantify the amount of the bacteria, but also reflect changes in community profiles to overcome the shortage in this aspect[8,11]. Our result showed that there were a group of very bright stripes and the number of the stripes in PN group was obviously much more than that in the normal group, EN group and probiotic group, suggesting the gut microflora of the rats only applied with PN were destructed, the species of normal microflora were reduced, and the abnormal intestinal flora increased. The stripes of probiotic group were obviously more than that of the EN group, indicating the species of the rat intestinal microflora increased after adding the probiotics. The 7 stripes in this group were more similar to each other and some specific stripes were identical to the normal group, implying the administration of probiotics was in favor of transferring the rat gut microflora with abdominal infection to the normal state. On the whole, these results suggest early EN can correct the gut microflora disorder caused by PN, and the balance of gut microflora can be improved after adding the probiotics.
Occludin[12-16] is one of the major tight junction structural proteins which determine the intestinal selective barrier function. The measurement of the intestinal epithelial transmembrane junction protein (occludin) could reflect the destruction of the gut tight junctions by the pathogen to some degree. Therefore, in our experiment, the expression of occludin on the enterocyte in distal intestine and the colon tissue were detected. The occludin of the PN group was the least in the 3 groups. After EN and probiotics were added, the occludin increased significantly; occludin level in the probiotic group increased more than that in the other two groups. In order to understand the ultrastructure of the tight junction, the samples were primarily observed by electron microscopy. Compared to the PN group, the tight junction form of the small and large intestine was more intact, the arrangement was tighter and the villi kept more integrity in EN group. And the small and large intestinal epithelial forms in probiotic group were more intact and the gut epithelial tight junctions were clearer than other groups. These results suggested the effect of the probiotics on the gut epithelia, especially the large intestine epithelia, was more significant. With improvement of the gut microflora, the expression of the occludin also increased, and the epithelial tight junction appeared more intact by electron microscopy.
There is increasing evidence that probiotic micro-organisms can interact with the gut associated lymphoid tissue (GALT) and influence mucosal and systemic immune systems[17]. We also studied IgA level of the intestinal tissue. Our result using the EN or probiotics showed that the recovery of the gut IgA expression was more significant than that of the PN group, in the small intestine of EN group and large intestine of probiotic group. Thus using the EN and probiotics could accelerate the expression of occludin and IgA. They could also affect bacterial translocation. After adding the probiotics and/or EN, the death rate, bacterial translocation rates in the blood, mesentery lymph nodes, liver and lung, and the endotoxin level were much lower than the PN group. The death rates and bacterial translocation rates were also decreased in turn.
In conclusion, the mechanism by which EN combined with probiotics improves the gut barrier of the rats with abdominal infection lies in: (1) inhibiting the growth of pathogen and correcting the gut microflora disorder; (2) increasing the expression of occludin and IgA, strengthening the gut epithelial tight junction and the immune state of the local intestine.
S- Editor Wang J L- Editor Zhu LH E- Editor Liu WF
| 1. | Sun JS, Hou SM, Liu TK, Lu KS. Analysis of neogenesis in rabbit skeletal muscles after chronic traction. Histol Histopathol. 1994;9:699-703. [PubMed] [Cited in This Article: ] |
| 2. | Eizaguirre I, Urkia NG, Asensio AB, Zubillaga I, Zubillaga P, Vidales C, Garcia-Arenzana JM, Aldazabal P. Probiotic supplementation reduces the risk of bacterial translocation in experimental short bowel syndrome. J Pediatr Surg. 2002;37:699-702. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 70] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 3. | Oláh A, Belágyi T, Issekutz A, Gamal ME, Bengmark S. Randomized clinical trial of specific lactobacillus and fibre supplement to early enteral nutrition in patients with acute pancreatitis. Br J Surg. 2002;89:1103-1107. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 315] [Cited by in F6Publishing: 286] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 4. | Rayes N, Seehofer D, Müller AR, Hansen S, Bengmark S, Neuhaus P. [Influence of probiotics and fibre on the incidence of bacterial infections following major abdominal surgery - results of a prospective trial]. Z Gastroenterol. 2002;40:869-876. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 59] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 5. | Lidington D, Ouellette Y, Li F, Tyml K. Conducted vasoconstriction is reduced in a mouse model of sepsis. J Vasc Res. 2003;40:149-158. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 27] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 6. | Qin HL, Su ZD, Hu LG, Ding ZX, Lin QT. Effect of early intrajejunal nutrition on pancreatic pathological features and gut barrier function in dogs with acute pancreatitis. Clin Nutr. 2002;21:469-473. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 59] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 7. | García-Martínez J, Acinas SG, Antón AI, Rodríguez-Valera F. Use of the 16S--23S ribosomal genes spacer region in studies of prokaryotic diversity. J Microbiol Methods. 1999;36:55-64. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 176] [Cited by in F6Publishing: 177] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 8. | Hang XM, Yang H, Corne W. [PCR amplification of anaerobic fungal 18S rDNA from landfill sites]. Shengwu Gongcheng Xuebao. 2001;17:515-519. [PubMed] [Cited in This Article: ] |
| 9. | Qin HL, Shen TY, Gao ZG, Fan XB, Hang XM, Jiang YQ, Zhang HZ. Effect of lactobacillus on the gut microflora and barrier function of the rats with abdominal infection. World J Gastroenterol. 2005;11:2591-2596. [PubMed] [Cited in This Article: ] |
| 10. | Bengmark S. Ecological control of the gastrointestinal tract. The role of probiotic flora. Gut. 1998;42:2-7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 285] [Cited by in F6Publishing: 296] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 11. | Tilsala-Timisjärvi A, Alatossava T. Characterization of the 16S-23S and 23S-5S rRNA intergenic spacer regions of dairy propionibacteria and their identification with species-specific primers by PCR. Int J Food Microbiol. 2001;68:45-52. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 29] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 12. | Hirano J, Yoshida T, Sugiyama T, Koide N, Mori I, Yokochi T. The effect of Lactobacillus rhamnosus on enterohemorrhagic Escherichia coli infection of human intestinal cells in vitro. Microbiol Immunol. 2003;47:405-409. [PubMed] [Cited in This Article: ] |
| 13. | Berkes J, Viswanathan VK, Savkovic SD, Hecht G. Intestinal epithelial responses to enteric pathogens: effects on the tight junction barrier, ion transport, and inflammation. Gut. 2003;52:439-451. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 409] [Cited by in F6Publishing: 415] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 14. | Resta-Lenert S, Barrett KE. Live probiotics protect intestinal epithelial cells from the effects of infection with enteroinvasive Escherichia coli (EIEC). Gut. 2003;52:988-997. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 455] [Cited by in F6Publishing: 406] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 15. | Yan F, Polk DB. Probiotic bacterium prevents cytokine-induced apoptosis in intestinal epithelial cells. J Biol Chem. 2002;277:50959-50965. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 369] [Cited by in F6Publishing: 362] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 16. | Michail S, Abernathy F. Lactobacillus plantarum reduces the in vitro secretory response of intestinal epithelial cells to enteropathogenic Escherichia coli infection. J Pediatr Gastroenterol Nutr. 2002;35:350-355. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 61] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 17. | Jenkins B, Holsten S, Bengmark S, Martindale R. Probiotics: a practical review of their role in specific clinical scenarios. Nutr Clin Pract. 2005;20:262-270. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 0.8] [Reference Citation Analysis (0)] |