Published online Jan 14, 2005. doi: 10.3748/wjg.v11.i2.289
Revised: March 18, 2004
Accepted: April 5, 2004
Published online: January 14, 2005
AIM: Codon 72 exon 4 polymorphism (Arg72Pro) of the p53 gene has been implicated in cancer risk. Our objective was to investigate the possible association between p53 Arg72Pro polymorphism and susceptibility to hepatocellular carcinoma (HCC) among Chinese population.
METHODS: The p53 Arg72Pro genotypes were determined by PCR-based restriction fragment length polymorphism (RFLP) analysis in 507 HCC cases and 541 controls. Odds ratios (ORs) for HCC and 95% confidence intervals (CIs) from unconditional logistic regression models were used to evaluate relative risks. Potential risk factors were included in the logistic regression models as covariates in the multivariate analyses on genotype and HCC.
RESULTS: The frequencies for Pro and Arg alleles were 44.5%, 55.5% in HCC cases, and 40.3% and 59.7% in controls, respectively. The Pro allele was significantly associated with the presence of HCC (P = 0.05) and had a higher risk for HCC (OR = 1.19, 95% CI 1.00-1.41) as compared with the Arg allele. After adjusted for potential risk factors, Arg/Pro heterozygotes had an 1.21-fold increased risk (95% CI 0.82-1.78, P = 0.34) of HCC compared with Arg homozygotes, whereas the risk for Pro homozygotes was 1.79 (95% CI 1.06-3.01, P = 0.03) times higher than that for Arg homozygotes. Pro-allele carriers had a higher relative risk of HCC than the Arg-only carriers (adjusted OR = 1.33, 95% CI 0.92-1.92, P = 0.13), although the difference was not statistically significant.
CONCLUSION: Homozygosity for Pro of p53 Arg72Pro is potentially one of the genetic risk factors for HCC in Chinese population. The p53 Arg72Pro polymorphism may be used as a stratification marker in screening individuals at a high risk of HCC.
- Citation: Zhu ZZ, Cong WM, Liu SF, Dong H, Zhu GS, Wu MC. Homozygosity for Pro of p53 Arg72Pro as a potential risk factor for hepatocellular carcinoma in Chinese population. World J Gastroenterol 2005; 11(2): 289-292
- URL: https://www.wjgnet.com/1007-9327/full/v11/i2/289.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i2.289
Hepatocellular carcinoma (HCC) is one of the most common malignant neoplasms worldwide with most cases exhibited in southeast Asia and tropical Africa[1]. In mainland China, the mortality rate of HCC is 337 for men and 123 for women per million people and both rank top in the world[2]. Furthermore, the age-adjusted death rate of HCC is still increasing in both rural and urban areas of mainland China[3].
Etiologically, HCC is a complex and multifactorial disease that is linked to both viral and chemical carcinogens. Major etiologic factors include infection with HBV and HCV, cigarette smoking, alcohol drinking and AFB1 exposure[4-9]. However, not all individuals with exposure to risk factors develop cancer even after a long-term follow-up indicating that susceptibility to HCC is mediated by genetically determined differences. Germline polymorphisms of several genes, most of which encode for xenobiotic metabolizing enzymes (cytochrome P450, glutathione S-transferase, N-acetyltransferase 2, microsomal epoxide hydrolase, and uridine 5’-diphosphate-glucuronosyltransferases)[10-14], have been studied as potential risk factors for HCC. However, the pathogenesis of human HCC is a multistage process with the involvement of a series of genes, including oncogenes and tumor suppressor genes. Germline polymorphisms of these genes may also determine individual susceptibility to HCC.
The p53 tumor suppressor gene is of critical importance for the regulation of cell cycle and maintenance of genomic integrity. Loss of p53 function has been suggested to be a critical step in multistage hepatocarcinogenesis[15]. The wild-type p53 gene exhibits a polymorphism at codon 72 in exon 4, with a single nucleotide change that causes a substitution of proline for arginine (Arg72Pro)[16]. The Arg72Pro polymorphism is located in a proline-rich region (residues 64-92) of the p53 protein, where the 72Pro amino acid constitutes one of five PXXP motifs resembling a SH3 binding domain. The region is required for the growth suppression and apoptosis mediated by p53 but not for cell cycle arrest. The two polymorphic variants of wild-type p53 have been shown to have some different biochemical and biological properties[17]. Since Storey et al (1998) established an association of p53 Arg72Pro with cervical cancer, the p53 polymorphism has been studied as a risk factor in various cancers with inconsistent results[18-29].
To investigate the possible association between p53 Arg72Pro polymorphism and susceptibility to HCC, we conducted a hospital-based case-control study in a large-size sample of Chinese population.
Five hundred and seven cases of HCC and 541 controls were recruited from the Eastern Hepatobiliary Surgery Hospital, Shanghai, China, during the period from February 1999 to May 2003. Controls were subjects with intrahepatic stones (n = 207), cavernous hemangioma (n = 238), and other benign liver diseases (n = 96). Informed consent was obtained from all study subjects. All individuals received surgeries on liver and liver tissues were available, including formalin-fixed and paraffin-embedded archival samples (193 HCC and 291 controls) and formalin-fixed samples (314 HCC and 250 controls), for DNA isolation. For each formalin-fixed and paraffin-embedded archival sample, five 7-μm-thick sections were obtained for microdissection-based extraction of genomic DNA. Two flanking sections, 4 μm, hematoxylin and eosin-stained, were prepared to ensure the composition of histological components. One pathologist (Dr. WM Cong) assessed all patients and assigned non-tumor and tumor areas on 4-μm slides (non-tumor areas were used in analyses). For each formalin-fixed sample, non-tumor liver tissues were obtained within one week after operation, and stored at -80 °C until examination. All pathological diagnoses were reviewed by the same pathologist. Genomic DNA was isolated from the liver tissues using standard phenol-chloroform methods.
Information on age, sex, cigarette smoking, alcohol drinking, HBsAg status, anti-HCV status and family history of HCC in first-degree relatives was obtained from the hospital registration.
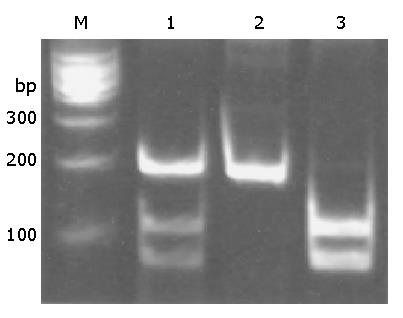
The genotypes of p53 Arg72Pro were determined using PCR-based restriction fragment length polymorphism (RFLP) method. The PCR primers used for amplifying the polymorphism region were: forward, 5’-TTGCCGTCCCAAGCAATGGATGA-3’; reverse, 5’-TCTGGGAAGGGACAGAAGATGAC-3’. PCR condition was 2 min at 94 °C, followed by 35 cycles of 30 s at 94 °C, 30 s at 60 °C, and 30 s at 72 °C, and with a final extension at 72 °C for 7 min. A 10-μL aliquot of PCR product was digested overnight at 60 °C in a 15-μL reaction volume containing 10 units of BstU I (New England BioLabs). After overnight digestion, the fragments were separated by electrophoresis on a vertical 90 g/L non-denaturing polyacrylamide gel at 120 V for 45 min, stained with ethidium bromide. Homozygotes for Pro were represented by a DNA band with the size of 199 bp, whereas Arg homozygotes were represented by DNA bands with sizes of 113 bp and 86 bp. Heterozygotes displayed a combination of both alleles (199, 113, and 86 bp) (Figure 1). Negative and positive controls were assessed during analysis to ensure that PCR products were not contaminated and that the enzyme digestion worked correctly. In addition, laboratory personnel were kept blind as to group status, and the extent of random misclassification was controlled through randomly genotyping 100 samples twice.
Odds ratios (ORs) for HCC and 95% confidence intervals (CIs) from unconditional logistic regression models were used to evaluate relative risks. t-test was used to evaluate the age difference between groups. In the multivariate analyses on genotype and HCC, potential risk factors including sex, age, cigarette smoking, alcohol drinking, HBsAg status, anti-HCV status and family history of HCC in first-degree relatives were included in the logistic regression models as covariates. All the above analyses were performed using SPSS 10.0 software (SPSS, Chicago, IL). Hardy-Weinberg equilibrium (HWE) tests and allele-specific OR were performed using website-based software at http://ihg.gsf.de/ihg/snps.html.
There were a total of 507 HCC cases and 541 controls in this study. The distribution of the p53 Arg72Pro genotypes in HCC cases and controls, and the genotype- and allele-specific ORs for HCC are shown in Table 1. Male, cigarette smoking, HBsAg-positive, anti-HCV-positive and family history of HCC in first-degree relatives were also significant risk factors for developing HCC. No significant association with HCC was found for alcohol drinking. The age (mean±SD) of HCC cases and controls were 50.3±11.6 and 44.7±10.8 years respectively (P<0.001). We have randomly genotyped 100 samples twice and got concordant results.
| Risk factor | Variable | HCC case | Control | Crude | Adjusted | P |
| n (%) | n (%) | OR (95% CI) | OR (95% CI)1 | |||
| p53 Arg72Pro | Arg | 563 (55.5) | 646 (59.7) | 1 | ||
| Pro | 451 (44.5) | 436 (40.3) | 1.19 (1.00-1.41) | 0.05 | ||
| Arg/Arg | 145 (28.6) | 188 (34.8) | 1 | 1 | ||
| Arg/Pro | 273 (53.8) | 270 (49.9) | 1.31 (1.00-1.73) | 1.21 (0.82-1.78) | 0.34 | |
| Pro/Pro | 89 (17.6) | 83 (15.3) | 1.39 (0.96-2.01) | 1.79 (1.06-3.01) | 0.03 | |
| Sex | Female | 73 (14.4) | 278 (51.4) | 1 | 1 | |
| Male | 434 (85.6) | 263 (48.6) | 6.28 (4.66-8.48) | 4.97 (3.28-7.52) | <0.001 | |
| Cigarette smoking | No | 288 (56.8) | 437 (80.8) | 1 | 1 | |
| Yes | 219 (43.2) | 104 (19.2) | 3.20 (2.42-4.21) | 1.90 (1.19-3.02) | <0.01 | |
| Alcohol drinking | No | 304 (60.0) | 446 (82.4) | 1 | 1 | |
| Yes | 203 (40.0) | 95 (17.6) | 3.13 (2.36-4.16) | 1.20 (0.75-1.93) | 0.44 | |
| HBsAg | Negative | 137 (27.0) | 445 (82.3) | 1 | 1 | |
| Positive | 370 (73.0) | 96 (17.7) | 12.52 (9.32-16.82) | 16.83 (11.53-24.56) | <0.001 | |
| Anti-HCV | Negative | 469 (92.5) | 527 (97.4) | 1 | 1 | |
| Positive | 38 (7.5) | 14 (2.6) | 3.05 (1.63-5.69) | 7.29 (3.17-16.80) | <0.001 | |
| Family history2 | No | 433 (85.4) | 532 (98.3) | 1 | 1 | |
| Yes | 74 (14.6) | 9 (1.7) | 10.07 (5.00-20.33) | 9.47 (3.92-22.90) | <0.001 |
The frequencies of the three genotypes were as follows: Arg/Arg 34.8%, Arg/Pro 49.9% and Pro/Pro 15.3% in controls and Arg/Arg 28.6%, Arg/Pro 53.8% and Pro/Pro 17.6% in HCC cases respectively. Based on these data, the frequencies for Pro and Arg alleles were 44.5%, 55.5% in HCC cases, and 40.3% and 59.7% in controls, respectively. The Pro allele was significantly associated with the presence of HCC (P = 0.05) and had a higher risk for HCC (OR = 1.19, 95% CI 1.00-1.41) as compared to the Arg allele. Genotype distributions were in HWE for controls, but not for HCC cases (P = 0.04) with marginally statistical significance. The genotype frequencies in our control group were similar to those reported by other authors in Chinese population[22,30].
No overall association between p53 Arg72Pro genotypes and HCC was observed. However, particular genotypic frequencies were different in HCC cases and controls. After adjusted for potential risk factors, Arg/Pro heterozygotes had an 1.21-fold increased risk of HCC compared to Arg homozygotes, whereas the risk for Pro homozygotes was 1.79 (95% CI 1.06-3.01, P = 0.03) times higher than that for Arg homozygotes. Pro-allele carriers had a higher relative risk of HCC compared to Arg-only carriers, although the difference was not statistically significant.
It is also important to elucidate the frequency of loss of heterozygosity (LOH) in HCC using tissues out of the liver as referencing materials since the surrounding non-cancerous liver tissues used as referencing materials might have already accumulated genetic alterations. Thus, we also examined genotypes of p53 Arg72Pro in blood DNA of a part of our HCC patients homozygous for the Arg or the Pro allele (33 and 21 cases, respectively). No LOH was detected in this locus.
The p53 gene is one of the most extensively studied human genes because of its role as a tumor suppressor gene. Its diverse functions include DNA binding, cell cycle control, DNA repair, differentiation, genomic plasticity, and apoptosis[31]. Thus, the overall function of p53 is to maintain genomic integrity as a whole, providing a protective effect against tumorigenesis. Wild type p53 is polymorphic at codon 72 in human populations and it has been reported that homozygosity for Pro of p53 Arg72Pro is potentially a risk factor for cancers of the lung, esophagus, stomach, breast, nasopharynx, urothelium, and prostate[23-29]. In this study, we used PCR-based RFLP method to analyze the p53 Arg72Pro polymorphism in HCC among the Chinese population.
We found that the Pro allele was significantly associated with the presence of HCC and that carriers of the Pro, or the “risk allele”, had an 1.33-fold increased risk of HCC compared to Arg-only carriers, without statistical significance. Arg/Pro heterozygotes of the p53 polymorphism had an 1.21-fold increased risk of HCC compared to Arg homozygotes. Pro homozygotes had an 1.79-fold increased risk of HCC with statistical significance. This large epidemiological study suggests that homozygosity for Pro of p53 Arg72Pro is potentially one of the genetic risk factors for HCC in Chinese population. The p53 Arg72Pro polymorphism may be used as a stratification marker in screening individuals at a high risk of HCC.
There have been two case-control studies conducted to examine the association between p53 Arg72Pro and HCC[32,33]. A significant association between Pro allele and HCC in HBsAg-positive males with chronic liver diseases or family history of HCC was reported in a Taiwanese case-control study conducted by Yu et al[32]. However, no overall increased frequency of the Pro allele was observed in HCC cases in that study. One possible explanation is that the effect of Pro allele of p53 Arg72Pro polymorphism might be masked by the stronger tumorigenic effect of chronic HBV infection. A recent case-control study performed in Spain by Anzola et al[33] failed to observe any association between p53 Arg72Pro and HCC. The inconsistency in the association could be attributable to ethnic difference, since the study by Anzola et al was performed in a Caucasian population, while the study by Yu et al, as well as the current study, were performed in an Asian population.
The hypothesized relationship of the p53 polymorphism to cancer susceptibility has been unclear. It was reported that these two polymorphic variants of wild type p53 differ in E6-mediated degradation, transcription activation and induction of apoptosis[17]. Recent studies indicated that the Arg allele is preferentially mutated and retained in various human cancers arising in Pro/Arg heterozygotes, and that the p53 mutant is a more potent inhibitor of p73 when p53 has Arg in codon 72 rather than Pro[33-36]. These findings suggest that this polymorphism acts as an intragenic modifier of mutant p53 behavior and has an effect on the biological activity of p53. However, results of other studies show that the polymorphism does not influence protein-protein interaction (p53mut-p53WT or p53mut-p73βWT)[37,38]. Thus, the implications of the p53 polymorphism in cancer development require further study. We must also consider the possibility that the p53 Arg72Pro is simply in a state of linkage disequilibrium (LD) with an as-yet-unidentified functional locus. The highest cancer risk figures have been reported in intronic polymorphisms (MspI RFLPs in intron 6 and a 16-bp duplication in intron 3) in p53 gene suggesting that the mechanism underlying these associations might be a locus which is in LD with p53 Arg72Pro polymorphism rather than direct functional involvement of the Arg/Pro substitution itself[20,21].
The p53 Arg72Pro polymorphism displays a similar association pattern in HCC compared with previously examined esophageal and lung cancer patients from the Chinese population, in which the risk of Pro homozygotes for cancer was about 2 times against Arg homozygotes[22,24,39]. Taken together, there might be, at least in Chinese population, a common genetic basis for the pathogenesis of these different cancers.
In summary, homozygosity for Pro of p53 Arg72Pro is potentially one of the genetic risk factors for HCC in Chinese population. The p53 Arg72Pro polymorphism may be used as a stratification marker in screening individuals at a high risk of HCC.
Assistant Editor Guo SY Edited by Zhang JZ and Ma JY
| 1. | Bosch FX, Ribes J, Borràs J. Epidemiology of primary liver cancer. Semin Liver Dis. 1999;19:271-285. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 671] [Cited by in F6Publishing: 722] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 2. | Zhang S, Li L, Lu F. Mortality of primary liver cancer in China from 1990 through 1992. Zhonghua ZhongLiu ZaZhi. 1999;21:245-249. [PubMed] [Cited in This Article: ] |
| 3. | Li L, Lu F, Zhang S. Analyses of variation trend and short-term detection of Chinese malignant tumor mortality during twenty years. Zhonghua ZhongLiu ZaZhi. 1997;19:3-9. [PubMed] [Cited in This Article: ] |
| 4. | Tang ZY. Hepatocellular carcinoma--cause, treatment and metastasis. World J Gastroenterol. 2001;7:445-454. [PubMed] [Cited in This Article: ] |
| 5. | Zhu ZZ, Cong WM. Roles of hepatitis B virus and hepatitis C virus in hepato-carcinogenesis. Zhonghua GanZangBing ZaZhi. 2003;11:574-576. [PubMed] [Cited in This Article: ] |
| 6. | Kuper H, Tzonou A, Kaklamani E, Hsieh CC, Lagiou P, Adami HO, Trichopoulos D, Stuver SO. Tobacco smoking, alcohol consumption and their interaction in the causation of hepatocellular carcinoma. Int J Cancer. 2000;85:498-502. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 3] [Reference Citation Analysis (0)] |
| 7. | Chen CJ, Wang LY, Lu SN, Wu MH, You SL, Zhang YJ, Wang LW, Santella RM. Elevated aflatoxin exposure and increased risk of hepatocellular carcinoma. Hepatology. 1996;24:38-42. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 107] [Cited by in F6Publishing: 117] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 8. | Ross RK, Yuan JM, Yu MC, Wogan GN, Qian GS, Tu JT, Groopman JD, Gao YT, Henderson BE. Urinary aflatoxin biomarkers and risk of hepatocellular carcinoma. Lancet. 1992;339:943-946. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 469] [Cited by in F6Publishing: 385] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 9. | Qian GS, Ross RK, Yu MC, Yuan JM, Gao YT, Henderson BE, Wogan GN, Groopman JD. A follow-up study of urinary markers of aflatoxin exposure and liver cancer risk in Shanghai, People's Republic of China. Cancer Epidemiol Biomarkers Prev. 1994;3:3-10. [PubMed] [Cited in This Article: ] |
| 10. | Silvestri L, Sonzogni L, De Silvestri A, Gritti C, Foti L, Zavaglia C, Leveri M, Cividini A, Mondelli MU, Civardi E. CYP enzyme polymorphisms and susceptibility to HCV-related chronic liver disease and liver cancer. Int J Cancer. 2003;104:310-317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 55] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 11. | Sun CA, Wang LY, Chen CJ, Lu SN, You SL, Wang LW, Wang Q, Wu DM, Santella RM. Genetic polymorphisms of glutathione S-transferases M1 and T1 associated with susceptibility to aflatoxin-related hepatocarcinogenesis among chronic hepatitis B carriers: a nested case-control study in Taiwan. Carcinogenesis. 2001;22:1289-1294. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 75] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 12. | Huang YS, Chern HD, Wu JC, Chao Y, Huang YH, Chang FY, Lee SD. Polymorphism of the N-acetyltransferase 2 gene, red meat intake, and the susceptibility of hepatocellular carcinoma. Am J Gastroenterol. 2003;98:1417-1422. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 34] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 13. | Sonzogni L, Silvestri L, De Silvestri A, Gritti C, Foti L, Zavaglia C, Bottelli R, Mondelli MU, Civardi E, Silini EM. Polymorphisms of microsomal epoxide hydrolase gene and severity of HCV-related liver disease. Hepatology. 2002;36:195-201. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 49] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 14. | Vogel A, Kneip S, Barut A, Ehmer U, Tukey RH, Manns MP, Strassburg CP. Genetic link of hepatocellular carcinoma with polymorphisms of the UDP-glucuronosyltransferase UGT1A7 gene. Gastroenterology. 2001;121:1136-1144. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 105] [Cited by in F6Publishing: 107] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 15. | Staib F, Hussain SP, Hofseth LJ, Wang XW, Harris CC. TP53 and liver carcinogenesis. Hum Mutat. 2003;21:201-216. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 169] [Cited by in F6Publishing: 185] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 16. | Buchman VL, Chumakov PM, Ninkina NN, Samarina OP, Georgiev GP. A variation in the structure of the protein-coding region of the human p53 gene. Gene. 1988;70:245-252. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 166] [Cited by in F6Publishing: 195] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 17. | Thomas M, Kalita A, Labrecque S, Pim D, Banks L, Matlashewski G. Two polymorphic variants of wild-type p53 differ biochemically and biologically. Mol Cell Biol. 1999;19:1092-1100. [PubMed] [Cited in This Article: ] |
| 18. | Storey A, Thomas M, Kalita A, Harwood C, Gardiol D, Mantovani F, Breuer J, Leigh IM, Matlashewski G, Banks L. Role of a p53 polymorphism in the development of human papillomavirus-associated cancer. Nature. 1998;393:229-234. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 645] [Cited by in F6Publishing: 700] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 19. | Minaguchi T, Kanamori Y, Matsushima M, Yoshikawa H, Taketani Y, Nakamura Y. No evidence of correlation between polymorphism at codon 72 of p53 and risk of cervical cancer in Japanese patients with human papillomavirus 16/18 infection. Cancer Res. 1998;58:4585-4586. [PubMed] [Cited in This Article: ] |
| 20. | Birgander R, Själander A, Rannug A, Alexandrie AK, Sundberg MI, Seidegård J, Tornling G, Beckman G, Beckman L. P53 polymorphisms and haplotypes in lung cancer. Carcinogenesis. 1995;16:2233-2236. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 83] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 21. | Birgander R, Själander A, Zhou Z, Fan C, Beckman L, Beckman G. p53 polymorphisms and haplotypes in nasopharyngeal cancer. Hum Hered. 1996;46:49-54. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 36] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 22. | Zhang JH, Li Y, Wang R, Wen DG, Wu ML, He M. p53 gene polymorphism with susceptibility to esophageal cancer and lung cancer in Chinese population. Zhonghua ZhongLiu ZaZhi. 2003;25:365-367. [PubMed] [Cited in This Article: ] |
| 23. | Irarrázabal CE, Rojas C, Aracena R, Márquez C, Gil L. Chilean pilot study on the risk of lung cancer associated with codon 72 polymorphism in the gene of protein p53. Toxicol Lett. 2003;144:69-76. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 30] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 24. | Lee JM, Lee YC, Yang SY, Shi WL, Lee CJ, Luh SP, Chen CJ, Hsieh CY, Wu MT. Genetic polymorphisms of p53 and GSTP1,but not NAT2,are associated with susceptibility to squamous-cell carcinoma of the esophagus. Int J Cancer. 2000;89:458-464. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 4] [Reference Citation Analysis (0)] |
| 25. | Hiyama T, Tanaka S, Kitadai Y, Ito M, Sumii M, Yoshihara M, Shimamoto F, Haruma K, Chayama K. p53 Codon 72 polymorphism in gastric cancer susceptibility in patients with Helicobacter pylori-associated chronic gastritis. Int J Cancer. 2002;100:304-308. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 56] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 26. | Själander A, Birgander R, Hallmans G, Cajander S, Lenner P, Athlin L, Beckman G, Beckman L. p53 polymorphisms and haplotypes in breast cancer. Carcinogenesis. 1996;17:1313-1316. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 146] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 27. | Tsai MH, Lin CD, Hsieh YY, Chang FC, Tsai FJ, Chen WC, Tsai CH. Prognostic significance of the proline form of p53 codon 72 polymorphism in nasopharyngeal carcinoma. Laryngoscope. 2002;112:116-119. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 39] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 28. | Kuroda Y, Tsukino H, Nakao H, Imai H, Katoh T. p53 Codon 72 polymorphism and urothelial cancer risk. Cancer Lett. 2003;189:77-83. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 42] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 29. | Suzuki K, Matsui H, Ohtake N, Nakata S, Takei T, Nakazato H, Okugi H, Koike H, Ono Y, Ito K. A p53 codon 72 polymorphism associated with prostate cancer development and progression in Japanese. J Biomed Sci. 2003;10:430-435. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 49] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 30. | Beckman G, Birgander R, Själander A, Saha N, Holmberg PA, Kivelä A, Beckman L. Is p53 polymorphism maintained by natural selection? Hum Hered. 1994;44:266-270. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 233] [Cited by in F6Publishing: 245] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 31. | Hofseth LJ, Hussain SP, Harris CC. p53: 25 years after its discovery. Trends Pharmacol Sci. 2004;25:177-181. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 296] [Cited by in F6Publishing: 306] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 32. | Yu MW, Yang SY, Chiu YH, Chiang YC, Liaw YF, Chen CJ. A p53 genetic polymorphism as a modulator of hepatocellular carcinoma risk in relation to chronic liver disease, familial tendency, and cigarette smoking in hepatitis B carriers. Hepatology. 1999;29:697-702. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 101] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 33. | Anzola M, Cuevas N, López-Martínez M, Saiz A, Burgos JJ, de Pancorbo MM. Frequent loss of p53 codon 72 Pro variant in hepatitis C virus-positive carriers with hepatocellular carcinoma. Cancer Lett. 2003;193:199-205. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 36] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 34. | Marin MC, Jost CA, Brooks LA, Irwin MS, O'Nions J, Tidy JA, James N, McGregor JM, Harwood CA, Yulug IG. A common polymorphism acts as an intragenic modifier of mutant p53 behaviour. Nat Genet. 2000;25:47-54. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 366] [Cited by in F6Publishing: 387] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 35. | Tada M, Furuuchi K, Kaneda M, Matsumoto J, Takahashi M, Hirai A, Mitsumoto Y, Iggo RD, Moriuchi T. Inactivate the remaining p53 allele or the alternate p73? Preferential selection of the Arg72 polymorphism in cancers with recessive p53 mutants but not transdominant mutants. Carcinogenesis. 2001;22:515-517. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 55] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 36. | Papadakis ED, Soulitzis N, Spandidos DA. Association of p53 codon 72 polymorphism with advanced lung cancer: the Arg allele is preferentially retained in tumours arising in Arg/Pro germline heterozygotes. Br J Cancer. 2002;87:1013-1018. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 67] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 37. | Monti P, Campomenosi P, Ciribilli Y, Iannone R, Aprile A, Inga A, Tada M, Menichini P, Abbondandolo A, Fronza G. Characterization of the p53 mutants ability to inhibit p73 beta transactivation using a yeast-based functional assay. Oncogene. 2003;22:5252-5260. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 35] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 38. | Gaiddon C, Lokshin M, Ahn J, Zhang T, Prives C. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol Cell Biol. 2001;21:1874-1887. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 344] [Cited by in F6Publishing: 346] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 39. | Zhang L, Xing D, He Z, Lin D. p53 gene codon 72 polymorphism and susceptibility to esophageal squamous cell carcinoma in a Chinese population. Zhonghua YiXue YiChuanXue ZaZhi. 2002;19:10-13. [PubMed] [Cited in This Article: ] |