Published online Mar 21, 2005. doi: 10.3748/wjg.v11.i11.1673
Revised: October 31, 2004
Accepted: December 9, 2004
Published online: March 21, 2005
AIM: To explore the clinicopathological and molecular genetic features of hereditary nonpolyposis colorectal cancer (HNPCC) in Chinese population.
METHODS: We collected 16 Chinese HNPCC families from Wenzhou, Zhejiang Province, China. Tumor tissues and peripheral white blood cells were studied using microdissection, microsatellite analysis, immunostaining of hMSH2 and hMLH1 proteins and direct DNA sequencing of hMSH2 and hMLH1 genes.
RESULTS: (1) A total of 50 patients had CRC. Average age at diagnosis of the first CRC was 45.7 years; 40.9% and 28.7% of the CRCs were located proximal to the splenic flexure and in the rectum, respectively. Thirty-eight percent of the colorectal cancer patients had synchronous and metachronous CRC. 34.4% and 25% of the CRCs were poor differentiation cancer and mucinous adenocarcinoma, respectively. Fourteen extracoloni tumors were found, and the hepatic cancer was the most common tumor type. Twenty-one patients whose median survival time was 5.7 years died during 1-23 years. Twenty-nine patients have survived for 1-28 years, 58.6%, 41.4% and 24.1% patients have survived for more than 5, 10 and 15 years, respectively; (2) All nine tumor-tissues showed microsatellite instability (MSI) at more than two loci. Four tumor-tissues lost hMSH2 protein expression and one lost hMLH1 protein expression. Three pathological germline mutations were identified from five genetically analyzed families; two of three mutations had not been reported previously as they were a transition from C to A in exon 14 (codon 743) of hMSH2 and a TTC deletion in exon 14 (codon 530) of hMLH1.
CONCLUSION: Chinese HNPCC have specific clinicopathological features, such as early onset, propensity to involve the proximal colon, and high frequency of multiple CRCs, liver cancer more frequent than endometrial cancer. Chinese HNPCC showed relatively frequent germline mutation of mismatch repair (MMR) genes that correlated closely with high-level MSI and loss of expression of MMR genes protein.
- Citation: Luo DC, Cai Q, Sun MH, Ni YZ, Ni SC, Chen ZJ, Li XY, Tao CW, Zhang XM, Shi DR. Clinicopathological and molecular genetic analysis of HNPCC in China. World J Gastroenterol 2005; 11(11): 1673-1679
- URL: https://www.wjgnet.com/1007-9327/full/v11/i11/1673.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i11.1673
In 1895, Warthin first described some families with an excess of colorectal, uterine and gastric cancers. In 1960s, Lynch accurately described these cancer-prone families. This condition was first termed the “cancer family syndrome” and was later renamed hereditary nonpolyposis colorectal cancer (HNPCC). According to the absence or presence of extracolonic malignancies, these families were divided into Lynch syndrome I (hereditary site-specific colorectal cancer) and Lynch II syndrome (colorectal cancer in association with extracolonic cancer)[1]; which accounts for 1-10% of the total colorectal cancer population[1-5]. Clinically, it is diagnosed by Amsterdam criteria[6]: (1) three or more relatives with histologically verified colorectal cancer, one of whom is the first-degree to the other two; (2) colorectal cancer affecting at least two generations; and (3) one or more colorectal cancer cases diagnosed before the age of 50. In addition, familial adenomatous polyposis (FAP) must be ruled out. As the criteria are too rigid for small families, they exclude extra-colonic cancers associated with HNPCC. In Asia, on the other hand, the Japan Research Society for Cancer of the Colon and Rectum developed the clinical criteria (Japanese criteria) for HNPCC in 1991[7]. The criteria include A: a case with three or more colorectal cancers within the first-degree relatives; B: a case with two or more colorectal cancers within the first-degree relatives meeting the following criteria: (1) age onset of colorectal cancers being earlier than 50 years old; (2) with right colon involvement; (3) with synchronous or metachronous multiple colorectal cancers; (4) associated with synchronous or metachronous extracolonic malignancies.
Germline mutations of six genes involved in DNA mismatch repair (MMR), i.e., hMSH2, hMLH1, PMS1, PMS2, MSH6 (also known as GTBP) and MLH3, have been identified in patients with the disease, and the former two genes account for the large majority of mutations found in families with HNPCC. Totally, these genes are now believed to account for about 50-70% of all families with HNPCC and over 90% of the identified mutations focused on the two genes, hMSH2 and hMLH1[8-14]. There are many studies about the procedures of genetic testing of HNPCC, such as microsatellite instability (MSI), immunohistochemistry (IHC) and direct DNA sequencing[15-18].
It is of no doubt that there is a large population of HNPCC in China[19]. In 1996, Mo et al[20] first reported the clinical features of HNPCC cases. Until now there have been only some case reports of HNPCC in China and no systemic study of molecular genetic aspects of HNPCC has been presented. In the present study, 16 Chinese HNPCC families are included of which nine families fulfilling the Amsterdam criteria and seven families fulfilling the Japanese criteria B. We conducted clinicopathological and molecular genetic analyses of Chinese HNPCC families.
From 1999 to 2004, 16 Chinese HNPCC families that were registered at the Department of Surgical Oncology in the Second People’s Hospital of Wenzhou and at the Department of Surgery in the First Affiliated Hospital of Wenzhou Medical College were collected, of which nine HNPCC families fulfilled the Amsterdam criteria and seven HNPCC families fulfilled the Japanese criteria B. When the probands were verified, we investigated the more detailed family history of patients in the hospital or through inquiry by telephone, mail or visit. The tree of each family pedigree was drawn. And these HNPCC patients are being followed up.
Tumor tissues of nine identified patients, peripheral white blood cells of probands and members (over the age of 18) in the five genetic analyzed kindreds were collected for the study.
Microdissection and DNA extraction The 7-μm paraffin-embedded sections were deparaffinized. They were lightly stained with hematoxylin for microdissection. The microdissection was performed under the dissection microscope with a scalpel. The microdissected tissues were transferred directly into a centrifugation tube with 150 μL cell lysis buffer (0.5 mol/L Tris, 20 mmol/L EDTA, 10 mmol/L NaCl, 10 g/L SDS, 0.5 g/L proteinase K). The subsequent DNA extraction was performed according to the protocol of the DNA extraction kit (Daxia Biotech Ltd, Shanghai). So was Genomic DNA from peripheral white blood cells.
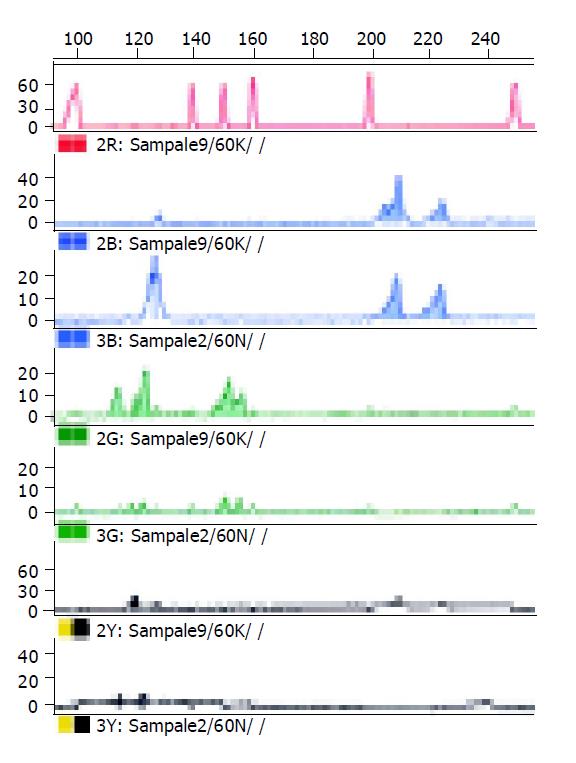
Microsatellite instability analysis Matched normal and tumor DNA were investigated with a panel of five microsatellite markers (including BAT26 and BAT25, D2S123, D5S346 and D17S250) that were recommended by the International Collaborative Group on Hereditary Non-Polyposis Cancer and the National Cancer Institute[21,22]. The primer sequences have been published elsewhere[22]. The primer pairs were synthesized by Shenyou Biotech Ltd. Each forward primer was labeled with a fluorescent dye at 5’ end. The mixture was denatured, snap cooled and electrophoresed on ABI 310 automated DNA sequencer according to the manufacturer’s instructions. The electrophoresis results were analyzed by GeneScan software. MSI was determined according to Gebert et al[23]. Additional peaks (bands) at a microsatellite locus in the tumor compared with the normal tissue from the same patient were interpreted as MSI. Cases with MSI in more than two of the five loci were interpreted as exhibiting high microsatellite instability (MSI-H).
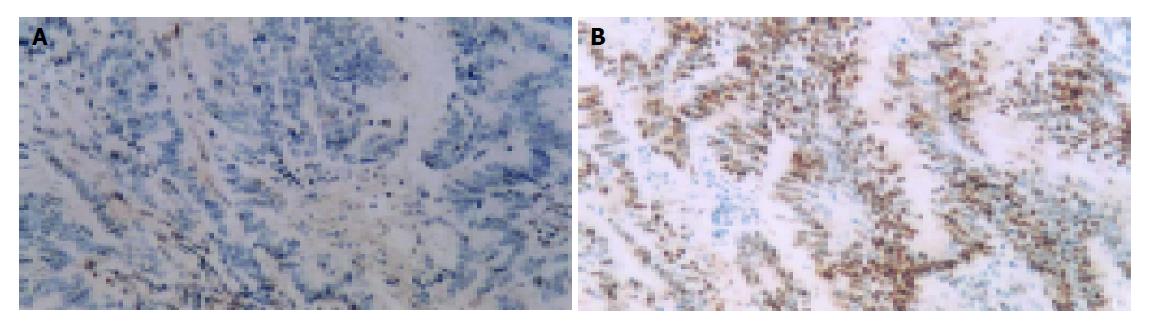
Immunostaining for hMSH2 and hMLH1 Immunostaining was performed using a monoclonal antibody against the hMSH2 (Oncogene Ltd) and a monoclonal antibody against the hMLH1 (Pharmingen Ltd) at 1:40 dilutions. The antibodies were detected by the Envison two-step method. The slides were counter stained with hematoxylin. Infiltrating lymphocytes as well as normal colonic crypt epithelium next to the tumor area served as internal positive controls. Diminished expression of hMSH2 or hMLH1 in cancer tissues were demonstrated when there was complete absence of detectable nuclear staining of neoplastic cells.
Sequencing analysis All 19 exons of hMLH1 gene and all 16 exons of hMSH2 gene (including all intron-exon borders) from proband’s genomic DNA were individually amplified. PCR reaction was set in 25 μL volume containing 100 ng template DNA, dNTPs 0.2 mmol/L, MgCl2 1.5 mmol/L, Taq polymerase 1 U, and then denatured for 5 min at 94 °C, cycled (45 s at 94 °C, 45 s at 55 °C and 45 s at 72 °C) 35 times and extended for 7 min at 72 °C. The PCR products were purified using the QIAquick-spin PCR purification kits (Qiagen Inc.) and then performed on an ABI 310 automated sequencer (ABS Inc.).
From 1999 to 2004, we investigated 16 Chinese HNPCC families of which nine families fulfilled the Amsterdam criteria and seven families fulfilled the Japanese criteria B. A total of 56 patients developed malignant tumors, 50 of whom had CRC. There were 37 male and 13 female, age ranging from 19 to 73 years (average 45.7), 36 patients (72%) developed CRC below 50 years, including 17 patients (34%) under 40 years of age, four patients (8%) under 30 years of age. 40.9% and 28.7% of the colorectal cancers’ loci were located in the colon proximal to spleen flexure and in rectum, respectively. The remaining loci were not clear. Synchronous and metachronous colorectal cancer occurred in 19 (38%) patients. We reviewed the HE of 32 cases available. From Table 1 we found 11 (34.4%) cases of poor differentiation, eight (25%) cases of mucinous adenocarcinoma. Fourteen extracoloni tumors of 13 patients were found in nine Lynch syndrome II families, including four hepatic cancers, three endometrial carcinomas, three breast cancers, two stomach cancers, one bladder cancer, and one neck cancer. Hepatic cancer is the most frequent extracolonic cancer in our series, accounting for 30.7%. Twenty-nine patients have survived for 1-28 years, 17 (58.6%) of 29 patients have survived for more than 5 years, 12 (41.4%) of 29 patients have survived for more than 10 years, 7 (24.1%) of 29 patients have survived for more than 15 years. Twenty-one patients whose median survival time was 5.7 years died during 1-23 years.
| Family | Criteria | Tumor | BAT26 | D2S123 | BAT25 | D3S346 | D17S250 | MSIstatus | Determination | hMSH2 protein | hMLH1 protein |
| H1 | AC | CRC | + | + | + | + | + | 5/5 | MSI-H | Negative | N |
| H2 | AC | CRC | + | + | + | + | + | 5/5 | MSI-H | N | Negative |
| H5-1 | AC | CRC | + | + | + | + | + | 5/5 | MSI-H | N | N |
| H5-2 | AC | CRC | + | - | + | + | + | 4/5 | MSI-H | N | N |
| H5-3 | AC | CRC | + | NR | + | NR | NR | 2/2 | MSI-H | N | N |
| H6 | JC | CRC | + | + | + | - | - | 3/5 | MSI-H | N | N |
| H7-1 | AC | CRC | + | + | + | + | + | 5/5 | MSI-H | Negative | N |
| H7-2 | AC | CRC | + | NR | + | + | + | 4/4 | MSI-H | Negative | N |
| H7-3 | AC | CRC | - | + | + | - | + | 3/5 | MSI-H | Negative | N |
All nine tumors in the five HNPCC families showed MSI at more than two loci (MSI-H). Four tumors showed MSI in 5/5 loci, two tumors displayed MSI in 4/5 loci, two tumors presented MSI in 3/5 loci, the other had MSI in 2/5 loci (Table 1, Figure 1).
Lack of hMLH1 immunostaining was observed in tumors from H2 proband. Tumors from probands of H1 and H7 were negative for hMSH2 immunostaining. Normal expression of hMSH2 and hMLH1 protein was observed in tumors from H5 and H6 proband (Table 1, Figure 2).
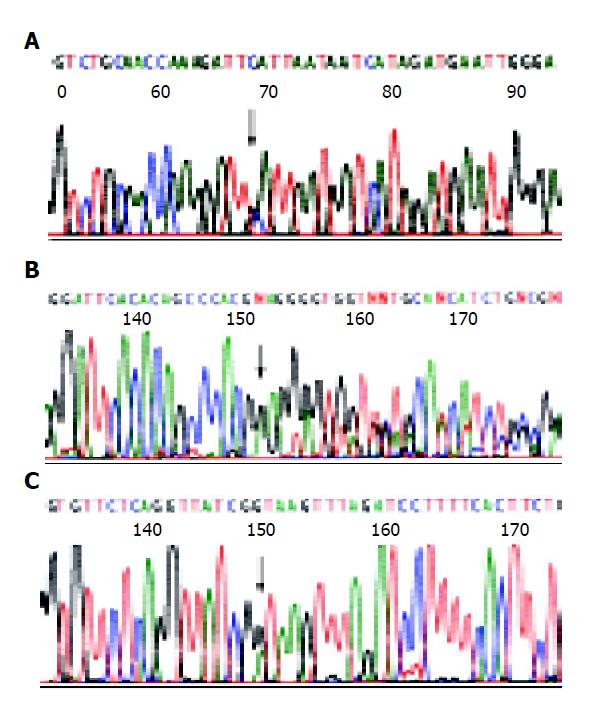
Germline mutations were found in three of the five HNPCC families (60%). The first pathological mutation was a transition from C to A in exon 14 (codon 743) of hMSH2 (family H7). The second mutation was a TTC deletion in exon 14 (codon 530) of hMLH1 (family H5). The third mutation was a transition from G to A in intron 15-exon 15 borders of hMLH1 (family H6)(Figure 3). All the three were definitely pathological mutations, of which the former two had not been reported previously. All three mutations give rise to protein truncation or protein structure alteration. In addition, the affected sister and father of H7 proband also carried the same mutation in exon 14 of hMSH2. Two sons of H5 proband also suffered from colorectal cancer at young age, both carrying the same germline mutation as their father did.
HNPCC is an autosomal, dominantly inherited disease characterized by the development of cancer at an early age[5,24-27], predominance of proximal colonic cancer[26,28-30], excess of multiple cancers[31-33], poorly differentiated cancer[34], an increased risk for selected extracolonic adenocarcinomas[35] and better prognosis[36,37].
Early age of cancer onset is one of the most striking features about HNPCC. The average age to develop colorectal cancer was 45 years, 20 years earlier than the sporadic colorectal cancer. The study involving 43 HNPCC kindreds and 140 HNPCC patients by Bertario et al[26], showed that the average age of onset was 49 years. Cai et al[19], reported the mean age of cancer onset in 30 Chinese HNPCC families, which was 44.1 years in 140 patients, with 74.6% under 50 years of age and 8.5% under 30. In the present study, the median age of onset of the first colorectal cancer was 45.7 years, with 72% under 50 years of age and 8% under 30. Moreover, we found that the age of onset of the first colorectal cancer decreased generation after generation. So to a relatively young colorectal cancer patient, especially younger than 50 years, special attention must be paid while inquiring about family history.
HNPCC is inclined to be located in the proximal colon. Colon cancers are more often right-sided, constituting 47.8% of total cancer and 74.6% of colorectal cancers[5]. A Swedish national investigation in 2001 showed that the proportion of cancers located in the proximal colon was 51% of the total[38]. In our study, the colorectal cancer is found proximal to the splenic flexure constituting 40.9% of the total cancer. Because of the early age of the onset of cancer and as most of the tumor would develop in the proximal colon, we recommend that colonoscopy be performed for the family members with HNPCC and repeated annually or biannually thereafter from the age of 25. In this series, we found three colon cancers and five adenomas by colonoscopy in the family members with HNPCC.
HNPCC showed tendencies of multiple synchronous and metachronous colon cancer[27,32,33]. Fitzgibbons reported that the percentage of synchronous and metachronous colorectal cancers were 18.1% and 24.2%, respectively. It was higher than sporadic colon cancer, being 4.8% and 7.2%, respectively. And the difference was statistically evident[5,27]. Cai et al, reported that 23 (19.5%) of 118 patients presented with multiple cancers in the colorectal cancer[19]. Zhao et al, reported that 39.5% of colorectal cancer patients developed metachronous colorectal cancer within 10 years after their initial colorectal cancer resection. In our series, 19 of 50 (38%) patients presented with multiple cancers in the colorectal cancer. The high incidence of multiple cancers implies that subtotal colectomy is an appropriate management when colon cancers are found in affected patients. It can reduce the chance of developing synchronous colorectal cancer and simplify the endoscopic examination[27]. But considering the effect on the quality of life after subtotal colectomy and the psychological attack on the patients, we usually chose segmental resection for colorectal carcinoma and gave intensive follow-up.
Extracolonic tumors were often seen in HNPCC kindred[27,35,39,40], such as carcinomas of endometrium, ovary, stomach, small bowel, urologic system, hepatobiliary system, breast, brain, larynx, pancreas and as well as leukemia, lymphoma, soft tissue sarcoma, and cutaneous tumors. According to the reports of Western countries, the endometrial and stomach cancers are the first and the second most common tumor in HNPCC, respectively[33,41,42]. In our series, 14 extracolonic tumors of the 13 patients in the nine Lynch syndrome II families were found, of which four were hepatic cancers, three endometrial carcinomas, three breast cancers, two stomach cancers, one bladder cancer, one neck cancer. Hepatic cancer is the most frequent extracolonic cancer in our series, accounting for 30.7%. Zhao et al, in China reported 34 cases of extracolonic cancer in 16 HNPCC families. He also found that stomach cancer was the most common extracolonic tumor in HNPCC (11 cases of stomach cancers) and endometrial cancer was less common (seven of 34) than gastric cancer. The difference in extracolonic tumor spectrum between China and Western countries may lie in many aspects. The small size of Chinese sample may be one of the reasons. Besides, life style, ethnicity and genotype may also contribute to the observed variation.
They were recognized by histopathological criteria in HNPCC: (1) mucinous histotype; (2) poorly differentiated tumors; (3) presence of peritumoral lymphocytic infiltrate, with Crohn’s-like lymphoid reaction. Jass et al[34], reported that there is an excess of poorly differentiated and mucinous tumors in HNPCC. Similarly, in our series, poor differentiation cancers and mucinous adenocarcinoma accounted for 34.4% and 25%, respectively. But, there are rarely mucinous histotypic presence of peritumoral lymphocytic infiltrate, with Crohn’s-like lymphoid reaction in our study, perhaps it was due to the small sample of our study or the Chinese racial difference.
Patients with HNPCC have been suggested to have a better prognosis than patients with common sporadic colorectal cancer. Sankila et al, compared the survival rates of 175 patients with HNPCC with those of 14000 patients with sporadic colorectal cancer diagnosed at <65 years of age in Finland from 1953 to 1993. They showed that the overall five-year cumulative relative survival rate was 65% for patients with HNPCC and 44% for patients with sporadic colorectal cancer[36]. In our study, 21 patients whose median survival time was 5.7 years died during 1-23 years. Twenty-nine patients have survived for 1-28 years, 58.6%, 41.4% and 24.1% patients have survived for more than 5, 10 and 15 years, respectively. The better survival rates may be caused by the heavy mutation burden affecting MMR-deficient tumor cells[36]. Takemoto et al[37], suggest that there might be a possibility of ITCIL having a role for a better prognosis after colorectal cancer surgery, which is closely related to MSI. But there is a conflicting data existing on the prognosis of hereditary colorectal cancer. Bertario et al[26], found no substantial survival advantage for HNPCC patients compared with the sporadic group, after adjustment for age, gender, stage and tumor location.
Microsatellite has been widely considered as an ideal genetic marker. MSI reveals loss of the function of MMR genes. It can serve as a reliable preliminary screening strategy of HNPCC family as several studies have shown that MSI occurs in about 80-90% of HNPCC tumors[43-47]. In the current study, we adopted a panel of five sensitive microsatellite markers accepted by the International Collaborative Group for HNPCC and the National Cancer Institute to detect the MSI status[48]. One hundred percent (9/9) of the nine tumors displayed high-level MSI, which showed high-level defection of MMR function in the affected patients from HNPCC families fulfilling Amsterdam or Japanese criteria B. Sixty percent of the families that have affected patients with high-level MSI were found with germline mutation of hMLH1 or hMSH2 gene, which showed that MSI status and germline mutation of hMLH1 and hMSH2 gene correlated closely with each other.
Ninety percent of HNPCC cases are associated with the mutation of hMSH2 and hMLH1 genes[49]. Recent studies showed that the immunostaining of proteins produced by these two genes could serve as a convenient, rapid and cheap approach in screening HNPCC families[50-52]. In our study, three tumors showed the lost expression of hMSH2 protein and a germline mutation of hMSH2 gene was identified, four tumors showed hMLH1 protein expression and a germline mutation of hMLH1 gene was identified. Two tumors showed that no pathological germline mutation had been detected, but one tumor displaying no expression of hMSH2 protein, one tumor showed the lost expression of hMLH1 protein. In general, immunohistochemical alteration of hMSH2 and hMLH1 proteins and germline mutation of hMLH1 and hMSH2 gene correlated closely with each other. IHC is also very useful in screening HNPCC families.
Direct gene sequencing remains the most reliable method for HNPCC diagnosis. Till now about 400 different predisposing mutations have been reported, mainly affecting the MMR genes hMLH1 (about half), hMSH2 (about 40%) and MSH6 (about 10%)[49]. There appeared no hot spot mutations among those found in these mutations. The three mutations (one in hMSH2 and two in hMLH1) found in five collective families are all pathological mutations. The rate of mutation is 60% (3/5). The first pathological mutation in H7 proband was a transition from C to A in exon 14 (codon 743) of hMSH2. The second mutation in H5 proband was a TTC deletion in exon 14 (codon 530) of hMLH1. The third mutation in H6 proband was a transition from G to A in intron 15-exon 15 borders of hMLH1. The former two pathological mutations had not been reported before (http://archive.uwcm.ac.uk/uwcm/mg/hgmd0.html). All three mutations gave rise to protein truncation or protein structure alteration. The mutations existed also in genomic DNA from other affected family members. For example, the affected sister of H5 proband also carried the same mutation in exon 14 of hMSH2. Two sons of H7 proband also suffered from colorectal cancer at the age of 37 and 38, both carrying the same germline mutation as their proband did, the other son was not found to have any tumor till the age of 43 and also his daughter till the age of 46, both the same germline mutation had not been identified. All new mutations appearing in our study demonstrates the wide spectrum of the mutation responsible for HNPCC. MMR gene mutation analysis will give both HNPCC proband and his family members better management and surveillance, and it will also support genetic counseling as well as gene therapy in the future. For the proband himself, it is helpful for us to conduct positive and effective therapy to reduce the occurrence of possible metachronous multiple colorectal cancer. To the mutation carriers in a family who have not yet suffered from colorectal cancer, close follow-up and early diagnosis are more likely to be performed. To the non-mutation carriers, we should free them from unnecessary psychological and economical burden[50]. But the mutation may be different in a variety of races and geographical regions. Mutations of hMSH2 and hMLH1 accounted for 25-86% of the total cases[40,53,54]. A deeper study of germline mutation remains a heavy assignment for us.
Science Editor Li WZ Language Editor Elsevier HK
| 1. | Lynch HT, Smyrk TC, Watson P, Lanspa SJ, Lynch JF, Lynch PM, Cavalieri RJ, Boland CR. Genetics, natural history, tumor spectrum, and pathology of hereditary nonpolyposis colorectal cancer: an updated review. Gastroenterology. 1993;104:1535-1549. [PubMed] [Cited in This Article: ] |
| 2. | Katballe N, Christensen M, Wikman FP, Ørntoft TF, Laurberg S. Frequency of hereditary non-polyposis colorectal cancer in Danish colorectal cancer patients. Gut. 2002;50:43-51. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 57] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 3. | Marra G, Boland CR. Hereditary nonpolyposis colorectal cancer: the syndrome, the genes, and historical perspectives. J Natl Cancer Inst. 1995;87:1114-1125. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 342] [Cited by in F6Publishing: 325] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 4. | Raedle J, Schaffner M, Esser N, Sahm S, Trojan J, Kriener S, Brieger A, Nier H, Bockhorn H, Berg PL. Frequency of the Amsterdam criteria in a regional German cohort of patients with colorectal cancer. Z Gastroenterol. 2002;40:561-568. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 5. | Fitzgibbons RJ, Lynch HT, Stanislav GV, Watson PA, Lanspa SJ, Marcus JN, Smyrk T, Kriegler MD, Lynch JF. Recognition and treatment of patients with hereditary nonpolyposis colon cancer (Lynch syndromes I and II). Ann Surg. 1987;206:289-295. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 161] [Cited by in F6Publishing: 160] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 6. | Vasen HF, Mecklin JP, Khan PM, Lynch HT. The International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC). Dis Colon Rectum. 1991;34:424-425. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1357] [Cited by in F6Publishing: 1248] [Article Influence: 37.8] [Reference Citation Analysis (0)] |
| 7. | Fujita S, Moriya Y, Sugihara K, Akasu T, Ushio K. Prognosis of hereditary nonpolyposis colorectal cancer (HNPCC) and the role of Japanese criteria for HNPCC. Jpn J Clin Oncol. 1996;26:351-355. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 40] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 8. | Peltomäki P, Aaltonen LA, Sistonen P, Pylkkänen L, Mecklin JP, Järvinen H, Green JS, Jass JR, Weber JL, Leach FS. Genetic mapping of a locus predisposing to human colorectal cancer. Science. 1993;260:810-812. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 599] [Cited by in F6Publishing: 633] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 9. | Leach FS, Nicolaides NC, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomäki P, Sistonen P, Aaltonen LA, Nyström-Lahti M. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215-1225. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1588] [Cited by in F6Publishing: 1470] [Article Influence: 47.4] [Reference Citation Analysis (0)] |
| 10. | Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, Kane M, Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027-1038. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1956] [Cited by in F6Publishing: 1845] [Article Influence: 59.5] [Reference Citation Analysis (0)] |
| 11. | Nicolaides NC, Papadopoulos N, Liu B, Wei YF, Carter KC, Ruben SM, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature. 1994;371:75-80. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1070] [Cited by in F6Publishing: 1003] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 12. | Nyström-Lahti M, Parsons R, Sistonen P, Pylkkänen L, Aaltonen LA, Leach FS, Hamilton SR, Watson P, Bronson E, Fusaro R. Mismatch repair genes on chromosomes 2p and 3p account for a major share of hereditary nonpolyposis colorectal cancer families evaluable by linkage. Am J Hum Genet. 1994;55:659-665. [PubMed] [Cited in This Article: ] |
| 13. | Papadopoulos N, Nicolaides NC, Wei YF, Ruben SM, Carter KC, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM, Adams MD. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625-1629. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1335] [Cited by in F6Publishing: 1249] [Article Influence: 41.6] [Reference Citation Analysis (0)] |
| 14. | Bronner CE, Baker SM, Morrison PT, Warren G, Smith LG, Lescoe MK, Kane M, Earabino C, Lipford J, Lindblom A. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258-261. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1448] [Cited by in F6Publishing: 1512] [Article Influence: 50.4] [Reference Citation Analysis (0)] |
| 15. | Wijnen J, Vasen H, Khan PM, Menko FH, van der Klift H, van Leeuwen C, van den Broek M, van Leeuwen-Cornelisse I, Nagengast F, Meijers-Heijboer A. Seven new mutations in hMSH2, an HNPCC gene, identified by denaturing gradient-gel electrophoresis. Am J Hum Genet. 1995;56:1060-1066. [PubMed] [Cited in This Article: ] |
| 16. | Ikenaga M, Tomita N, Sekimoto M, Ohue M, Yamamoto H, Miyake Y, Mishima H, Nishisho I, Kikkawa N, Monden M. Use of microsatellite analysis in young patients with colorectal cancer to identify those with hereditary nonpolyposis colorectal cancer. J Surg Oncol. 2002;79:157-165. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 17] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 17. | Holinski-Feder E, Müller-Koch Y, Friedl W, Moeslein G, Keller G, Plaschke J, Ballhausen W, Gross M, Baldwin-Jedele K, Jungck M. DHPLC mutation analysis of the hereditary nonpolyposis colon cancer (HNPCC) genes hMLH1 and hMSH2. J Biochem Biophys Methods. 2001;47:21-32. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 71] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 18. | Wahlberg SS, Schmeits J, Thomas G, Loda M, Garber J, Syngal S, Kolodner RD, Fox E. Evaluation of microsatellite instability and immunohistochemistry for the prediction of germ-line MSH2 and MLH1 mutations in hereditary nonpolyposis colon cancer families. Cancer Res. 2002;62:3485-3492. [PubMed] [Cited in This Article: ] |
| 19. | Cai SJ, Xu Y, Cai GX, Lian P, Guan ZQ, Mo SJ, Sun MH, Cai Q, Shi DR. Clinical characteristics and diagnosis of patients with hereditary nonpolyposis colorectal cancer. World J Gastroenterol. 2003;9:284-287. [PubMed] [Cited in This Article: ] |
| 20. | Mo SJ, Cai H, Cai SJ. Hereditary non-polyposis colorectal cancer: A report of 10 Chinese families. Zhonghua Xiaohua Zazhi. 1996;16:326-328. [Cited in This Article: ] |
| 21. | Bocker T, Rüschoff J, Fishel R. Molecular diagnostics of cancer predisposition: hereditary non-polyposis colorectal carcinoma and mismatch repair defects. Biochim Biophys Acta. 1999;1423:O1-O10. [PubMed] [Cited in This Article: ] |
| 22. | Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248-5257. [PubMed] [Cited in This Article: ] |
| 23. | Gebert J, Sun M, Ridder R, Hinz U, Lehnert T, Möller P, Schackert HK, Herfarth C, von Knebel Doeberitz M. Molecular profiling of sporadic colorectal tumors by microsatellite analysis. Int J Oncol. 2000;16:169-179. [PubMed] [Cited in This Article: ] |
| 24. | Wei SC, Wang MH, Shieh MC, Wang CY, Wong JM. Clinical characteristics of Taiwanese hereditary non-polyposis colorectal cancer kindreds. J Formos Med Assoc. 2002;101:206-209. [PubMed] [Cited in This Article: ] |
| 25. | Guillem JG, Puig-La Calle J, Cellini C, Murray M, Ng J, Fazzari M, Paty PB, Quan SH, Wong WD, Cohen AM. Varying features of early age-of-onset "sporadic" and hereditary nonpolyposis colorectal cancer patients. Dis Colon Rectum. 1999;42:36-42. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 27] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 26. | Bertario L, Russo A, Sala P, Eboli M, Radice P, Presciuttini S, Andreola S, Rodriguez-Bigas MA, Pizzetti P, Spinelli P. Survival of patients with hereditary colorectal cancer: comparison of HNPCC and colorectal cancer in FAP patients with sporadic colorectal cancer. Int J Cancer. 1999;80:183-187. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 2] [Reference Citation Analysis (0)] |
| 27. | Lynch HT, Smyrk T. Hereditary nonpolyposis colorectal cancer (Lynch syndrome). An updated review. Cancer. 1996;78:1149-1167. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 5] [Reference Citation Analysis (0)] |
| 28. | Lynch HT, Lynch JF. Hereditary nonpolyposis colorectal cancer. Semin Surg Oncol. 2000;18:305-313. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 6] [Reference Citation Analysis (0)] |
| 29. | Bernstein IT, Bisgaard ML, Myrhøj T. Registration of hereditary non-polyposis colorectal cancer. Ugeskr Laeger. 1999;161:6174-6178. [PubMed] [Cited in This Article: ] |
| 30. | Ponz de Leon M, Benatti P, Percesepe A, Rossi G, Viel A, Santarosa M, Pedroni M, Roncucci L. Clinical and molecular diagnosis of hereditary non-polyposis colorectal cancer: problems and pitfalls in an extended pedigree. Ital J Gastroenterol Hepatol. 1999;31:476-480. [PubMed] [Cited in This Article: ] |
| 31. | Box JC, Rodriguez-Bigas MA, Weber TK, Petrelli NJ. Clinical implications of multiple colorectal carcinomas in hereditary nonpolyposis colorectal carcinoma. Dis Colon Rectum. 1999;42:717-721. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 27] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 32. | Hemminki K, Li X, Dong C. Second primary cancers after sporadic and familial colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2001;10:793-798. [PubMed] [Cited in This Article: ] |
| 33. | Lin KM, Shashidharan M, Ternent CA, Thorson AG, Blatchford GJ, Christensen MA, Lanspa SJ, Lemon SJ, Watson P, Lynch HT. Colorectal and extracolonic cancer variations in MLH1/MSH2 hereditary nonpolyposis colorectal cancer kindreds and the general population. Dis Colon Rectum. 1998;41:428-433. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 78] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 34. | Jass JR, Smyrk TC, Stewart SM, Lane MR, Lanspa SJ, Lynch HT. Pathology of hereditary non-polyposis colorectal cancer. Anticancer Res. 1994;14:1631-1634. [PubMed] [Cited in This Article: ] |
| 35. | Rodriguez-Bigas MA, Vasen HF, Lynch HT, Watson P, Myrhøj T, Järvinen HJ, Mecklin JP, Macrae F, St John DJ, Bertario L. Characteristics of small bowel carcinoma in hereditary nonpolyposis colorectal carcinoma. International Collaborative Group on HNPCC. Cancer. 1998;83:240-244. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 2] [Reference Citation Analysis (0)] |
| 36. | Sankila R, Aaltonen LA, Järvinen HJ, Mecklin JP. Better survival rates in patients with MLH1-associated hereditary colorectal cancer. Gastroenterology. 1996;110:682-687. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 252] [Cited by in F6Publishing: 232] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 37. | Takemoto N, Konishi F, Yamashita K, Kojima M, Furukawa T, Miyakura Y, Shitoh K, Nagai H. The correlation of microsatellite instability and tumor-infiltrating lymphocytes in hereditary non-polyposis colorectal cancer (HNPCC) and sporadic colorectal cancers: the significance of different types of lymphocyte infiltration. Jpn J Clin Oncol. 2004;34:90-98. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 47] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 38. | Hemminki K, Li X. Familial colorectal adenocarcinoma and hereditary nonpolyposis colorectal cancer: a nationwide epidemiological study from Sweden. Br J Cancer. 2001;84:969-974. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 29] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 39. | Love RR. Small bowel cancers, B-cell lymphatic leukemia, and six primary cancers with metastases and prolonged survival in the cancer family syndrome of Lynch. Cancer. 1985;55:499-502. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 2] [Reference Citation Analysis (0)] |
| 40. | Yuan Y, Ye J, Zheng S. Clinical and genetic features of International Collaborative Group-hereditary nonpolyposis colorectal cancer families and suspected hereditary nonpolyposis colorectal cancer families. Chin Med J (Engl). 2004;117:748-752. [PubMed] [Cited in This Article: ] |
| 41. | Scaife CL, Rodriguez-Bigas MA. Lynch syndrome: implications for the surgeon. Clin Colorectal Cancer. 2003;3:92-98. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 42. | Watson P, Lynch HT. The tumor spectrum in HNPCC. Anticancer Res. 1994;14:1635-1639. [PubMed] [Cited in This Article: ] |
| 43. | Lamberti C, Kruse R, Ruelfs C, Caspari R, Wang Y, Jungck M, Mathiak M, Malayeri HR, Friedl W, Sauerbruch T. Microsatellite instability-a useful diagnostic tool to select patients at high risk for hereditary non-polyposis colorectal cancer: a study in different groups of patients with colorectal cancer. Gut. 1999;44:839-843. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 69] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 44. | Terdiman JP, Gum JR, Conrad PG, Miller GA, Weinberg V, Crawley SC, Levin TR, Reeves C, Schmitt A, Hepburn M. Efficient detection of hereditary nonpolyposis colorectal cancer gene carriers by screening for tumor microsatellite instability before germline genetic testing. Gastroenterology. 2001;120:21-30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 109] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 45. | Loukola A, Eklin K, Laiho P, Salovaara R, Kristo P, Järvinen H, Mecklin JP, Launonen V, Aaltonen LA. Microsatellite marker analysis in screening for hereditary nonpolyposis colorectal cancer (HNPCC). Cancer Res. 2001;61:4545-4549. [PubMed] [Cited in This Article: ] |
| 46. | Aaltonen LA, Peltomäki P, Mecklin JP, Järvinen H, Jass JR, Green JS, Lynch HT, Watson P, Tallqvist G, Juhola M. Replication errors in benign and malignant tumors from hereditary nonpolyposis colorectal cancer patients. Cancer Res. 1994;54:1645-1648. [PubMed] [Cited in This Article: ] |
| 47. | Craanen ME, Blok P, Offerhaus GJ, Tytgat GN. Recent developments in hereditary nonpolyposis colorectal cancer. Scand J Gastroenterol Suppl. 1996;218:92-97. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 9] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 48. | Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248-5257. [Cited in This Article: ] |
| 49. | Peltomäki P. Deficient DNA mismatch repair: a common etiologic factor for colon cancer. Hum Mol Genet. 2001;10:735-740. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 336] [Cited by in F6Publishing: 364] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 50. | Chaves P, Cruz C, Lage P, Claro I, Cravo M, Leitão CN, Soares J. Immunohistochemical detection of mismatch repair gene proteins as a useful tool for the identification of colorectal carcinoma with the mutator phenotype. J Pathol. 2000;191:355-360. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 3] [Reference Citation Analysis (0)] |
| 51. | Chiaravalli AM, Furlan D, Facco C, Tibiletti MG, Dionigi A, Casati B, Albarello L, Riva C, Capella C. Immunohistochemical pattern of hMSH2/hMLH1 in familial and sporadic colorectal, gastric, endometrial and ovarian carcinomas with instability in microsatellite sequences. Virchows Arch. 2001;438:39-48. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 83] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 52. | Debniak T, Kurzawski G, Gorski B, Kladny J, Domagala W, Lubinski J. Value of pedigree/clinical data, immunohistochemistry and microsatellite instability analyses in reducing the cost of determining hMLH1 and hMSH2 gene mutations in patients with colorectal cancer. Eur J Cancer. 2000;36:49-54. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 72] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 53. | Peltomäki P, Vasen HF. Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. The International Collaborative Group on Hereditary Nonpolyposis Colorectal Cancer. Gastroenterology. 1997;113:1146-1158. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 487] [Cited by in F6Publishing: 518] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 54. | Wagner A, Barrows A, Wijnen JT, van der Klift H, Franken PF, Verkuijlen P, Nakagawa H, Geugien M, Jaghmohan-Changur S, Breukel C. Molecular analysis of hereditary nonpolyposis colorectal cancer in the United States: high mutation detection rate among clinically selected families and characterization of an American founder genomic deletion of the MSH2 gene. Am J Hum Genet. 2003;72:1088-1100. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 148] [Cited by in F6Publishing: 160] [Article Influence: 7.6] [Reference Citation Analysis (0)] |