Abstract
Hunting for tumoral material in body fluids, traditionally in blood, the so-called liquid biopsy is set to revolutionize the diagnosis and management of oncological patients. However, other biofluids can also be considered as alternative sources of biomarkers to provide clinically valuable information for multiple diseases. This is the case of bile, a fluid produced in the liver, stored in the gallbladder, and excreted to the duodenum, which complex composition is known to change in different pathological conditions. Remarkably, different works have demonstrated that the identification of mutations in bile cell-free DNA (cfDNA) can outperform blood analysis for the early diagnosis of biliopancreatic tumors causing biliary strictures. Here, the literature in which bile has been tested as a liquid biopsy matrix where lipids, metabolites, proteins, and cfDNA among other analytes were measured is reviewed. Moreover, the clinical situations and procedures where bile can be available, discussing the possible applications and limitations of bile analysis are summarized. The scientific relevance and clinical potential of bile harvesting, biobanking, and analysis are put forward. All this evidence supports the value of bile as a liquid biopsy matrix for the management of patients beyond cancer, and perhaps also beyond “blood, sweat, and tears”.
Keywords
Hepatopancreatobiliary tumors, diagnosis, biomarkers, bile collection, biliary disease, liquid biopsyIntroduction
Liquid biopsy (LB) represents the sampling and analysis of human body fluids including blood, urine, saliva, cerebrospinal or pleural fluid, ascites, tears, sweat, and bile as a minimally invasive method for the diagnosis and prognosis of different diseases [1]. The rationale implies that biofluids are loaded with a variety of analytes (DNA, proteins, metabolites, extracellular vesicles, or cells) where the detection of specific biomarkers can distinguish between health and disease or even inform about disease progression and response to treatment. Accordingly, the number of clinical trials for LB in different therapeutic areas is growing dramatically, with more than 1,500 trials using blood in the oncology field [2] for early disease detection, to guide therapy, and to monitor outcome and residual disease [2]. However, sensitivity and specificity represent key challenges in the development of LB strategies for cancer screening, and the selection of more reliable biomarkers and/or the development of sample enrichment procedures represent areas of intense research [1, 3]. In addition, the use of biofluids other than blood could overcome some limitations, including blood biomarker concentration and specificity avoiding for instance clonal hematopoiesis mutations [4].
In this sense, as recently reviewed [5–10], bile has emerged as a promising LB matrix for the management of patients with pancreaticobiliary tumors causing biliary strictures. Here, the most recent findings on the utility of bile circulating cell-free DNA (cfDNA) analyses in those oncological conditions are reviewed. Moreover, different clinical situations where bile could be available, the current bile collection procedures, and the potential clinical applications of bile analyses are presented.
Limitations and advantages of bile as LB in oncology
Bile is constantly produced by hepatocytes, stored in the gallbladder, and secreted into the duodenum through the biliary tract. Bile is a complex fluid composed of bile acids, phospholipids, cholesterol, bilirubin, proteins, inorganic salts, extracellular vesicles, DNA, and RNA [11–14]. Bile composition can change in different pathological conditions; therefore, the identification of these changes can be harnessed into good diagnostic tools, can inform about pathological processes, or can even help in directing treatments [5, 9, 14–16]. Useful analytes include small molecules and metabolites usually analyzed and measured by nuclear magnetic resonance (NMR) spectroscopy or liquid chromatography tandem mass spectrometry (LC-MS), proteins mainly explored and measured by proteomics and enzyme-linked immunosorbent assay (ELISA), and nucleic acids (RNA and DNA) analyzed by next generation sequencing (NGS) or droplet digital polymerase chain reaction (ddPCR) [9, 15–18]. In the case of the hepatobiliopancreatic (HBP) tumors, due to their anatomical and physiological characteristics, bile may be in direct contact with the lesions, and therefore higher concentrations of tumor biomarkers may be present in bile compared to plasma or urine, increasing the sensitivity of this LB strategy [19]. Moreover, its relatively confined nature and location reduce the possibility of detecting aberrant biomarkers coming from another diseased organ, increasing the specificity of the results. In fact, in this context, as a tumor-adjacent fluid, several studies have shown that the analysis of bile outperforms that of blood, and in addition has the potential to recapitulate tumor heterogeneity [20–23].
However, the use of bile as LB matrix has some limitations, for instance, its availability. LB strategies are generally minimally invasive, however, most methods used to obtain bile (see below) cannot have this consideration. Nevertheless, accumulating evidence demonstrates that patients undergoing interventions for therapeutic or diagnostic purposes in which the biliary tract is accessible can benefit from bile analysis. For instance, and as discussed below, the mutational status of bile cfDNA can help to diagnose strictures of unknown etiology, identifying and anticipating the presence of malignancies [20]. Thus, given the informative potential of bile, its collection should be considered when procedures in which the biliary tract is accessed are performed. Accordingly, the Nouvelle-Aquitaine, Euskadi, and Navarre Euroregion funded a project for the creation of a bile biobank named Bilebank (https://bilebank.org/) where bile and patients’ associated clinical data are available for analytical and research purposes.
Procedures in the clinical practice where bile can be collected
Several approaches, surgical, percutaneous (interventional radiology), or endoscopic [24–26] are used in different clinical situations for biliary drainage (Table 1). In those situations, bile could be collected and used to perform different analyses to help with the diagnosis, prognosis, and treatment of patients.
Clinical approaches for biliary drainage
| Method type | Specific clinical approaches |
|---|---|
| Surgical approaches | Gallbladder puncture |
| Bile duct puncture | |
| Maintained bile drainage (T-tube) | |
| Percutaneous approaches | Percutaneous gallbladder drainage (PGBD) |
| Percutaneous gallbladder aspiration (PGBA) | |
| Percutaneous biliary drainage (PBD) | |
| Endoscopic approaches | Endoscopic retrograde cholangiopancreatography (ERCP) |
| Endoscopic ultrasound-guided biliary drainage (EUS-BD) | |
| Endoscopic nasogallbladder drainage (ENGBD) | |
| Endoscopic nasobiliary drainage (ENBD) | |
| Nasoduodenal and nasobiliary intubation |
Surgical procedures
Before percutaneous and endoscopic approaches for biliary drainage were developed and extensively implemented, surgical management was the predominant method [27–29]. Consequently, in classical studies, bile was collected during cholecystectomy (open or laparoscopic), surgery for liver resection and transplantation, or surgical duct exploration procedures, using techniques such as gallbladder puncture, bile duct puncture, or T-tube placement [29–32]. Nowadays these approaches might be useful to obtain bile in patients with early-stage biliopancreatic tumors (BPT) undergoing straight surgery without prior drainage requirements.
Gallbladder puncture
At surgery, bile can be aspirated completely from the gallbladder with a sterile needle and syringe. When gallbladder has to be removed, sampling should be obtained at the beginning of the operation before gallbladder manipulation and cystic artery ligation in order to prevent potential contamination due to mucosal healing or ischemia. If gallbladder is not being removed, post-operative bile leak should be minimized by using a thin needle, creating a self-sealing tunnel, and suturing the needle hole [33–35].
Bile duct puncture
Samples of hepatic bile are very difficult to obtain if the patient is not undergoing bile duct or gallbladder surgery because there is a high risk of bile leak after direct puncture. In gallbladder surgery and stone disease, hepatic bile can be obtained through the cholangiogram catheter introduced through the cystic duct before contrast injection [36, 37]. In patients undergoing pancreatic surgery involving bile duct transection (pancreaticoduodenectomy or total pancreatectomy) biliary swab has been used to detect biliary colonization [38].
Maintained bile drainage (T-tube)
Some patients require a temporary bile shunt (T-tube) that diverts part of the bile flowing from the liver to a transcutaneous port for external collection [33]. T-tubes allow for partial collection of liver bile during the time that the shunt is in place. Nowadays, there is no justification for the routine use of T-tube drainage after open or laparoscopic common bile duct exploration in patients with common bile duct stones [39].
Percutaneous procedures
Percutaneous gallbladder or bile duct drainage is used in patients with cholecystitis or cholangitis, but also in patients with BPT, who need a biliary drainage and endoscopic approaches are not clinically indicated, technically possible, or available [40–43]. Usually, this drainage is maintained for some days until clinical improvement or a more definitive approach is decided. Mainly two percutaneous techniques are used and biliary sampling and molecular analysis have been performed in multiple studies [9].
Percutaneous gallbladder drainage (PGBD) or gallbladder aspiration (PGBA)
PGBD or PGBA is traditionally the first-line approach in high-risk surgical acute cholecystitis patients [44].
Percutaneous bile duct drainage (PBD) or bile duct puncture
PBD or bile duct puncture is a minimally invasive method used for treating malignant biliary obstruction. Collected bile has been used for instance for culture and antimicrobial susceptibility tests [45] or to assess post-hepatectomy liver function in patients with biliary tract disease [31, 32].
Endoscopic procedures
Nowadays, endoscopic retrograde cholangiopancreatography (ERCP) is the first-line intervention for biliary drainage in patients with BPT, when it is available and technically possible [24, 25, 46]. ERCP achieves success in 90% of cases, but it fails in cases with altered anatomy or when duodenum access is obstructed [47]. Therapeutic endoscopic ultrasound (EUS) techniques [48, 49] are indicated when ERCP is not feasible, and its use has become more extensive in the last years [50, 51].
Transpapillary ERCP
ERCP is performed with a duodenoscope that permits the wire-guided cannulation of the ampulla of Vater and then biliary drainage and bile collection in patients with an obstruction of the bile duct [52, 53]. A catheter and a syringe can be used to collect bile, but also other devices [54] have been developed helping to collect tissue and cell samples in addition to bile aspiration. In this context, several studies have performed molecular analyses of bile collected by ERCP [9, 20, 55, 56].
ENGBD or ENBD is performed by insertion of a temporary nasobiliary tube can be performed in order to assure biliary drainage and continuous or intermittent flushing of the bile duct. In patients with symptomatic cholelithiasis awaiting cholecystectomy it allows bile collection in different periods before gallbladder removal [34]. It has also been used for bile collection in pharmacokinetic studies [57, 58].
Endoscopic ultrasound (EUS)
Nowadays, laparoscopic cholecystectomy is the first-line procedure indicated when cholecystitis occurs. However, non-operable patients usually require gallbladder drainage, and the EUS approach through the stomach or duodenum EUS guidance gallbladder drainage (EUS-GBD) is recommended [46, 51, 59]. EUS-assisted and EUS-guided techniques (cholecystoduodenostomy, cholecystogastrostomy, hepaticogastrostomy, or choledochoduodenostomy) are also available for biliary drainage in patients with unresectable distal malignant biliary obstruction in cases of failed ERCP or PBD because of altered anatomy, previous surgery or duodenum obstruction [43, 51, 59–62].
Other possible strategies
Given the important clinical information that different bile analyses can provide, it is possible to envision the development of new, or the optimization of old strategies for bile collection in a less invasive or risk-free manner in a broader spectrum of patients.
Collection of duodenal bile (spontaneous or after stimulation with a choleretic agent)
Pancreatic juice has been collected from the duodenal lumen using the endoscope suction channel or a catheter upon secretin stimulation [63]. Both methods could also be used for duodenal bile collection, and choleretic agents such as secretin or cholecystokinin (CCK)-pancreozymin used to perform bile drainage tests [64] could be used to stimulate bile secretion.
Clinical applications of the molecular analysis of bile
There are multiple pathological conditions (Table 2) in which bile might be collected and its analysis could be clinically relevant (Table 3) [16, 28]. However, in most of these situations, bile composition has not been extensively interrogated, and therefore the utility of measuring changes in bile acids, metabolites, microbiota, proteins, or nucleic acids remains unknown (Figure 1).
Pathological situations where bile is available and its analysis could be helpful
| Pathological situations |
|---|
| Benign or indeterminate biliary stenoses |
| Primary sclerosing cholangitis (PSC) |
| Chronic cholangitis at risk of developing cholangiocarcinoma (CCA) [repetitive chronic secondary cholangitis, immunoglobulin G4 (IgG4)-related cholangitis, etc.] |
| Chronic pancreatitis with biliary strictures |
| Pancreatic cysts with biliary strictures |
| Pancreatic ductal adenocarcinoma (PDAC), CCA, bile duct carcinoma (BDC), hepatocellular carcinoma (HCC), and ampullary cancer presenting biliary strictures and/or obstructive jaundice |
| Pancreatic surgery (pancreaticoduodenectomy or total pancreatectomy) |
| Acute cholangitis or cholecystitis |
| Patients with jaundice |
| Pathologies associated with biliary microcrystals: idiopathic acute pancreatitis, recurrent acute pancreatitis, unexplained biliary pain, and post-cholecystectomy biliary pain |
| Liver surgery: hepatectomy and transplantation |
Possible applications of bile sampling and analysis
| Bile applications | Reference(s) |
|---|---|
| Bile culture and targeted antibiotic testing | [16, 45] |
| Differential diagnosis of patients with jaundice | [65] |
| Early biliopancreatic cancer diagnosis | [9] |
| Diagnosis of indeterminate biliary strictures | [10, 20] |
| Clinical conditions of patients with choledocholithiasis or bile microlithiasis | [66] |
| Pharmacokinetic analyses and biliary excretion studies | [67, 68] |
| Progression/severity of hepatobiliary diseases | [28] |
| Early detection of post-liver resection and transplantation failure | [69] |
| Regenerative medicine | [70] |
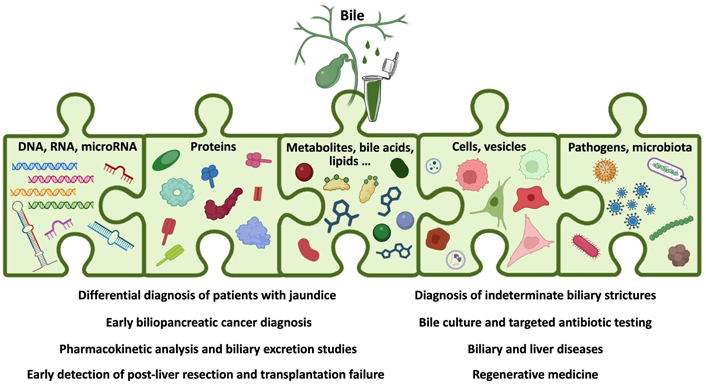
Schematic representation of the analytes present in bile and the clinical situations where bile molecular analyses could be informative. Created with BioRender.com
However, different studies already support bile analysis as a valuable tool. Indeed, changes in bile composition have been correlated with clinical severity, disease progression, and final outcome in patients with different hepatobiliary diseases including choledochal cysts, extrahepatic portal venous obstruction, and infantile obstructive cholangiopathy [28]. Moreover, changes in bile composition have been proposed to help predict hepatic function and liver failure following liver surgery. For instance, monitoring the levels of interleukin 6 (IL-6) in bile could help detect acute rejection after liver transplantation [71] or liver failure after resection [32], and the sequential monitoring of bile salt composition has been suggested to be useful to discriminate functional versus non-functional grafts during liver transplantation [72].
Biliary crystals are associated with gallstone disease, idiopathic pancreatitis, sphincter of Oddi dysfunction, unexplained biliary pain, and post-cholecystectomy biliary pain [73]. Microscopic bile examination, especially when EUS is not available, can be useful for the diagnosis of these pathologies.
During cholangitis and biliary infections, bile culture is required to guide antimicrobial choice as in intraoperative bile cultures of patients undergoing pancreaticoduodenectomy or total pancreatectomy [38, 45, 74].
Biliary excretion evaluation is another field where bile collection would be of interest, as bile is an important route of elimination for many drugs, with a relevant impact on pharmacokinetics and drug-drug interactions [67, 75].
Further studies are required to elucidate whether bile analysis could also help to diagnose or to identify the mechanisms of disease in patients with jaundice, for instance, alterations of bilirubin metabolism or hepatocellular dysfunction [65]. In the case of obstructive jaundice, bile LB has demonstrated its extraordinary performance in the discrimination between benign and malignant strictures caused by BPT [20]. Although a minority of biliary strictures are benign [76], their accurate diagnosis is a dilemma and the sensitivity of current diagnostic methods does not exceed 60% [77]. As discussed below, the analysis of mutations in the cfDNA isolated from bile obtained during ERCP in patients with biliary strictures can diagnose the presence of BPT with 90% sensitivity [20], without the need for further explorations, and shortening not only the time for diagnosis but for oncological therapy. BPT have a very poor prognosis because they are often diagnosed in advanced stages, mainly due to nonspecific symptoms, and have ineffective oncological treatments. Therefore, biliary molecular analyses would benefit the management and outcome of these patients. In this context, the increased understanding of carcinogenic mechanisms in BPT is leading to new molecular classifications that facilitate the selection of targeted treatments and appropriate prognostic guidance. These molecular studies are commonly conducted on histological samples [78–81]. The use of bile or pancreatic juice obtained during ERCP, or collected after secretin-induced secretion [9, 63] in addition to enhancing diagnostic sensitivity might also guide treatment. Similarly, for biliary strictures developing in patients with potentially preneoplastic conditions (such as PSC, chronic pancreatitis, or pancreatic cysts) requiring ERCP, molecular analyses of bile obtained during the procedure can increase the diagnostic sensitivity for malignancy in cases where the anatomopathological study was inconclusive, or even anticipate the presence of tumors, which may impact transplant decisions [82].
Informative bile analytes
Informative analytes present in bile include bile acids, lipids, metabolites, proteins, cell-free DNA, cell-free microRNAs, extracellular vesicles, and circulating tumor cells. Different recent publications have reviewed the utility of these analytes, extracellular vesicles, and cells present in the bile [5–7, 9]. In the last section of this review, data regarding the identification of mutations and DNA methylation marks in bile cfDNA and the use of bile as a source material for the generation of organoids are summarized.
Lipids and metabolites
Bile acids, phospholipids, and cholesterol are the main lipid components in human bile and different strategies have been reported for their analyses [83–87]. Alterations in bile lipids composition have been described in different hepatopancreatobiliary diseases, so their eventual analysis in bile might help in the management of these patients. For instance, compared with controls, patients with cholangitis and/or jaundice with biliary obstruction present a reduction in total bile acids, cholesterol, phosphatidylcholine, and inorganic phosphate [88]. Moreover, the level of different lipids and their metabolites such as specific oxidized phospholipids (oxPLs) [89], the lipid peroxidation product 4-hydroxynonenal (4-HNE) [90], or a panel of ten lipid species [91], have been reported altered in patients with BPT.
Proteins
Changes in the levels of different proteins in bile have been reported to be associated with different pathologies [6, 7, 9]. Moreover, protocols to improve the methodology and to increase bile proteome coverage have been published over the years [92–95]. Nowadays, a number of reports propose that the biliary levels of different proteins may serve as cancer biomarkers, such as neutrophil gelatinase-associated lipocalin (NGAL), carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6), or proteins of the mucin family, among others [9, 96]. Moreover, bile levels of insulin-like growth factor-I (IGF-I) in patients undergoing ERCP for biliary obstruction discriminate extrahepatic CCA from either pancreatic cancer or benign strictures [97]. The identification in human bile of a combination of five proteins was also able to discriminate malignant strictures [91]. As mentioned above, biliary levels of IL-6 or hepatocyte growth factor (HGF) have been proposed to predict liver failure after partial hepatectomy in patients with biliary tract carcinoma, outperforming serum analyses [31, 32]. Although this is a rapidly growing and changing field, the role of biliary proteins as biomarkers in different pathologies remains to be explored in prospective clinical studies. This could include for instance, the panel of proteins recently identified in serum extracellular vesicles for the prediction, early diagnosis, and prognostication of CCA in PSC patients [98], or components of the extracellular matrix which undergoes both quantitative and qualitative modifications after biliary damage, including transformation [99]. Therefore, further studies are required to demonstrate whether bile can serve as a matrix that enables the study of such changes and whether biomarkers in this context can help in the management of patients with hepatobiliopancreatic diseases.
Vesicles and microRNAs
Bile contains microRNAs that are stable and therefore have potential clinical utility as disease marker panels [100]. Moreover, bile extracellular vesicles contain abundant microRNA species [100], so could they be a source for their analyses. Changes in bile microRNA levels in different hepatopancreatobiliary diseases have been described and reviewed [6]. Altered content of microRNAs was reported in the bile from liver transplant recipients [101]. Moreover, concentrations of three microRNAs (miR-517a, miR-892a, and miR-106a*) are increased in the bile of patients with biliary complications (ischemic-type biliary lesions) after liver transplantation [102]. Patients with CCA showed a significantly distinct microRNA profile in bile compared with patients with benign biliary strictures [103] or with PSC patients [104]. All these findings indicate the potential diagnostic value of bile microRNAs.
Bile circulating tumor DNA and BPT
As in other biofluids, DNA molecules circulate freely in the bile of healthy and diseased individuals. In the case of cancer patients, a fraction of these cfDNA molecules corresponds to circulating tumor DNA (ctDNA) [19, 105]. These molecules released by tumor cells preserve characteristics of the tissue of origin [1] and therefore could be used to diagnose the presence of pre-malignant or malignant cells anywhere in the biliary tract, or even in the pancreas.
Mutations
Several studies have demonstrated the feasibility and usefulness of detecting genetic mutations in bile cfDNA, surpassing the limited sensitivity of other reference diagnostic tools such as cytology or intraductal tissue biopsies. Thus, articles published more than twenty years ago already reported the detection of specific mutations in tumor protein 53 (TP53) and Kirsten rat sarcoma (KRAS), the two most prevalently mutated genes in patients with BPT, in the bile of patients with malignant strictures [106–110]. Although conventional sequencing technologies applied in these studies lacked the sensitivity of current amplification and sequencing tools, making sensitivity highly variable between reports, they already highlighted the detection of mutations in bile cfDNA samples as a promising strategy to improve the diagnosis of malignant diseases. Furthermore, it was also noted that although the presence of KRAS mutations could not be used as a diagnosis of CCA in PSC patients, it should be considered as a risk factor for malignancy development, which might have implications for the timing of liver transplantation [111].
More recent work has shifted to the use of NGS panels, thus broadening the range of genomic alterations studied, and highlighting the inclusion of mutations for which targeted therapies exist. Firstly, in 2018 Kinugasa and colleagues [56] analyzed 49 genes in bile samples of patients with gallbladder cancer using a custom enrichment panel and demonstrated its better performance compared to cytology (58.3% versus 45.8%) [56]. Similar results using other commercialized or custom cancer related-gene panels were obtained for patients with different pancreato-biliary tumors, reaching sensitivities ranging from 53% to 96.2% [23, 55, 112–114]. These panels include genes frequently mutated in multiple cancer types, including biliopancreatic malignancies such as TP53, KRAS, cyclin-dependent kinase inhibitor 2A (CDKN2A), erb-b2 receptor tyrosine kinase 2 (ERBB2), erb-b2 receptor tyrosine kinase 3 (ERBB3), mothers against decapentaplegic homolog 4 (SMAD4), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA), among others.
Moreover, Arechederra et al. [20] demonstrated that the detection of mutations in bile cfDNA is a very sensitive and specific method to early diagnose malignancy in patients with an initial clinical-pathological diagnosis of biliary stricture of benign or indeterminate origin. When these patients had their bile cfDNA analyzed at the time of initial diagnosis with an NGS panel called the Bilemut assay, a sensitivity of 100% sensitivity is achieved. Interestingly, of the four patients that harboured mutations but did not develop malignancy after the 1-year follow-up, one of them developed PDAC after extended follow-up and another died without a definitive diagnosis [20].
Most of the studies reported above compare the performance obtained with paired tissue and bile samples, and all concur in that more genomic alterations could be detected in bile than in the corresponding tissue, in agreement with the fact that LB recapitulates tumor heterogeneity better than tissue biopsy [22, 55, 56, 112]. In addition, the better sensitivity for cancer detection obtained in bile than in plasma is well demonstrated [21, 22, 55]. All in all, the increased sensitivity of new technologies, as well as the possibility to assess the presence of druggable-mutations, has boosted the potential utility of bile for both diagnosis of malignancy and targeting therapy.
DNA methylations
Together with the genetic landscape of mutations, the rewiring of the epigenomic features is a common and early event in carcinogenesis. In particular, the methylation landscape is generally characterized by diffuse DNA hypomethylation and a focal CpG island hypermethylation [115]. Importantly, DNA methylation patterns show cell type specificity, are highly stable as covalent modifications, and can be detected in cfDNA [116–119]. However, to date, few studies have focused on the differential detection of DNA methylation markers in bile.
The first report from 2003 showed a high hypermethylation of cyclin-dependent kinase inhibitor 2A (CDKN2A) (p16INK4a and p14ARF) promoter region in bile samples from patients with malignant biliary diseases compared with those harboring benign biliary disorders (53.5% and 6%, respectively) [120]. Of note, this study included 11 patients with PSC and showed a similar prevalence of methylation than in patients with malignancy [120]. These results, as those regarding KRAS mutation in PSC patients [111], suggest that the presence of these methylations in patients with PSC should be treated with caution as it may indicate an increased risk but not a diagnosis of CCA. Shin et al. [121] identified and validated that the methylation status of five genes [cyclin D2 (CCND2), cadherin 13 (CDH13), glutamate ionotropic receptor NMDA type subunit 2B (GRIN2B), runt-related transcription factor 3 (RUNX3) and twist-related protein 1 (TWIST1)] in bile detected the presence of extrahepatic CCA (eCCA) with a sensitivity of 83%, which was far higher than that of bile citology (46%). Similarly, the methylation of dickkopf WNT signaling pathway inhibitor 3 (DKK3), p16, secreted frizzled-related protein 2 (SFRP2), dickkopf WNT signaling pathway inhibitor 2 (DKK2), neuronal pentraxin 2 (NPTX2) and preproenkephalin (ppENK) was more frequently detected in the bile from patients with malignant than in bile from patients with benign biliary strictures [122]. We have also recently identified a DNA methylation marker (HOXD8) which accurately detects biliary tract cancers both in tissue and bile samples [123]. When combining the detection of mutations in 23 genes and methylation levels in 44 genes, an assay named BileScreen, a 90% sensitivity and 80% specificity in detecting pancreatobiliary tract cancer was obtained [124]. Finally, Vedeld and colleagues [82] aimed to establish a DNA methylation panel for the early and specific detection of CCA in PSC patients. Remarkably, these authors reported that the methylation status of cysteine dioxygenase type 1 (CDO1), cannabinoid receptor interacting protein 1 (CNRIP1), septin 9 (SEPT9), and vimentin (VIM) reached a sensibility of 100% for detecting CCA in patients with underlying PSC up to 12 months prior to a confirmed CCA diagnosis [82].
Organoids BPT
As from other tissue origins, liver organoids have emerged as a novel in vitro tool with multiple applications, from the study of liver pathophysiology to drug discovery, personalized drug recommendation, toxicity assessment, or regenerative medicine [125]. Liver organoids can be derived from different sources including tissue biopsies, pluripotent stem cells, and even bile samples [125]. Importantly, in the context of the biliary tree, the difficulty in accessing healthy cholangiocytes and the limited samples from end-stage disease, make this model even more important. In 2019, a detailed protocol to establish patients-derived organoids from human bile collected via ERCP of patients with PSC was published [126]. The authors demonstrated that these bile-derived organoids could be expanded and maintained long-term in vitro and that they retained a biliary phenotype [126]. Shortly after, Roos and colleagues [70] demonstrated a successful organoid establishment using bile collected via not only ERCP but also from gallbladder after resection and also by percutaneous transhepatic cholangiopathy drainage (PTCD). Interestingly, organoids initiating cells in bile are likely of extrahepatic origin and not intrahepatic, and although all organoids initiated from these three sources of bile show features similar to cholangiocytes, regional-specific characteristics (common bile duct or gallbladder) were highlighted depending on the procedure employed to obtain the bile [70]. Importantly, these bile-cholangiocyte organoids were capable of repopulating human extrahepatic bile duct scaffolds [70], suggesting their future application in the field of regenerative medicine. Shifting to patient-derived organoids, transcriptomic analysis of PSC organoids revealed an immune-reactive phenotype compared to non-PSC organoids, demonstrating that these organoids preserve disease characteristics [126–128]. Although not much information exists on bile-derived tumor organoids, when available, they will be a very useful tool for molecular and pharmacological studies. In the context of bile collected from patients with cancer, and similar to when working with tissue samples [129], a mixture of organoids derived from both normal bile duct epithelium and bile duct cancer cells could be obtained. Kinoshita and colleagues [130] established organoids from the bile of biliary cancer patients and proposed three protocols to enrich the culture in cancer organoids: by repeat passage, by xenografting, or by selection with a TP53 inhibitor when harboring a TP53 mutation.
Conclusions
Many studies and other published evidence, including clinical trials, are demonstrating the potential of LB strategies in the management of patients mainly in the oncology field. Although several biofluids are initially available as LB matrices, most studies are based on blood, due to its ease of collection and informative potential. However, other biofluids such as bile, given its complex and specific composition, which has been shown to vary in pathological conditions, and its anatomical location, in a confined and lesion adjacent space, can provide increased sensitivity and specificity in different clinical situations.
Although much remains to be done, evidence compiled in this review demonstrates the potential of bile analysis to provide molecular information useful for the diagnosis, prognosis, and therapeutic guidance of different diseases, the outcome of pharmacological studies, or the establishment of cellular disease models and regenerative medicine strategies.
In this scenario, bile collection could be implemented in clinical practice in those patients undergoing procedures in which the biliary tract is exposed or accessible. Furthermore, it is not unreasonable to imagine that new strategies might be developed to collect bile in those patients in whom it could be informative to improve their management.
In the near future, standardized LB protocols and sensitive methodologies will revolutionize the management of cancer patients. Bile analysis may be part of this revolution and contribute also to the management of other diseases.
Abbreviations
| BPT: |
biliopancreatic tumors |
| CCA: |
cholangiocarcinoma |
| cfDNA: |
circulating cell-free DNA |
| ERCP: |
endoscopic retrograde cholangiopancreatography |
| EUS: |
endoscopic ultrasound |
| KRAS : |
Kirsten rat sarcoma |
| LB: |
liquid biopsy |
| NGS: |
next generation sequencing |
| PBD: |
percutaneous biliary drainage |
| PGBA: |
percutaneous gallbladder aspiration |
| PGBD: |
percutaneous gallbladder drainage |
| PSC: |
primary sclerosing cholangitis |
| TP53 : |
tumor protein 53 |
Declarations
Author contributions
MA: Conceptualization, Investigation, Writing—original draft, Writing—review & editing, Supervision. MR, DO, MAÁ, and JU: Investigation, Writing—original draft, Writing—review & editing. CB: Conceptualization, Investigation, Writing—original draft, Writing—review & editing, Supervision. All authors read and approved the submitted version.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent to publication
Not applicable.
Availability of data and materials
Not applicable.
Funding
This work was supported by European Union Horizon 2020 Transcan project [2022-784-024] to MA, MAÁ, and CB; Thermo Fisher 2022 Oncomine Clinical Research Grant to MAÁ and CB; AECT Eurorregión Nueva Aquitania Euskadi Navarra “Innovación Eurorregional” [2020/101] and [2023/2] to MAÁ and CB; MCIN/AEI/10.13039/501100011033 grants [PID2019-104265RB-I00] and [PID2022-137181OB-I00] to CB; Instituto de Salud Carlos III (ISCIII), [PI22/00471] which co-funded by the European Union to MA; the Scientific Foundation of the Spanish Association Against Cancer [INVES223049AREC] to MA; Departamento de Salud Gobierno de Navarra [42/2021] to JU. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Copyright
© The Author(s) 2024.