Biodegradation of Fumonisins by the Consecutive Action of a Fusion Enzyme
Abstract
:1. Introduction
2. Results
2.1. Cloning of the Fusion Gene FUMDI and Construction of Recombinant Plasmids
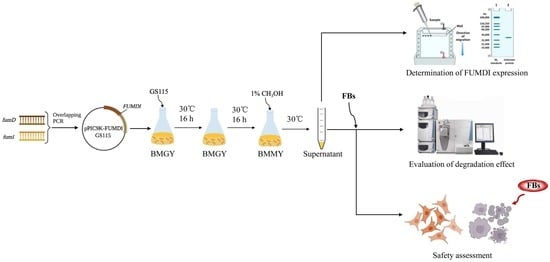
2.2. Expression and Purification of the FUMDI Enzyme
2.3. FB1 Degradation by the FUMDI Enzyme
2.4. Reverse Effect of Fusion Enzyme FUMDI on FB1-Induced Cell Viability Decrease
2.5. Effect of Fusion Enzyme and Its Final Products on Cell Apoptosis, Oxidative Stress, and Endoplasmic Reticulum (ER) Stress
2.6. Fusion Enzyme FUMDI Degradation Efficiency on Other FBs
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Chemicals and Reaction System
5.2. Plasmids, Strains, Cultural Conditions, and DNA Manipulations
5.3. Construction of Fusion Genes and Recombinant Plasmids
5.4. Expression of Fusion Enzyme FUMDI in Recombinant P. pastoris GS115
5.5. Degradation of FB1 and HFB1 by FUMDI
5.6. Mycotoxin Extraction and LC–MS/MS Analysis
5.7. In Vitro Cell Cytotoxicity Assay
5.7.1. Measurement of Cell Viability
5.7.2. Total RNA Extraction and qRT–PCR Determination of Genes Related to Apoptosis and ER Stress
5.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CCK-8 | Cell Counting Kit-8 |
| F. verticillioides | Fusarium verticillioides |
| F. proliferatum | Fusarium proliferatum |
| P. pastoris | Pichia pastoris |
| ER | endoplasmic reticulum |
| FBs | fumonisin Bs |
| FB1 | Fumonisin B1 |
| FB2 | Fumonisin B2 |
| FB3 | Fumonisin B3 |
| GES-1 | human gastric epithelial cell line |
| HFBs | Hydrolyzed Fumonisin Bs |
| HFB1 | Hydrolyzed Fumonisin B1 |
| LC–MS/MS | liquid chromatography–tandem mass spectrometry |
| PCR | double-polymerase chain reaction |
References
- Marín, S.; Magan, N.; Ramos, A.J.; Sanchis, V. Fumonisin-producing strains of fusarium: A review of their ecophysiology. J. Food Prot. 2004, 67, 1792–1805. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wei, Z.; Wang, Y.; Long, M.; Wu, W.; Kuca, K. Fumonisin B(1): Mechanisms of toxicity and biological detoxification progress in animals. Food Chem. Toxicol. 2021, 149, 111977. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, S.Z.; Rehman, B.; Selamat, J.; Akram, N.; Ahmad, M.N.; Sanny, M.; Sukor, R.; Samsudin, N.I. Assessment of Fumonisin B1 Concentrations in Wheat and Barley Products in the Punjab Region of Pakistan. J. Food Prot. 2020, 83, 1284–1288. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, T.F.A.; Dalla Lana, D.F.; Quintana, L.D.; Schmitt, E.G.; Kaminski, T.A.; Paula, F.R.; Fuentefria, A.M.; Machado, M.M.; Souza de Oliveira, L.F. Fumonisin B(1) induces toxicity in human leukocytes at low concentrations: Are computational studies effective to determine biosafety? Toxicon 2020, 182, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wei, Y.; Xie, Y.; Yan, C.; Du, H.; Li, Z. Ochratoxin A and fumonisin B(1) exhibit synergistic cytotoxic effects by inducing apoptosis on rat liver cells. Toxicon 2020, 181, 19–27. [Google Scholar] [CrossRef]
- Yu, S.; Jia, B.; Liu, N.; Yu, D.; Zhang, S.; Wu, A. Fumonisin B1 triggers carcinogenesis via HDAC/PI3K/Akt signalling pathway in human esophageal epithelial cells. Sci. Total Environ. 2021, 787, 147405. [Google Scholar] [CrossRef]
- Müller, S.; Dekant, W.; Mally, A. Fumonisin B1 and the kidney: Modes of action for renal tumor formation by fumonisin B1 in rodents. Food Chem. Toxicol. 2012, 50, 3833–3846. [Google Scholar] [CrossRef]
- Voss, K.A.; Plattner, R.D.; Riley, R.T.; Meredith, F.I.; Norred, W.P. In vivo effects of fumonisin B1-producing and fumonisin B1-nonproducing Fusarium moniliforme isolates are similar: Fumonisins B2 and B3 cause hepato- and nephrotoxicity in rats. Mycopathologia 1998, 141, 45–58. [Google Scholar] [CrossRef]
- Yu, S.; Jia, B.; Liu, N.; Yu, D.; Wu, A. Evaluation of the Individual and Combined Toxicity of Fumonisin Mycotoxins in Human Gastric Epithelial Cells. Int. J. Mol. Sci. 2020, 21, 5917. [Google Scholar] [CrossRef]
- Cendoya, E.; Nichea, M.J.; Monge, M.P.; Sulyok, M.; Chiacchiera, S.M.; Ramirez, M.L. Fumonisin occurrence in wheat-based products from Argentina. Food Addit. Contam. Part B Surveill. 2019, 12, 31–37. [Google Scholar] [CrossRef]
- Aboagye-Nuamah, F.; Kwoseh, C.K.; Maier, D.E. Toxigenic mycoflora, aflatoxin and fumonisin contamination of poultry feeds in Ghana. Toxicon 2021, 198, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Gilbert-Sandoval, I.; Wesseling, S.; Rietjens, I. Occurrence and probabilistic risk assessment of fumonisin b1, fumonisin b2 and deoxynivalenol in Nixtamalized Maize in Mexico City. Toxins 2020, 12, 644. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Jiang, H.; Xu, J.; Zhang, J.; Li, F. Dynamic fumonisin B(2) production by aspergillus niger intented used in food industry in China. Toxins 2017, 9, 217. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.A.; Wang, Y.; Shen, Z.; Li, X.; Saleemi, M.K.; He, C. Mycotoxin contamination and control strategy in human, domestic animal and poultry: A review. Microb. Pathog. 2020, 142, 104095. [Google Scholar] [CrossRef] [PubMed]
- Buszewska-Forajta, M. Mycotoxins, invisible danger of feedstuff with toxic effect on animals. Toxicon 2020, 182, 34–53. [Google Scholar] [CrossRef]
- Voss, K.; Ryu, D.; Jackson, L.; Riley, R.; Gelineau-van Waes, J. Reduction of fumonisin toxicity by extrusion and nixtamalization (alkaline cooking). J. Agric. Food Chem. 2017, 65, 7088–7096. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, Y.; Liu, Z.; Jin, S.; Pan, K.; Liu, H.; Liu, T.; Li, X.; Zhang, C.; Luo, X.; et al. Biological detoxification of fumonisin by a novel carboxylesterase from Sphingomonadales bacterium and its biochemical characterization. Int. J. Biol. Macromol. 2021, 169, 18–27. [Google Scholar] [CrossRef]
- Alberts, J.; Schatzmayr, G.; Moll, W.D.; Davids, I.; Rheeder, J.; Burger, H.M.; Shephard, G.; Gelderblom, W. Detoxification of the fumonisin mycotoxins in Maize: An enzymatic approach. Toxins 2019, 11, 523. [Google Scholar] [CrossRef]
- Hahn, I.; Nagl, V.; Schwartz-Zimmermann, H.E.; Varga, E.; Schwarz, C.; Slavik, V.; Reisinger, N.; Malachová, A.; Cirlini, M.; Generotti, S.; et al. Effects of orally administered fumonisin B₁ (FB₁), partially hydrolysed FB₁, hydrolysed FB₁ and N-(1-deoxy-D-fructos-1-yl) FB₁ on the sphingolipid metabolism in rats. Food Chem. Toxicol. 2015, 76, 11–18. [Google Scholar] [CrossRef]
- Harrer, H.; Laviad, E.L.; Humpf, H.U.; Futerman, A.H. Identification of N-acyl-fumonisin B1 as new cytotoxic metabolites of fumonisin mycotoxins. Mol. Nutr. Food Res. 2013, 57, 516–522. [Google Scholar] [CrossRef]
- Seiferlein, M.; Humpf, H.U.; Voss, K.A.; Sullards, M.C.; Allegood, J.C.; Wang, E.; Merrill, A.H., Jr. Hydrolyzed fumonisins HFB1 and HFB2 are acylated in vitro and in vivo by ceramide synthase to form cytotoxic N-acyl-metabolites. Mol. Nutr. Food Res. 2007, 51, 1120–1130. [Google Scholar] [CrossRef] [PubMed]
- Harrer, H.; Humpf, H.U.; Voss, K.A. In vivo formation of N-acyl-fumonisin B1. Mycotoxin Res. 2015, 31, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Caloni, F.; Spotti, M.; Pompa, G.; Zucco, F.; Stammati, A.; De Angelis, I. Evaluation of fumonisin B(1) and its metabolites absorption and toxicity on intestinal cells line Caco-2. Toxicon 2002, 40, 1181–1188. [Google Scholar] [CrossRef]
- Yu, S.; Jia, B.; Lin, H.; Zhang, S.; Yu, D.; Liu, N.; Wu, A. Effects of fumonisin B and hydrolyzed fumonisin B on growth and intestinal microbiota in broilers. Toxins 2022, 14, 163. [Google Scholar] [CrossRef] [PubMed]
- Ji, C.; Fan, Y.; Zhao, L. Review on biological degradation of mycotoxins. Anim. Nutr. 2016, 2, 127–133. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, I.C.; Seamons, T.R.; Emmons, J.C.; Javdan, S.B.; Deans, T.L. Enhanced regulation of prokaryotic gene expression by a eukaryotic transcriptional activator. Nat. Commun. 2021, 12, 4109. [Google Scholar] [CrossRef]
- Chen, M.; Wang, J.; Lin, L.; Wei, W.; Shen, Y.; Wei, D. High-level expression of a beta-mannanase (manB) in Pichia pastoris GS115 for mannose production with Penicillium brevicompactum fermentation pretreatment of soybean meal. Bioprocess Biosyst. Eng. 2021, 44, 549–561. [Google Scholar] [CrossRef]
- Sun, T.; Yan, P.; Zhan, N.; Zhang, L.; Chen, Z.; Zhang, A.; Shan, A. The optimization of fermentation conditions for Pichia pastoris GS115 producing recombinant xylanase. Eng. Life Sci. 2020, 20, 216–228. [Google Scholar] [CrossRef]
- Zhang, S.; Zhou, S.; Lyu, B.; Qiu, N.; Li, J.; Zhao, Y.; Wu, Y. Dietary exposure to fumonisins and ochratoxins in the Chinese general population during 2007–2020: Results from three consecutive total diet studies. Food Chem. Toxicol. 2022, 159, 112768. [Google Scholar] [CrossRef]
- Keawmanee, P.; Rattanakreetakul, C.; Pongpisutta, R. Microbial reduction of fumonisin B1 by the new isolate serratia marcescens 329-2. Toxins 2021, 13, 638. [Google Scholar] [CrossRef]
- Li, W.; Zhao, H.; Zhuang, R.; Wang, Y.; Cao, W.; He, Y.; Jiang, Y.; Rui, R.; Ju, S. Fumonisin B(1) exposure adversely affects porcine oocyte maturation in vitro by inducing mitochondrial dysfunction and oxidative stress. Theriogenology 2021, 164, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Feijó Corrêa, J.A.; Orso, P.B.; Bordin, K.; Hara, R.V.; Luciano, F.B. Toxicological effects of fumonisin B(1) in combination with other Fusarium toxins. Food Chem. Toxicol. 2018, 121, 483–494. [Google Scholar] [CrossRef] [PubMed]
- Masching, S.; Naehrer, K.; Schwartz-Zimmermann, H.E.; Sărăndan, M.; Schaumberger, S.; Dohnal, I.; Nagl, V.; Schatzmayr, D. Gastrointestinal degradation of fumonisin B₁ by carboxylesterase FumD prevents fumonisin induced alteration of sphingolipid metabolism in Turkey and Swine. Toxins 2016, 8, 84. [Google Scholar] [CrossRef] [PubMed]
- Schertz, H.; Dänicke, S.; Frahm, J.; Schatzmayr, D.; Dohnal, I.; Bichl, G.; Schwartz-Zimmermann, H.E.; Colicchia, S.; Breves, G.; Teifke, J.P.; et al. Biomarker evaluation and toxic effects of an acute oral and systemic fumonisin exposure of pigs with a special focus on dietary fumonisin esterase supplementation. Toxins 2018, 10, 296. [Google Scholar] [CrossRef] [PubMed]
- Grenier, B.; Bracarense, A.P.; Schwartz, H.E.; Lucioli, J.; Cossalter, A.M.; Moll, W.D.; Schatzmayr, G.; Oswald, I.P. Biotransformation approaches to alleviate the effects induced by fusarium mycotoxins in swine. J. Agric. Food Chem. 2013, 61, 6711–6719. [Google Scholar] [CrossRef]
- Xu, X.; Lai, Y.; Hua, Z.-C. Apoptosis and apoptotic body: Disease message and therapeutic target potentials. Biosci. Rep. 2019, 39, BSR20180992. [Google Scholar] [CrossRef]
- Yu, S.; Jia, B.; Yang, Y.; Liu, N.; Wu, A. Involvement of PERK-CHOP pathway in fumonisin B1- induced cytotoxicity in human gastric epithelial cells. Food Chem. Toxicol. 2020, 136, 111080. [Google Scholar] [CrossRef]
- Arumugam, T.; Ghazi, T.; Chuturgoon, A.A. Molecular and epigenetic modes of Fumonisin B(1) mediated toxicity and carcinogenesis and detoxification strategies. Crit. Rev. Toxicol. 2021, 51, 76–94. [Google Scholar] [CrossRef]






| Mycotoxin | Precursor Ion (m/z) | Retention Time (min) | Product Ion (m/z) | Collision Energy (eV) |
|---|---|---|---|---|
| FB1 | 722.4 | 5.16 | 352.3 * 334.0 704.5 | 40 34 18 |
| FB2 | 706.3 | 6.32 | 336.5 * 354.5 318.3 | 29 36 35 |
| FB3 | 706.3 | 5.63 | 336.5 * 354.5 318.3 | 29 36 35 |
| HFB1 | 406.3 | 4.94 | 370.6 * 388.6 | 17 15 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, K.; Yu, S.; Yu, D.; Lin, H.; Liu, N.; Wu, A. Biodegradation of Fumonisins by the Consecutive Action of a Fusion Enzyme. Toxins 2022, 14, 266. https://doi.org/10.3390/toxins14040266
Li K, Yu S, Yu D, Lin H, Liu N, Wu A. Biodegradation of Fumonisins by the Consecutive Action of a Fusion Enzyme. Toxins. 2022; 14(4):266. https://doi.org/10.3390/toxins14040266
Chicago/Turabian StyleLi, Kailin, Song Yu, Dianzhen Yu, Huikang Lin, Na Liu, and Aibo Wu. 2022. "Biodegradation of Fumonisins by the Consecutive Action of a Fusion Enzyme" Toxins 14, no. 4: 266. https://doi.org/10.3390/toxins14040266