Insights into the Impact of Rosmarinic Acid on CHO Cell Culture Improvement through Transcriptomics Analysis
Abstract
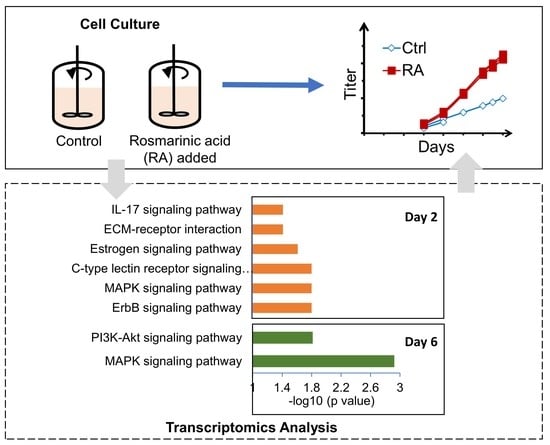
:1. Introduction
2. Materials and Methods
2.1. Cell line, Media, and Upstream Cell Culture Processes
2.2. Upstream Cell Culture Titer and Other In-Process Parameter Assays
2.3. RNA Isolation and Microarray Analysis
2.4. Metabolic Pathway Analysis
3. Results
3.1. RA Enhances Cell Growth and Protein Production in CHO Cells
3.2. RA Treatment Alters the Transcriptional Profile of CHO Cells
3.3. Gene Expression Analysis Reveals RA Modulates Pathways Associated with Apoptosis and Survival
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lu, R.-M.; Hwang, Y.-C.; Liu, I.J.; Lee, C.-C.; Tsai, H.-Z.; Li, H.-J.; Wu, H.-C. Development of therapeutic antibodies for the treatment of diseases. J. Biomed. Sci. 2020, 27, 1. [Google Scholar] [CrossRef] [PubMed]
- Tuccori, M.; Ferraro, S.; Convertino, I.; Cappello, E.; Valdiserra, G.; Blandizzi, C.; Maggi, F.; Focosi, D. Anti-SARS-CoV-2 neutralizing monoclonal antibodies: Clinical pipeline. MAbs 2020, 12, 1854149. [Google Scholar] [CrossRef] [PubMed]
- Kelley, B. Developing therapeutic monoclonal antibodies at pandemic pace. Nat. Biotechnol. 2020, 38, 540–545. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Rehmann, M.S.; Xu, X.; Huang, C.; Tian, J.; Qian, N.-X.; Li, Z.J. Improving titer while maintaining quality of final formulated drug substance via optimization of CHO cell culture conditions in low-iron chemically defined media. mAbs 2018, 10, 488–499. [Google Scholar] [CrossRef]
- Huang, Y.-M.; Hu, W.; Rustandi, E.; Chang, K.; Yusuf-Makagiansar, H.; Ryll, T. Maximizing productivity of CHO cell-based fed-batch culture using chemically defined media conditions and typical manufacturing equipment. Biotechnol. Prog. 2010, 26, 1400–1410. [Google Scholar] [CrossRef]
- Handlogten, M.W.; Zhu, M.; Ahuja, S. Intracellular response of CHO cells to oxidative stress and its influence on metabolism and antibody production. Biochem. Eng. J. 2018, 133, 12–20. [Google Scholar] [CrossRef]
- Yun, Z.; Takagi, M.; Yoshida, T. Combined addition of glutathione and iron chelators for decrease of intracellular level of reactive oxygen species and death of chinese hamster ovary cells. J. Biosci. Bioeng. 2003, 95, 124–127. [Google Scholar] [CrossRef]
- Xu, J.; Rehmann, M.S.; Tian, J.; He, Q.; Chen, J.; Lee, J.; Borys, M.C.; Li, Z.J. Rosmarinic acid, a new raw material, doubled monoclonal antibody titer in cell culture manufacturing. Biochem. Eng. J. 2020, 160. [Google Scholar] [CrossRef]
- Lee, A.P.; Kok, Y.J.; Lakshmanan, M.; Leong, D.; Zheng, L.; Lim, H.L.; Chen, S.; Mak, S.Y.; Ang, K.S.; Templeton, N.; et al. Multi-omics profiling of a CHO cell culture system unravels the effect of culture pH on cell growth, antibody titer and product quality. Biotechnol. Bioeng. 2021, 118, 4305–4316. [Google Scholar] [CrossRef]
- Xu, N.; Ma, C.; Ou, J.; Sun, W.W.; Zhou, L.; Hu, H.; Liu, X.M. Comparative Proteomic Analysis of Three Chinese Hamster Ovary (CHO) Host Cells. Biochem. Eng. J. 2017, 124, 122–129. [Google Scholar] [CrossRef]
- Chen, C.; Le, H.; Follstad, B.; Goudar, C.T. A Comparative Transcriptomics Workflow for Analyzing Microarray Data From CHO Cell Cultures. Biotechnol. J. 2018, 13, 1700228. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Dhara, V.G.; Orzolek, L.D.; Hao, H.; More, A.J.; Lau, E.C.; Betenbaugh, M.J. Elucidating the impact of cottonseed hydrolysates on CHO cell culture performance through transcriptomic analysis. Appl. Microbiol. Biotechnol. 2021, 105, 271–285. [Google Scholar] [CrossRef] [PubMed]
- Datta, P.; Linhardt, R.J.; Sharfstein, S.T. An 'omics approach towards CHO cell engineering. Biotechnol. Bioeng. 2013, 110, 1255–1271. [Google Scholar] [CrossRef] [PubMed]
- Sharfstein, S.T. Omics insights into production-scale bioreactors. Biotechnol. J. 2016, 11, 1124–1125. [Google Scholar] [CrossRef] [Green Version]
- Huang, Z.; Xu, J.; Yongky, A.; Morris, C.S.; Polanco, A.L.; Reily, M.; Borys, M.C.; Li, Z.J.; Yoon, S. CHO cell productivity improvement by genome-scale modeling and pathway analysis: Application to feed supplements. Biochem. Eng. J. 2020, 160, 107638. [Google Scholar] [CrossRef]
- Lakshmanan, M.; Kok, Y.J.; Lee, A.P.; Kyriakopoulos, S.; Lim, H.L.; Teo, G.; Poh, S.L.; Tang, W.Q.; Hong, J.; Tan, A.H.-M.; et al. Multi-omics profiling of CHO parental hosts reveals cell line-specific variations in bioprocessing traits. Biotechnol. Bioeng. 2019, 116, 2117–2129. [Google Scholar] [CrossRef]
- Ali, A.S.; Raju, R.; Kshirsagar, R.; Ivanov, A.R.; Gilbert, A.; Zang, L.; Karger, B.L. Multi-Omics Study on the Impact of Cysteine Feed Level on Cell Viability and mAb Production in a CHO Bioprocess. Biotechnol. J. 2019, 14, 1800352. [Google Scholar] [CrossRef]
- Ali, A.S.; Chen, R.; Raju, R.; Kshirsagar, R.; Gilbert, A.; Zang, L.; Karger, B.L.; Ivanov, A.R. Multi-Omics Reveals Impact of Cysteine Feed Concentration and Resulting Redox Imbalance on Cellular Energy Metabolism and Specific Productivity in CHO Cell Bioprocessing. Biotechnol. J. 2020, 15, 1900565. [Google Scholar] [CrossRef]
- Gao, Y.; Ray, S.; Dai, S.; Ivanov, A.R.; Abu-Absi, N.R.; Lewis, A.M.; Huang, Z.; Xing, Z.; Borys, M.C.; Li, Z.J.; et al. Combined metabolomics and proteomics reveals hypoxia as a cause of lower productivity on scale-up to a 5000-liter CHO bioprocess. Biotechnol. J. 2016, 11, 1190–1200. [Google Scholar] [CrossRef]
- Vodopivec, M.; Lah, L.; Narat, M.; Curk, T. Metabolomic profiling of CHO fed-batch growth phases at 10, 100, and 1000 L. Biotechnol. Bioeng. 2019, 116, 2720–2729. [Google Scholar] [CrossRef]
- Bertrand, V.; Karst, D.J.; Bachmann, A.; Cantalupo, K.; Soos, M.; Morbidelli, M. Transcriptome and proteome analysis of steady-state in a perfusion CHO cell culture process. Biotechnol. Bioeng. 2019, 116, 1959–1972. [Google Scholar] [CrossRef]
- Orellana, C.A.; Martínez, V.S.; MacDonald, M.A.; Henry, M.N.; Gillard, M.; Gray, P.P.; Nielsen, L.K.; Mahler, S.; Marcellin, E. ‘Omics driven discoveries of gene targets for apoptosis attenuation in CHO cells. Biotechnol. Bioeng. 2021, 118, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Jin, M.; Song, H.; Huang, C.; Xu, X.; Tian, J.; Qian, N.-X.; Steger, K.; Lewen, N.S.; Tao, L.; et al. Brown drug substance color investigation in cell culture manufacturing using chemically defined media: A case study. Process. Biochem. 2014, 49, 130–139. [Google Scholar] [CrossRef]
- Jing, Y.; Qian, Y.; Ghandi, M.; He, A.; Borys, M.C.; Pan, S.H.; Li, Z.J. A mechanistic study on the effect of dexamethasone in moderating cell death in Chinese Hamster Ovary cell cultures. Biotechnol. Prog. 2012, 28, 490–496. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Xing, Z.; Lee, S.; Mackin, N.A.; He, A.; Kayne, P.S.; He, Q.; Qian, N.-X.; Li, Z.J. Hypoxia influences protein transport and epigenetic repression of CHO cell cultures in shake flasks. Biotechnol. J. 2014, 9, 1413–1424. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Society. Ser. B (Methodological) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Sha, S.; Bhatia, H.; Yoon, S. An RNA-seq based transcriptomic investigation into the productivity and growth variants with Chinese hamster ovary cells. J. Biotechnol. 2018, 271, 37–46. [Google Scholar] [CrossRef]
- Courtes, F.C.; Lin, J.; Lim, H.L.; Ng, S.W.; Wong, N.S.; Koh, G.; Vardy, L.; Yap, M.G.; Loo, B.; Lee, D.Y. Translatome analysis of CHO cells to identify key growth genes. J. Biotechnol. 2013, 167, 215–224. [Google Scholar] [CrossRef]
- Chen, C.; Le, H.; Goudar, C.T. An automated RNA-Seq analysis pipeline to identify and visualize differentially expressed genes and pathways in CHO cells. Biotechnol. Prog. 2015, 31, 1150–1162. [Google Scholar] [CrossRef]
- Sommeregger, W.; Mayrhofer, P.; Steinfellner, W.; Reinhart, D.; Henry, M.; Clynes, M.; Meleady, P.; Kunert, R. Proteomic differences in recombinant CHO cells producing two similar antibody fragments. Biotechnol. Bioeng. 2016, 113, 1902–1912. [Google Scholar] [CrossRef]
- Sha, S.; Kuang, B.; Yoon, S. Characterization of dynamic regulation in Chinese hamster ovary (CHO) cell cultures in the late exponential phase. Biochem. Eng. J. 2021, 167, 107897. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39 (Suppl. 2), W316–W322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamano, N.; Omasa, T. EGCG improves recombinant protein productivity in Chinese hamster ovary cell cultures via cell proliferation control. Cytotechnology 2018, 70, 1697–1706. [Google Scholar] [CrossRef] [PubMed]
- Toronjo Urquiza, L.; James, D.C.; Nagy, T.; Falconer, R.J. Screening Naturally Occurring Phenolic Antioxidants for Their Suitability as Additives to CHO Cell Culture Media Used to Produce Monoclonal Antibodies. Antioxidants 2019, 8, 159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toronjo-Urquiza, L.; Acosta-Martin, A.E.; James, D.C.; Nagy, T.; Falconer, R.J. Resveratrol addition to Chinese hamster ovary cell culture media: The effect on cell growth, monoclonal antibody synthesis, and its chemical modification. Biotechnol. Prog. 2020, 36, e2940. [Google Scholar] [CrossRef]
- Toronjo-Urquiza, L.; Acosta-Martin, A.E.; James, D.C.; Nagy, T.; Falconer, R.J. The use of catechins in Chinese hamster ovary cell media for the improvement of monoclonal antibody yields and a reduction of acidic species. Biotechnol. Prog. 2020, 36, e2980. [Google Scholar] [CrossRef] [PubMed]
- Ha, T.K.; Hansen, A.H.; Kol, S.; Kildegaard, H.F.; Lee, G.M. Baicalein Reduces Oxidative Stress in CHO Cell Cultures and Improves Recombinant Antibody Productivity. Biotechnol. J. 2018, 13, e1700425. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.; Tian, J.; Tan, Z.; Chen, J.; Zhang, N.; Huang, Y.; Vandermark, E.; Lee, J.; Borys, M.; Li, Z.J. Modulating cell culture oxidative stress reduces protein glycation and acidic charge variant formation. MAbs 2019, 11, 205–216. [Google Scholar] [CrossRef] [Green Version]
- Hossler, P.; Wang, M.; McDermott, S.; Racicot, C.; Chemfe, K.; Zhang, Y.; Chumsae, C.; Manuilov, A. Cell culture media supplementation of bioflavonoids for the targeted reduction of acidic species charge variants on recombinant therapeutic proteins. Biotechnol. Prog. 2015, 31, 1039–1052. [Google Scholar] [CrossRef]
- Hartley, F.; Walker, T.; Chung, V.; Morten, K. Mechanisms driving the lactate switch in Chinese hamster ovary cells. Biotechnol. Bioeng. 2018, 115, 1890–1903. [Google Scholar] [CrossRef]
- Goswami, J.; Sinskey, A.J.; Steller, H.; Stephanopoulos, G.N.; Wang, D.I.C. Apoptosis in batch cultures of Chinese Hamster Ovary cells. Biotechnol. Bioeng. 1999, 62, 632–640. [Google Scholar] [CrossRef] [Green Version]
- Mastrangelo, A.J.; Betenbaugh, M.J. Overcoming apoptosis: New methods for improving protein-expression systems. Trends Biotechnol. 1998, 16, 88–95. [Google Scholar] [CrossRef]
- Grilo, A.L.; Mantalaris, A. Apoptosis: A mammalian cell bioprocessing perspective. Biotechnol Adv. 2019, 37, 459–475. [Google Scholar] [CrossRef] [PubMed]
- Sekine, Y.; Hatanaka, R.; Watanabe, T.; Sono, N.; Iemura, S.; Natsume, T.; Kuranaga, E.; Miura, M.; Takeda, K.; Ichijo, H. The Kelch repeat protein KLHDC10 regulates oxidative stress-induced ASK1 activation by suppressing PP5. Mol. Cell 2012, 48, 692–704. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Quan, J.-H.; Lee, Y.-H.; Shin, D.-W.; Cha, G.-H. Toxoplasma gondii Proliferation Require Down-Regulation of Host Nox4 Expression via Activation of PI3 Kinase/Akt Signaling Pathway. PLoS ONE 2013, 8, e66306. [Google Scholar] [CrossRef] [Green Version]
- Redza-Dutordoir, M.; Averill-Bates, D.A. Activation of apoptosis signalling pathways by reactive oxygen species. Biochim. Biophys. Acta 2016, 1863, 2977–2992. [Google Scholar] [CrossRef]
- Reczek, C.R.; Chandel, N.S. The Two Faces of Reactive Oxygen Species in Cancer. Annu. Rev. Cancer Biol. 2017, 1, 79–98. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, X.; Hu, D. Furanodienone induces G0/G1 arrest and causes apoptosis via the ROS/MAPKs-mediated caspase-dependent pathway in human colorectal cancer cells: A study in vitro and in vivo. Cell Death Dis. 2017, 8, e2815. [Google Scholar] [CrossRef] [Green Version]
- Perillo, B.; Di Donato, M.; Pezone, A.; Di Zazzo, E.; Giovannelli, P.; Galasso, G.; Castoria, G.; Migliaccio, A. ROS in cancer therapy: The bright side of the moon. Exp. Mol. Med. 2020, 52, 192–203. [Google Scholar] [CrossRef]
- Birben, E.; Sahiner, U.M.; Sackesen, C.; Erzurum, S.; Kalayci, O. Oxidative Stress and Antioxidant Defense. World Allergy Organ. J. 2012, 5, 9–19. [Google Scholar] [CrossRef] [Green Version]
- Cost, G.J.; Freyvert, Y.; Vafiadis, A.; Santiago, Y.; Miller, J.C.; Rebar, E.; Collingwood, T.N.; Snowden, A.; Gregory, P.D. BAK and BAX deletion using zinc-finger nucleases yields apoptosis-resistant CHO cells. Biotechnol. Bioeng. 2010, 105, 330–340. [Google Scholar] [CrossRef] [PubMed]
- Chiang, G.G.; Sisk, W.P. Bcl-x(L) mediates increased production of humanized monoclonal antibodies in Chinese hamster ovary cells. Biotechnol. Bioeng. 2005, 91, 779–792. [Google Scholar] [CrossRef]
- Meents, H.; Enenkel, B.; Eppenberger, H.M.; Werner, R.G.; Fussenegger, M. Impact of coexpression and coamplification of sICAM and antiapoptosis determinants bcl-2/bcl-x(L) on productivity, cell survival, and mitochondria number in CHO-DG44 grown in suspension and serum-free media. Biotechnol. Bioeng. 2002, 80, 706–716. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Han, L.; Zong, H.; Ding, K.; Yuan, Y.; Bai, J.; Zhou, Y.; Zhang, B.; Zhu, J. Enhanced production of anti-PD1 antibody in CHO cells through transient co-transfection with anti-apoptotic genes Bcl-x L and Mcl-1. Bioprocess. Biosyst Eng. 2018, 41, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Sung, Y.H.; Lee, J.S.; Park, S.H.; Koo, J.; Lee, G.M. Influence of co-down-regulation of caspase-3 and caspase-7 by siRNAs on sodium butyrate-induced apoptotic cell death of Chinese hamster ovary cells producing thrombopoietin. Metab. Eng. 2007, 9, 452–464. [Google Scholar] [CrossRef]
- Lee, J.S.; Ha, T.K.; Park, J.H.; Lee, G.M. Anti-cell death engineering of CHO cells: Co-overexpression of Bcl-2 for apoptosis inhibition, Beclin-1 for autophagy induction. Biotechnol. Bioeng. 2013, 110, 2195–2207. [Google Scholar] [CrossRef]
- Yuan, L.; Wang, J.; Xiao, H.; Xiao, C.; Wang, Y.; Liu, X. Isoorientin induces apoptosis through mitochondrial dysfunction and inhibition of PI3K/Akt signaling pathway in HepG2 cancer cells. Toxicol. Appl. Pharmacol. 2012, 265, 83–92. [Google Scholar] [CrossRef]
- Chang, F.; Lee, J.T.; Navolanic, P.M.; Steelman, L.S.; Shelton, J.G.; Blalock, W.L.; Franklin, R.A.; McCubrey, J.A. Involvement of PI3K/Akt pathway in cell cycle progression, apoptosis, and neoplastic transformation: A target for cancer chemotherapy. Leukemia 2003, 17, 590–603. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.M.; Ma, J.Q.; Sun, Y.Z. Puerarin protects rat kidney from lead-induced apoptosis by modulating the PI3K/Akt/eNOS pathway. Toxicol. Appl. Pharmacol. 2012, 258, 330–342. [Google Scholar] [CrossRef]
- Granado-Serrano, A.B.; Angeles Martín, M.; Izquierdo-Pulido, M.; Goya, L.; Bravo, L.; Ramos, S. Molecular Mechanisms of (−)-Epicatechin and Chlorogenic Acid on the Regulation of the Apoptotic and Survival/Proliferation Pathways in a Human Hepatoma Cell Line. J. Agric. Food Chem. 2007, 55, 2020–2027. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Wang, H.-G. The protein kinase PKB/Akt regulates cell survival and apoptosis by inhibiting Bax conformational change. Oncogene 2001, 20, 7779–7786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, S.O.; Lee, G.M. Effect of Akt overexpression on programmed cell death in antibody-producing Chinese hamster ovary cells. J. Biotechnol. 2009, 139, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.-Z. Prox1 Facilitates Transfected CHO Cell Proliferation through Activation of the AKT Signaling Pathway. Int. J. Biomed. Sci. 2010, 6, 49–59. [Google Scholar]
- Coleman, O.; Suda, S.; Meiller, J.; Henry, M.; Riedl, M.; Barron, N.; Clynes, M.; Meleady, P. Increased growth rate and productivity following stable depletion of miR-7 in a mAb producing CHO cell line causes an increase in proteins associated with the Akt pathway and ribosome biogenesis. J. Proteom. 2019, 195, 23–32. [Google Scholar] [CrossRef] [PubMed]



| Pathway | Category | Adjusted p-Value | |
|---|---|---|---|
| RA-D2 vs. Control-D2 | ErbB signaling pathway | Environmental Information Processing/Signal transduction | 0.0240 |
| Parathyroid hormone synthesis, secretion and action | Organismal Systems/Endocrine system | 0.0240 | |
| Hepatitis B | Human Diseases/Infectious disease: viral | 0.0240 | |
| Colorectal cancer | Human Diseases/Cancer: specific types | 0.0240 | |
| MAPK signaling pathway | Environmental Information Processing/Signal transduction | 0.0240 | |
| Axon guidance | Organismal Systems/Development and regeneration | 0.0240 | |
| C-type lectin receptor signaling pathway | Organismal Systems/Immune system | 0.0240 | |
| Bladder cancer | Human Diseases/Cancer: specific types | 0.0240 | |
| MicroRNAs in cancer | Human Diseases/Cancer: overview | 0.0299 | |
| Estrogen signaling pathway | Organismal Systems/Endocrine system | 0.0335 | |
| Human T-cell leukemia virus 1 infection | Human Diseases/Infectious disease: viral | 0.0350 | |
| Fluid shear stress and atherosclerosis | Human Diseases/Cardiovascular disease | 0.0374 | |
| ECM-receptor interaction | Environmental Information Processing/Signaling molecules and interaction | 0.0479 | |
| IL-17 signaling pathway | Organismal Systems/Immune system | 0.0479 | |
| RA-D6 vs. Control-D6 | MAPK signaling pathway | Environmental Information Processing/Signal transduction | 0.0012 |
| MicroRNAs in cancer | Human Diseases/Cancer: overview | 0.0028 | |
| Proteoglycans in cancer | Human Diseases/Cancer: overview | 0.0089 | |
| PI3K-Akt signaling pathway | Environmental Information Processing/Signal transduction | 0.0153 | |
| Pathways in cancer | Human Diseases/Cancer: overview | 0.0153 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Z.; Xu, J.; Tian, J.; Aron, K.L.; Qian, Y.; Borys, M.C.; Li, Z.; Pendse, G. Insights into the Impact of Rosmarinic Acid on CHO Cell Culture Improvement through Transcriptomics Analysis. Processes 2022, 10, 533. https://doi.org/10.3390/pr10030533
Huang Z, Xu J, Tian J, Aron KL, Qian Y, Borys MC, Li Z, Pendse G. Insights into the Impact of Rosmarinic Acid on CHO Cell Culture Improvement through Transcriptomics Analysis. Processes. 2022; 10(3):533. https://doi.org/10.3390/pr10030533
Chicago/Turabian StyleHuang, Zhuangrong, Jianlin Xu, Jun Tian, Kathryn L. Aron, Yueming Qian, Michael C. Borys, Zhengjian Li, and Girish Pendse. 2022. "Insights into the Impact of Rosmarinic Acid on CHO Cell Culture Improvement through Transcriptomics Analysis" Processes 10, no. 3: 533. https://doi.org/10.3390/pr10030533