Direct Determination of Chitosan–Mucin Interactions Using a Single-Molecule Strategy: Comparison to Alginate–Mucin Interactions
Abstract
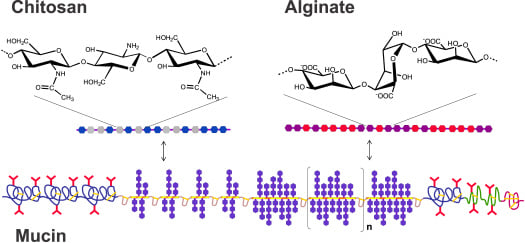
:1. Introduction

2. Experimental Section
2.1. Biopolymer Samples
| Polysaccharide | Properties & | |||||||
|---|---|---|---|---|---|---|---|---|
| Alginates | Source | FG | FM | FGG | FMG | NG > 1 | [η] (mL/g) | Mw (kg/mol) |
| L. hyperborea, stipe | 0.65 | 0.35 | 0.53 | 0.12 | 11 | 585 | 114 | |
| M. pyriferia | 0.38 | 0.62 | 0.19 | 0.20 | 5 | 1005 | 220 | |
| Chitosans | FA | FD | Mw (kg/mol) | |||||
| 0.01 | 0.99 | 610 | 162 | |||||
| 0.49 | 0.51 | 1090 | 250 | |||||
2.2. Immobilization of Mucin, Chitosans and Alginates
2.3. Determination of Molecular Pair Interactions
2.4. Analysis of Chitosan–Mucin and Alginate–Mucin Unbinding Data
3. Results and Discussion
3.1. Fraction of Force-Retraction Cycles with Molecular Interactions

3.2. Mucin-Chitosan Forced Unbinding Curves


3.3. Energy Landscape of the Mucin–Chitosan Interactions

| Polymer | Solvent pH | Range of rf (nN/s) | <xβ> (nm) | <τ0> (s) | <ΔG> (kBT) | Pred. R2 |
|---|---|---|---|---|---|---|
| Chitosan (FA = 0.49) | 5.5 | 0.64–1.89 | 1.18 ± 0.08 | 3.41 ± 1.01 | 7.26 ± 0.33 | 0.80–0.95 |
| 5.5 | 2.01–7.35 | 0.20 ± 0.02 | 0.07 ± 0.005 | 2.79 ± 0.07 | 0.90–0.97 | |
| 6.9 | 0.68–1.89 | 0.98 ± 0.06 | 1.07 ± 0.22 | 5.96 ± 0.22 | 0.80–0.94 | |
| 6.9 | 2.09–9.46 | 0.12 ± 0.01 | 0.06 ± 0.002 | 2.49 ± 0.04 | 0.89–0.97 | |
| Chitosan (FA = 0.01) | 5.5 | 1.02–2.57 | 1.27 ± 0.05 | 7.84 ± 1.78 | 8.69 ± 0.28 | 0.80–0.90 |
| 5.5 | 2.84–7.60 | 0.21 ± 0.01 | 0.07 ± 0.003 | 2.82 ± 0.08 | 0.87–0.98 | |
| 6.9 | 0.55–1.90 | 1.20 ± 0.08 | 10.5 ± 3.2 | 8.77 ± 0.36 | 0.74–0.83 | |
| 6.9 | 2.08–5.36 | 0.15 ± 0.02 | 0.07 ± 0.004 | 2.52 ± 0.06 | 0.84–0.98 | |
| Alginate (FG = 0.65) | 6.9 | 0.32–4.96 | 0.48 ± 0.06 | 2.78 ± 1.01 | 7.64 ± 0.18 | 0.92–0.98 |

3.4. Mucin–Alginate Interactions


3.5. Nature of Mucin–Chitosan and Mucin–Alginate Interactions
3.6. Relevance for Mucoadhesion
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Khutoryanskiy, V.V. Advances in mucoadhesion and mucoadhesive polymers. Macromol. Biosci. 2011, 11, 748–764. [Google Scholar] [CrossRef] [PubMed]
- Sogias, I.A.; Williams, A.C.; Khutoryanskiy, V.V. Why is chitosan mucoadhesive? Biomacromolecules 2008, 9, 1837–1842. [Google Scholar] [CrossRef] [PubMed]
- Alain, D.; Monique, D. Chitosan, Polymeric Biomaterials, Revised and Expanded; CRC Press: Boca Raton, FL, USA, 2001. [Google Scholar]
- Minke, R.; Blackwell, J. The structure of α-chitin. J. Mol. Biol. 1978, 120, 167–181. [Google Scholar] [CrossRef] [PubMed]
- Sikorski, P.; Hori, R.; Wada, M. Revisit of α-chitin crystal structure using high resolution X-ray diffraction data. Biomacromolecules 2009, 10, 1100–1105. [Google Scholar] [CrossRef] [PubMed]
- Fukamizo, T. Chitinolytic enzymes: Catalysis, substrate binding, and their application. Curr. Protein Pept. Sci. 2000, 1, 105–124. [Google Scholar] [CrossRef] [PubMed]
- Ohnuma, T.; Umemoto, N.; Kondo, K.; Numata, T.; Fukamizo, T. Complete subsite mapping of a “loopful” GH19 chitinase from rye seeds based on its crystal structure. FEBS Lett. 2013, 587, 2691–2697. [Google Scholar] [CrossRef] [PubMed]
- Stokke, B.; Varum, K.; Holme, H.; Hjerde, R.; Smidsrod, O. Sequence specificities for lysozyme depolymerization of partially N-acetylated chitosans. Can. J. Chem. 1995, 73, 1972–1981. [Google Scholar] [CrossRef]
- Horn, S.J.; Sorbotten, A.; Synstad, B.; Sikorski, P.; Sorlie, M.; Varum, K.M.; Eijsink, V.G.H. Endo/exo mechanism and processivity of family 18 chitinases produced by serratia marcescens. FEBS J. 2006, 273, 491–503. [Google Scholar] [CrossRef] [PubMed]
- Caridade, S.G.; Monge, C.; Gilde, F.; Boudou, T.; Mano, J.F.; Picart, C. Free-standing polyelectrolyte membranes made of chitosan and alginate. Biomacromolecules 2013, 14, 1653–1660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maurstad, G.; Mørch, Y.A.; Bausch, A.R.; Stokke, B.T. Polyelectrolyte layer interpenetration and swelling of alginate-chitosan multilayers studied by dual wavelength reflection interference contrast microscopy. Carbohydr. Polym. 2008, 71, 672–681. [Google Scholar] [CrossRef]
- Picart, C.; Schneider, A.; Etienne, O.; Mutterer, J.; Schaaf, P.; Egles, C.; Jessel, N.; Voegel, J.C. Controlled degradability of polysaccharide multilayer films in vitro and in vivo. Adv. Funct. Mater. 2005, 15, 1771–1780. [Google Scholar] [CrossRef]
- Schneider, A.; Richert, L.; Francius, G.; Voegel, J.C.; Picart, C. Elasticity, biodegradability and cell adhesive properties of chitosan/hyaluronan multilayer films. Biomed. Mater. 2007, 2. [Google Scholar] [CrossRef]
- Neto, A.I.; Cibrao, A.C.; Correia, C.R.; Carvalho, R.R.; Luz, G.M.; Ferrer, G.G.; Botelho, G.; Picart, C.; Alves, N.M.; Mano, J.F. Nanostructured polymeric coatings based on chitosan and dopamine-modified hyaluronic acid for biomedical applications. Small 2014, 10, 2459–2469. [Google Scholar] [CrossRef] [PubMed]
- Almodovar, J.; Place, L.W.; Gogolski, J.; Erickson, K.; Kipper, M.J. Layer-by-layer assembly of polysaccharide-based polyelectrolyte multilayers: A spectroscopic study of hydrophilicity, composition, and ion pairing. Biomacromolecules 2011, 12, 2755–2765. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Thormann, E.; Claesson, P.M.; Tyrode, E. Surface grafted chitosan gels. Part I. Molecular insight into the formation of chitosan and poly(acrylic acid) multi layers. Langmuir 2014, 30, 8866–8877. [Google Scholar] [CrossRef] [PubMed]
- Koping-Hoggard, M.; Varum, K.M.; Issa, M.; Danielsen, S.; Christensen, B.E.; Stokke, B.T.; Artursson, P. Improved chitosan-mediated gene delivery based on easily dissociated chitosan polyplexes of highly defined chitosan oligomers. Gene Ther. 2004, 11, 1441–1452. [Google Scholar] [CrossRef] [PubMed]
- Strand, S.P.; Danielsen, S.; Christensen, B.E.; Varum, K.M. Influence of chitosan structure on the formation and stability of DNA-chitosan polyelectrolyte complexes. Biomacromolecules 2005, 6, 3357–3366. [Google Scholar] [CrossRef] [PubMed]
- Rantala, J.K.; Makela, R.; Aaltola, A.R.; Laasola, P.; Mpindi, J.P.; Nees, M.; Saviranta, P.; Kallioniemi, O. A cell spot microarray method for production of high density sirna transfection microarrays. BMC Genomics 2011, 12. [Google Scholar] [CrossRef] [PubMed]
- MacLaughlin, F.C.; Mumper, R.J.; Wang, J.; Tagliaferri, J.M.; Gill, I.; Hinchchliffe, M.; Rolland, A.P. Chitosan and depolymerized chitosan oligomers as condensing carriers for in vivo plasmid delivery. J. Controll. Release 1998, 56, 259–272. [Google Scholar] [CrossRef]
- Malmstrom, J.; Lovmand, J.; Kristensen, S.; Sundh, M.; Duch, M.; Sutherland, D.S. Focal complex maturation and bridging on 200 nm vitronectin but not fibronectin patches reveal different mechanisms of focal adhesion formation. Nano Lett. 2011, 11, 2264–2271. [Google Scholar] [CrossRef] [PubMed]
- Agirre, M.; Zarate, J.; Ojeda, E.; Puras, G.; Desbrieres, J.; Pedraz, J.L. Low molecular weight chitosan (lmwc)-based polyplexes for pdna delivery: From bench to bedside. Polymers 2014, 6, 1727–1755. [Google Scholar] [CrossRef]
- Köping-Höggård, M.; Mel’nikova, Y.S.; Vårum, K.M.; Lindman, B.; Artursson, P. Relationship between the physical shape and the efficiency of oligomeric chitosan as a gene delivery system in vitro and in vivo. J. Gene Med. 2003, 5, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Khong, T.T.; Aarstad, O.A.; Skjak-Braek, G.; Draget, K.I.; Varum, K.M. Gelling concept combining chitosan and alginate-proof of principle. Biomacromolecules 2013, 14, 2765–2771. [Google Scholar] [CrossRef] [PubMed]
- Bernkop-Schnurch, A.; Dunnhaupt, S. Chitosan-based drug delivery systems. Eur. J. Pharm. Biopharm. 2012, 81, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Sigurdsson, H.H.; Loftsson, T.; Lehr, C.M. Assessment of mucoadhesion by a resonant mirror biosensor. Int. J. Pharm. 2006, 325, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Smart, J.D. The basics and underlying mechanisms of mucoadhesion. Adv. Drug Deliv. Rev. 2005, 57, 1556–1568. [Google Scholar] [CrossRef] [PubMed]
- Das Neves, J.; Bahia, M.F.; Amiji, M.M.; Sarmento, B. Mucoadhesive nanomedicines: Characterization and modulation of mucoadhesion at the nanoscale. Exp. Opin. Drug Deliv. 2011, 8, 1085–1104. [Google Scholar]
- Haas, J.; Lehr, C.M. Developments in the area of bioadhesive drug delivery systems. Exp. Opin. Biol. Ther. 2002, 2, 287–298. [Google Scholar] [CrossRef]
- Roy, S.; Pal, K.; Anis, A.; Pramanik, K.; Prabhakar, B. Polymers in mucoadhesive drug-delivery systems: A brief note. Des. Monomers Polym. 2009, 12, 483–495. [Google Scholar] [CrossRef]
- Chickering, D.E.; Mathiowitz, E. Definitions mechanisms and theories of bioadhesion. In Bioadhesive Drug Delivery Systems: Fundamentals, Novel Approaches, and Developments; Mathiowitz, E., Chickering, D.E., Lehr, C.-M., Eds.; Marcel Dekker: New York, NY, USA, 1999. [Google Scholar]
- Bshara, H.; Osman, R.; Mansour, S.; El-Shamy, A.A. Chitosan and cyclodextrin in intranasal microemulsion for improved brain buspirone hydrochloride pharmacokinetics in rats. Carbohydr. Polym. 2014, 99, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Sun, Y.G.; Jarboe, E.A.; Soisson, A.P.; Dodson, M.K.; Gaffney, D.K.; Peterson, C.M.; Janat-Amsbury, M.M. Mucoadhesive hybrid gel improves intraperitoneal platinum delivery. Int. J. Pharm. 2013, 458, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Gradauer, K.; Barthelmes, J.; Vonach, C.; Almer, G.; Mangge, H.; Teubl, B.; Roblegg, E.; Dunnhaupt, S.; Frohlich, E.; Bernkop-Schnurch, A.; et al. Liposomes coated with thiolated chitosan enhance oral peptide delivery to rats. J. Controll. Release 2013, 172, 872–878. [Google Scholar] [CrossRef]
- Luessen, H.L.; deLeeuw, B.J.; Langemeyer, M.W.E.; de Boer, A.G.; Verhoef, J.C.; Junginger, H.E. Mucoadhesive polymers in peroral peptide drug delivery 6. Carbomer and chitosan improve the intestinal absorption of the peptide drug buserelin in vivo. Pharm. Res. 1996, 13, 1668–1672. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.T.; Steinbach, J.M.; Liu, J.C.; Shimizu, S.; Kaimakliotis, H.Z.; Wheeler, M.A.; Hittelman, A.B.; Saltzman, W.M.; Weiss, R.M. Surface-modified nanoparticles enhance transurothelial penetration and delivery of survivin sirna in treating bladder cancer. Mol. Cancer Ther. 2014, 13, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Mouez, M.A.; Zaki, N.M.; Mansour, S.; Geneidi, A.S. Bioavailability enhancement of verapamil HCL via intranasal chitosan microspheres. Eur. J. Pharm. Sci. 2014, 51, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Nafee, N.A.; Ismail, F.A.; Boraie, N.A.; Mortada, L.M. Mucoadhesive buccal patches of miconazole nitrate: In vitro/in vivo performance and effect of ageing. Int. J. Pharm. 2003, 264, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Deacon, M.P.; McGurk, S.; Roberts, C.J.; Williams, P.M.; Tendler, S.J.B.; Davies, M.C.; Davis, S.S.; Harding, S.E. Atomic force microscopy of gastric mucin and chitosan mucoadhesive systems. Biochem. J. 2000, 348, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Fiebrig, I.; Harding, S.E.; Rowe, A.J.; Hyman, S.C.; Davis, S.S. Transmission electron microscopy studies on pig gastric mucin and its interactions with chitosan. Carbohydr. Polym. 1995, 28, 239–244. [Google Scholar] [CrossRef]
- Pettersson, T.; Dedinaite, A. Normal and friction forces between mucin and mucin–chitosan layers in absence and presence of sds. J. Colloid Interface Sci. 2008, 324, 246–256. [Google Scholar] [CrossRef] [PubMed]
- Rossi, S.; Ferrari, F.; Bonferoni, M.C.; Caramella, C. Characterization of chitosan hydrochloride–mucin interaction by means of viscosimetric and turbidimetric measurements. Eur. J. Pharm. Sci. 2000, 10, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Rossi, S.; Ferrari, F.; Bonferoni, M.C.; Caramella, C. Characterization of chitosan hydrochloride–mucin rheological interaction: Influence of polymer concentration and polymer: Mucin weight ratio. Eur. J. Pharm. Sci. 2001, 12, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.A.; Nobre, T.M.; Pavinatto, F.J.; Oliveira, O.N. Interaction of chitosan and mucin in a biomembrane model environment. J. Colloid Interface Sci. 2012, 376, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Svensson, O.; Lindh, L.; Cardenas, M.; Arnebrant, T. Layer-by-layer assembly of mucin and chitosan–influence of surface properties, concentration and type of mucin. J. Colloid Interface Sci. 2006, 299, 608–616. [Google Scholar] [CrossRef] [PubMed]
- Svensson, O.; Thuresson, K.; Arnebrant, T. Interactions between chitosan-modified particles and mucin–coated surfaces. J. Colloid Interface Sci. 2008, 325, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Dalmoro, A.; Barba, A.A.; Lamberti, G.; Grassi, M.; d’Amore, M. Pharmaceutical applications of biocompatible polymer blends containing sodium alginate. Adv. Polym. Technol. 2012, 31, 219–230. [Google Scholar] [CrossRef]
- Skaugrud, O.; Hagen, A.; Borgersen, B.; Dornish, M. Biomedical and pharmaceutical applications of alginate and chitosan. Biotechnol. Genet. Eng. Rev. 1999, 16, 23–40. [Google Scholar] [CrossRef] [PubMed]
- Sletmoen, M.; Maurstad, G.; Nordgard, C.T.; Draget, K.I.; Stokke, B.T. Oligoguluronate induced competitive displacement of mucin–alginate interactions: Relevance for mucolytic function. Soft Matter 2012, 8, 8413–8421. [Google Scholar] [CrossRef]
- Fuongfuchat, A.; Jamieson, A.M.; Blackwell, J.; Gerken, T.A. Rheological studies of the interaction of mucins with alginate and polyacrylate. Carbohydr. Res. 1996, 284, 85–99. [Google Scholar] [CrossRef] [PubMed]
- Nordgard, C.T.; Draget, K.I. Oligosaccharides as modulators of rheology in complex mucous systems. Biomacromolecules 2011, 12, 3084–3090. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.; Pearson, J.P.; Draget, K.I.; Dettmar, P.W.; Smidsrod, O. Rheological characterisation of mixed gels of mucin and alginate. Carbohydr. Polym. 2005, 59, 189–195. [Google Scholar] [CrossRef]
- Varum, K.M.; Ottoy, M.H.; Smidsrod, O. Water-solubility of partially N-acetylated chitosans as a function of pH: Effect of chemical-composition and depolymerization. Carbohydr. Polym. 1994, 25, 65–70. [Google Scholar] [CrossRef]
- Schatz, C.; Pichot, C.; Delair, T.; Viton, C.; Domard, A. Static light scattering studies on chitosan solutions: From macromolecular chains to colloidal dispersions. Langmuir 2003, 19, 9896–9903. [Google Scholar] [CrossRef]
- Taylor, C.; Draget, K.I.; Pearson, J.P.; Smidsrod, O. Mucous systems show a novel mechanical response to applied deformation. Biomacromolecules 2005, 6, 1524–1530. [Google Scholar] [CrossRef] [PubMed]
- Fogg, F.J.J.; Hutton, D.A.; Jumel, K.; Pearson, J.P.; Harding, S.E.; Allen, A. Characterization of pig colonic mucins. Biochem. J. 1996, 316, 937–942. [Google Scholar] [PubMed]
- Vårum, K.M.; Anthonsen, M.W.; Grasdalen, H.; Smidsrød, O. Determination of the degree of N-acetylation and the distribution of N-acetyl groups in partially N-deacetylated chitins (chitosans) by high-field n.M.R. Spectroscopy. Carbohydr. Res. 1991, 211, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Grasdalen, H.; Larsen, B.; Smidsrød, O. Study of the composition and sequence of uronate residues in alginate. Carbohydr. Res. 1979, 68, 23–31. [Google Scholar] [CrossRef]
- Anthonsen, M.W.; Vårum, K.M.; Smidsrød, O. Solution properties of chitosans: Conformation and chain stiffness of chitosans with different degrees of N-acetylation. Carbohydr. Polym. 1993, 22, 193–201. [Google Scholar] [CrossRef]
- Aarstad, O.; Strand, B.L.; Klepp-Andersen, L.M.; Skjåk-Bræk, G. Analysis of G-block distributions and their impact on gel properties of in vitro epimerized mannuronan. Biomacromolecules 2013, 14, 3409–3416. [Google Scholar] [CrossRef] [PubMed]
- Sletmoen, M.; Skjak-Braek, G.; Stokke, B.T. Single-molecular pair unbinding studies of mannuronan C-5 epimerase AlgE4 and its polymer substrate. Biomacromolecules 2004, 5, 1288–1295. [Google Scholar] [CrossRef] [PubMed]
- Haugstad, K.E.; Gerken, T.A.; Stokke, B.T.; Dam, T.K.; Brewer, C.F.; Sletmoen, M. Enhanced self-association of mucins possessing the T and Tn carbohydrate cancer antigens at the single-molecule level. Biomacromolecules 2012, 13, 1400–1409. [Google Scholar] [CrossRef] [PubMed]
- Hutter, J.L.; Bechhoefer, J. Calbiration of atomic–force micoscope tips. Rev. Sci. Instrum. 1993, 64, 1868–1873. [Google Scholar] [CrossRef]
- Te Riet, J.; Katan, A.J.; Rankl, C.; Stahl, S.W.; van Buul, A.M.; Phang, I.Y.; Gomez-Casado, A.; Schon, P.; Gerritsen, J.W.; Cambi, A.; et al. Inter laboratory round robin on cantilever calibration for afm force spectroscopy. Ultramicroscopy 2011, 111, 1659–1669. [Google Scholar]
- Bell, G.I. Models for the specific adhesion of cells to cells. Science 1978, 200, 618–627. [Google Scholar] [CrossRef] [PubMed]
- Evans, E.; Ritchie, K. Dynamic strength of molecular adhesion bonds. Biophys. J. 1997, 72, 1541–1555. [Google Scholar] [CrossRef] [PubMed]
- Evans, E. Probing the relation between force–lifetime–and chemistry in single molecular bonds. Annu. Rev. Biophys. Biomol. Struct. 2001, 30, 105–128. [Google Scholar] [CrossRef] [PubMed]
- Dudko, O.K.; Hummer, G.; Szabo, A. Intrinsic rates and activation free energies from single-molecule pulling experiments. Phys. Rev. Lett. 2006, 96. [Google Scholar] [CrossRef]
- Dudko, O.K.; Hummer, G.; Szabo, A. Theory, analysis, and interpretation of single-molecule force spectroscopy experiments. Proc. Natl. Acad. Sci. USA 2008, 105, 15755–15760. [Google Scholar] [CrossRef] [PubMed]
- Grandbois, M.; Beyer, M.; Rief, M.; Clausen-Schaumann, H.; Gaub, H.E. How strong is a covalent bond? Science 1999, 283, 1727–1730. [Google Scholar] [CrossRef] [PubMed]
- Hugel, T.; Grosholz, M.; Claussen-Schaumann, H.; Pfau, A.; Gaub, H.; Seitz, M. Elasticity of single polyelectrolyte chains and their desorption from solid supports studied by afm based single molecule force spectroscopy. Macromolecules 2001, 34, 1039–1047. [Google Scholar] [CrossRef]
- Friedsam, C.; Gaub, H.E.; Netz, R.R. Probing surfaces with single-polymer atomic force microscope experiments. Biointerphases 2006, 1, MR1–MR21. [Google Scholar] [CrossRef] [PubMed]
- Kocun, M.; Grandbois, M.; Cuccia, L.A. Single molecule atomic force microscopy and force spectroscopy of chitosan. Colloid Surf. B Biointerfaces 2011, 82, 470–476. [Google Scholar] [CrossRef]
- Spruijt, E.; van den Berg, S.A.; Cohen Stuart, M.A.; van der Gucht, J. Direct measurement of the strength of single ionic bonds between hydrated charges. ACS Nano 2012, 6, 5297–5303. [Google Scholar] [CrossRef] [PubMed]
- Rief, M.; Gautel, M.; Oesterhelt, F.; Fernandez, J.M.; Gaub, H.E. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science 1997, 276, 1109–1112. [Google Scholar] [CrossRef] [PubMed]
- Noy, A.; Vezenov, D.V.; Kayyem, J.F.; Meade, T.J.; Lieber, C.M. Stretching and breaking duplex DNA by chemical force microscopy. Chem. Biol. 1997, 4, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Ritzefeld, M.; Walhorn, V.; Anselmetti, D.; Sewald, N. Analysis of DNA interactions using single-molecule force spectroscopy. Amino Acids 2013, 44, 1457–1475. [Google Scholar] [CrossRef] [PubMed]
- Baumann, C.G.; Bloomfield, V.A.; Smith, S.B.; Bustamante, C.; Wang, M.D.; Block, S.M. Stretching of single collapsed DNA molecules. Biophys. J. 2000, 78, 1965–1978. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.M. Analytical descriptions of dynamic force spectroscopy: Behaviour of multiple connections. Anal. Chim. Acta 2003, 479, 107–115. [Google Scholar] [CrossRef]
- Noy, A. Force spectroscopy 101: How to design, perform, and analyze an afm-based single molecule force spectroscopy experiment. Curr. Opin. Chem. Biol. 2011, 15, 710–718. [Google Scholar] [CrossRef] [PubMed]
- Noy, A.; Friddle, R.W. Practical single molecule force spectroscopy: How to determine fundamental thermodynamic parameters of intermolecular bonds with an atomic force microscope. Methods 2013, 60, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Schatz, C.; Viton, C.; Delair, T.; Pichot, C.; Domard, A. Typical physicochemical behaviors of chitosan in aqueous solution. Biomacromolecules 2003, 4, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Hanes, J.; Demeester, J. Drug and gene delivery to mucosal tissues: The mucus barrier preface. Adv. Drug Deliv. Rev. 2009, 61, 73–74. [Google Scholar] [CrossRef] [PubMed]
- Lai, S.K.; Wang, Y.Y.; Hanes, J. Mucus-penetrating nanoparticles for drug and gene delivery to mucosal tissues. Adv. Drug Deliv. Rev. 2009, 61, 158–171. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.; Allen, A.; Dettmar, P.W.; Pearson, J.P. The gel matrix of gastric mucus is maintained by a complex interplay of transient and nontransient associations. Biomacromolecules 2003, 4, 922–927. [Google Scholar] [CrossRef] [PubMed]
- Laffleur, F.; Bernkop-Schnurch, A. Strategies for improving mucosal drug delivery. Nanomedicine 2013, 8, 2061–2075. [Google Scholar] [CrossRef] [PubMed]
- Crater, J.S.; Carrier, R.L. Barrier properties of gastrointestinal mucus to nanoparticle transport. Macromol. Biosci. 2010, 10, 1473–1483. [Google Scholar] [CrossRef] [PubMed]
- Lieleg, O.; Vladescu, I.; Ribbeck, K. Characterization of particle translocation through mucin hydrogels. Biophys. J. 2010, 98, 1782–1789. [Google Scholar] [CrossRef] [PubMed]
- Sigurdsson, H.H.; Kirch, J.; Lehr, C.M. Mucus as a barrier to lipophilic drugs. Int. J. Pharm. 2013, 453, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Ensign, L.M.; Schneider, C.; Suk, J.S.; Cone, R.; Hanes, J. Mucus penetrating nanoparticles: Biophysical tool and method of drug and gene delivery. Adv. Mater. 2012, 24, 3887–3894. [Google Scholar] [CrossRef] [PubMed]
- Lai, S.K.; Suk, J.S.; Pace, A.; Wang, Y.Y.; Yang, M.; Mert, O.; Chen, J.; Kim, J.; Hanes, J. Drug carrier nanoparticles that penetrate human chronic rhinosinusitis mucus. Biomaterials 2011, 32, 6285–6290. [Google Scholar] [CrossRef] [PubMed]
- Mert, O.; Lai, S.K.; Ensign, L.; Yang, M.; Wang, Y.Y.; Wood, J.; Hanes, J. A poly(ethylene glycol)-based surfactant for formulation of drug-loaded mucus penetrating particles. J. Controll. Release 2012, 157, 455–460. [Google Scholar] [CrossRef]
- Yang, M.; Lai, S.K.; Wang, Y.Y.; Zhong, W.X.; Happe, C.; Zhang, M.; Fu, J.; Hanes, J. Biodegradable nanoparticles composed entirely of safe materials that rapidly penetrate human mucus. Angew. Chem. Int. Ed. 2011, 50, 2597–2600. [Google Scholar] [CrossRef]
- Jabbari, E.; Peppas, N.A. Polymer-polymer interdiffusion and adhesion. J. Macromol. Sci. C Polym. Rev. 1994, 34, 205–241. [Google Scholar] [CrossRef]
- Shah, A.J.; Donovan, M.D. Rheological characterization of neutral and anionic polysaccharides with reduced mucociliary transport rates. AAPS PharmSciTech 2007, 8, E40–E47. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haugstad, K.E.; Håti, A.G.; Nordgård, C.T.; Adl, P.S.; Maurstad, G.; Sletmoen, M.; Draget, K.I.; Dias, R.S.; Stokke, B.T. Direct Determination of Chitosan–Mucin Interactions Using a Single-Molecule Strategy: Comparison to Alginate–Mucin Interactions. Polymers 2015, 7, 161-185. https://doi.org/10.3390/polym7020161
Haugstad KE, Håti AG, Nordgård CT, Adl PS, Maurstad G, Sletmoen M, Draget KI, Dias RS, Stokke BT. Direct Determination of Chitosan–Mucin Interactions Using a Single-Molecule Strategy: Comparison to Alginate–Mucin Interactions. Polymers. 2015; 7(2):161-185. https://doi.org/10.3390/polym7020161
Chicago/Turabian StyleHaugstad, Kristin E., Armend G. Håti, Catherine T. Nordgård, Patricia S. Adl, Gjertrud Maurstad, Marit Sletmoen, Kurt I. Draget, Rita S. Dias, and Bjørn T. Stokke. 2015. "Direct Determination of Chitosan–Mucin Interactions Using a Single-Molecule Strategy: Comparison to Alginate–Mucin Interactions" Polymers 7, no. 2: 161-185. https://doi.org/10.3390/polym7020161