Molecularly Imprinted Solid Phase Extraction Strategy for Quinic Acid
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Materials
2.2. Computational Modelling: Monomers Molar Ratio Screening
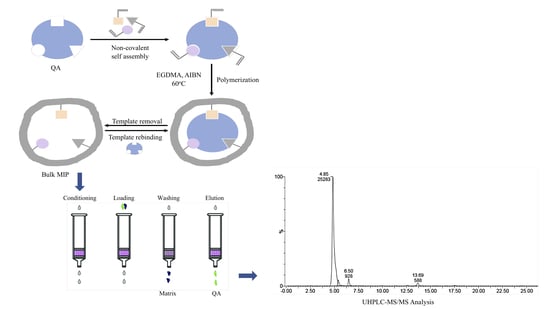
2.3. Bulk Polymers Preparation
2.4. Morphology Characterization
2.5. Equilibrium Rebinding Studies
2.6. Adsorption Kinetics
2.7. Binding Isotherm
2.8. MISPE Procedure Optimization
2.9. UHPLC-MS/MS Measurements
2.10. Method Validation
2.11. MIP Cartridge Reusability
2.12. Selectivity Study
2.13. MISPE Application on Coffee Extract
2.13.1. UHPLC Method for QA Quantification in Coffee Extract
2.13.2. Method Validation
2.13.3. Preparation of Aqueous Coffee Extract
2.13.4. Application of MISPE for Extraction of QA from Total Aqueous Coffee Extract
3. Results and Discussion
3.1. Computational Modelling: Monomers Molar Ratio Screening
3.2. Morphology Characterization
3.3. Rebinding Studies
3.4. Adsorption Kinetics
3.5. Binding Isotherm
3.6. MISPE Procedure Optimization
3.6.1. Loading Step Optimization
3.6.2. Washing Step Optimization
3.6.3. Elution Step Optimization
3.7. UHPLC-MS/MS Method Validation
3.8. MIP Cartridge Reusability
3.9. MISPE Selectivity
3.10. MISPE Application on Coffee Extract
3.10.1. UHPLC-MS/MS Method Validation
3.10.2. MISPE Application on Coffee Extract
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Madkour, L. Oxidative stress and oxidative damage-induced cell death. Reactive Oxygen Species (ROS), Nanoparticles, and Endoplasmic Reticulum (ER) Stress-Induced Cell Death Mechanisms; Academic Press: Cambridge, MA, USA, 2020; pp. 175–197. [Google Scholar]
- Jiang, S.; Liu, H.; Li, C. Dietary Regulation of Oxidative Stress in Chronic Metabolic Diseases. Foods 2021, 10, 1854. [Google Scholar] [CrossRef] [PubMed]
- Toghyani Khorasgani, A.; Amini-Khoei, H.; Shadkhast, M.; Salimian, S.; Majidian, M.; Dehkordi, S.H. Quinic acid through mitigation of oxidative stress in the hippocampus exerts analgesic effect in male mice. Future Nat. Prod. 2021, 7, 1–11. [Google Scholar]
- Magaña, A.; Kamimura, N.; Soumyanath, A.; Stevens, J.; Maier, C. Caffeoylquinic acids: Chemistry, biosynthesis, occurrence, analytical challenges, and bioactivity. Plant J. 2021, 107, 1299–1319. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, J.; Zhao, B.; Zhao-Wilson, X. Quinic acid could be a potential rejuvenating natural compound by improving survival of Caenorhabditis elegans under deleterious conditions. Rejuvenation Res. 2012, 15, 573–583. [Google Scholar] [CrossRef]
- Kwon, Y.K.; Choi, S.J.; Kim, C.R.; Kim, J.K.; Kim, Y.-J.; Choi, J.H.; Song, S.-W.; Kim, C.-J.; Park, G.G.; Park, C.-S.; et al. Antioxidant and cognitive-enhancing activities of Arctium lappa L. roots in Aβ1-42-induced mouse model. Appl. Biol. Chem. 2016, 59, 553–565. [Google Scholar] [CrossRef]
- Pero, R.W. Health consequences of catabolic synthesis of hippuric acid in humans. Curr. Clin. Pharmacol. 2010, 5, 67–73. [Google Scholar] [CrossRef]
- Fricker, R.A.; Green, E.L.; Jenkins, S.I.; Griffin, S.M. The Influence of Nicotinamide on Health and Disease in the Central Nervous System. Int. J. Tryptophan Res. 2018, 11, 1178646918776658. [Google Scholar] [CrossRef]
- Kansara, K.; Gupta, S.S. Chapter Fourteen-DNA Damage, Repair, and Maintenance of Telomere Length: Role of Nutritional Supplements. In Mutagenicity: Assays and Applications; Kumar, A., Ed.; Academic Press: Cambridge, MA, USA, 2018; pp. 287–307. [Google Scholar]
- Hulme, A.C. The Isolation of l-Quinic Acid from the Apple Fruit. J. Exp. Bot. 1951, 2, 298–315. [Google Scholar] [CrossRef]
- Yazdi, S.E.; Prinsloo, G.; Heyman, H.M.; Oosthuizen, C.B.; Klimkait, T.; Meyer, J.J.M. Anti-HIV-1 activity of quinic acid isolated from Helichrysum mimetes using NMR-based metabolomics and computational analysis. S. Afr. J. Bot. 2019, 126, 328–339. [Google Scholar] [CrossRef]
- Upadhyay, R.; Rao, L.J.M. An outlook on chlorogenic acids-occurrence, chemistry, technology, and biological activities. Crit. Rev. Food Sci. Nutr. 2013, 53, 968–984. [Google Scholar] [CrossRef]
- Tuyun, A.F.; Uslu, H. Extraction of D-(−)-Quinic Acid Using an Amine Extractant in Different Diluents. J. Chem. Eng. Data 2012, 57, 190–194. [Google Scholar] [CrossRef]
- Fischer, E. Einfluss der Configuration auf die Wirkung der Enzyme. Eur. J. Inorg. Chem. 1894, 27, 2985–2993. [Google Scholar]
- Saad, E.M.; Madbouly, A.; Ayoub, N.; el Nashar, R.M. Preparation and application of molecularly imprinted polymer for isolation of chicoric acid from Chicorium intybus L. medicinal plant. Anal. Chim. Acta. 2015, 877, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Wackerlig, J.; Schirhagl, R. Applications of Molecularly Imprinted Polymer Nanoparticles and Their Advances toward Industrial Use: A Review. Anal. Chem. 2016, 88, 250–261. [Google Scholar] [CrossRef]
- Uzun, L.; Turner, A.P.F. Molecularly-imprinted polymer sensors: Realising their potential. Biosens. Bioelectron. 2016, 76, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Speltini, A.; Scalabrini, A.; Maraschi, F.; Sturini, M.; Profumo, A. Newest applications of molecularly imprinted polymers for extraction of contaminants from environmental and food matrices: A review. Anal. Chim. Acta 2017, 974, 1–26. [Google Scholar] [CrossRef]
- Malik, M.I.; Shaikh, H.; Mustafa, G.; Bhanger, M.I. Recent Applications of Molecularly Imprinted Polymers in Analytical Chemistry. Sep. Purif. Rev. 2019, 48, 179–219. [Google Scholar] [CrossRef]
- Piletsky, S.; Turner, A. Imprinted polymers and their application in optical sensors. In Optical Biosensors: Today and Tomorrow; Elsevier: Amsterdam, The Netherlands, 2008; pp. 543–581. [Google Scholar]
- Piletsky, S.A.; Turner, A.P.F. Electrochemical Sensors Based on Molecularly Imprinted Polymers. Electroanalysis 2002, 14, 317–323. [Google Scholar] [CrossRef]
- Elfadil, D.; Lamaoui, A.; della Pelle, F.; Amine, A.; Compagnone, D. Molecularly Imprinted Polymers Combined with Electrochemical Sensors for Food Contaminants Analysis. Molecules 2021, 26, 4607. [Google Scholar] [CrossRef]
- Xu, X.-Y.; Tian, X.-G.; Cai, L.-G.; Xu, Z.-L.; Lei, H.-T.; Wang, H.; Sun, Y.-M. Molecularly imprinted polymer based surface plasmon resonance sensors for detection of Sudan dyes. Anal. Methods 2014, 6, 3751–3757. [Google Scholar] [CrossRef]
- Tsuru, N.; Kikuchi, M.; Kawaguchi, H.; Shiratori, S. A quartz crystal microbalance sensor coated with MIP for “Bisphenol A” and its properties. Thin Solid Film. 2006, 499, 380–385. [Google Scholar] [CrossRef]
- Eskandari, H.; Amirzehni, M.; Hassanzadeh, J.; Vahid, B. Mesoporous MIP-capped luminescent MOF as specific and sensitive analytical probe: Application for chlorpyrifos. Microchim. Acta 2020, 187, 673. [Google Scholar] [CrossRef] [PubMed]
- Antipchik, M.; Reut, J.; Ayankojo, A.G.; Öpik, A.; Syritski, V. MIP-based electrochemical sensor for direct detection of hepatitis C virus via E2 envelope protein. Talanta 2022, 250, 123737. [Google Scholar] [CrossRef] [PubMed]
- Mazouz, Z.; Mokni, M.; Fourati, N.; Zerrouki, C.; Barbault, F.; Seydou, M.; Kalfat, R.; Yaakoubi, N.; Omezzine, A.; Bouslema, A.; et al. Computational approach and electrochemical measurements for protein detection with MIP-based sensor. Biosens. Bioelectron. 2020, 151, 111978. [Google Scholar] [CrossRef]
- Lee, M.-H.; Chen, Y.-C.; Ho, M.-H.; Lin, H.-Y. Optical recognition of salivary proteins by use of molecularly imprinted poly(ethylene-co-vinyl alcohol)/quantum dot composite nanoparticles. Anal. Bioanal. Chem. 2010, 397, 1457–1466. [Google Scholar] [CrossRef]
- Rouhani, S.; Nahavandifard, F. Molecular imprinting-based fluorescent optosensor using a polymerizable 1,8-naphthalimide dye as a florescence functional monomer. Sens. Actuators B Chem. 2014, 197, 185–192. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Hardwick, S.A.; Sun, T.; Grattan, K.T.V. Intrinsic Fluorescence-Based Optical Fiber Sensor for Cocaine Using a Molecularly Imprinted Polymer as the Recognition Element. IEEE Sens. J. 2012, 12, 255–260. [Google Scholar] [CrossRef]
- Sun, H.; Lai, J.-P.; Lin, D.-S.; Huang, X.-X.; Zuo, Y.; Li, Y.-L. A novel fluorescent multi-functional monomer for preparation of silver ion-imprinted fluorescent on–off chemosensor. Sens. Actuators B Chem. 2016, 224, 485–491. [Google Scholar] [CrossRef]
- Ng, S.M.; Narayanaswamy, R. Fluorescence sensor using a molecularly imprinted polymer as a recognition receptor for the detection of aluminium ions in aqueous media. Anal. Bioanal. Chem. 2006, 386, 1235–1244. [Google Scholar] [CrossRef]
- Sener, G.; Ozgur, E.; Rad, A.Y.; Uzun, L.; Say, R.; Denizli, A. Rapid real-time detection of procalcitonin using a microcontact imprinted surface plasmon resonance biosensor. Analyst 2013, 138, 6422–6428. [Google Scholar] [CrossRef]
- Sener, G.; Uzun, L.; Say, R.; Denizli, A. Use of molecular imprinted nanoparticles as biorecognition element on surface plasmon resonance sensor. Sens. Actuators B Chem. 2011, 160, 791–799. [Google Scholar] [CrossRef]
- Zhao, N.; Chen, C.; Zhou, J. Surface plasmon resonance detection of ametryn using a molecularly imprinted sensing film prepared by surface-initiated atom transfer radical polymerization. Sens. Actuators B Chem. 2012, 166–167, 473–479. [Google Scholar] [CrossRef]
- Shaikh, H.; Şener, G.; Memon, N.; Bhanger, M.; Nizamani, S.; Üzek, R.; Denizli, A. Molecularly imprinted surface plasmon resonance (SPR) based sensing of bisphenol A for its selective detection in aqueous systems. Anal. Methods 2015, 7, 4661–4670. [Google Scholar] [CrossRef]
- Hammam, M.A.; Abdel-Halim, M.; Madbouly, A.; Wagdy, H.A.; el Nashar, R.M. Computational design of molecularly imprinted polymer for solid phase extraction of moxifloxacin hydrochloride from Avalox® tablets and spiked human urine samples. Microchem. J. 2019, 148, 51–56. [Google Scholar] [CrossRef]
- Omran, N.H.; Wagdy, H.A.; Abdel-Halim, M.; Nashar, R.M.E. Validation and Application of Molecularly Imprinted Polymers for SPE/UPLC–MS/MS Detection of Gemifloxacin Mesylate. Chromatographia 2019, 82, 1617–1631. [Google Scholar] [CrossRef]
- Stevenson, D.; El-Sharif, H.F.; Reddy, S.M. Selective extraction of proteins and other macromolecules from biological samples using molecular imprinted polymers. Bioanalysis 2016, 8, 2255–2263. [Google Scholar] [CrossRef] [PubMed]
- Pereira, I.; Rodrigues, M.F.; Chaves, A.R.; Vaz, B.G. Molecularly imprinted polymer (MIP) membrane assisted direct spray ionization mass spectrometry for agrochemicals screening in foodstuffs. Talanta 2018, 178, 507–514. [Google Scholar] [CrossRef]
- Voros, V.; Drioli, E.; Fonte, C.; Szekely, G. Process Intensification via Continuous and Simultaneous Isolation of Antioxidants: An Upcycling Approach for Olive Leaf Waste. ACS Sustain. Chem. Eng. 2019, 7, 18444–18452. [Google Scholar] [CrossRef]
- Szekely, G. Valorisation of agricultural waste with adsorption/nanofiltration hybrid process: From materials to sustainable process design. Green Chem. 2017, 19, 3116–3125. [Google Scholar]
- Martins, R.O.; Gomes, I.C.; Telles, A.D.M.; Kato, L.; Souza, P.S.; Chaves, A.R. Molecularly imprinted polymer as solid phase extraction phase for condensed tannin determination from Brazilian natural sources. J. Chromatogr. A 2020, 1620, 460977. [Google Scholar] [CrossRef]
- Kanao, E.; Tsuchiya, Y.; Tanaka, K.; Masuda, Y.; Tanigawa, T.; Naito, T.; Sano, T.; Kubo, T.; Otsuka, K. Poly(ethylene glycol) Hydrogels with a Boronic Acid Monomer via Molecular Imprinting for Selective Removal of Quinic Acid Gamma-Lactone in Coffee. ACS Appl. Polym. Mater. 2021, 3, 226–232. [Google Scholar] [CrossRef]
- Saad, E.M.; el Gohary, N.A.; Abdel-Halim, M.; Handoussa, H.; el Nashar, R.M.; Mizaikoff, B. Molecularly imprinted polymers for selective extraction of rosmarinic acid from Rosmarinus officinalis L. Food Chem. 2021, 335, 127644. [Google Scholar] [CrossRef] [PubMed]
- Mujahid, A.; Iqbal, N.; Afzal, A. Bioimprinting strategies: From soft lithography to biomimetic sensors and beyond. Biotechnol. Adv. 2013, 31, 1435–1447. [Google Scholar] [CrossRef] [PubMed]
- Hosny, H.; el Gohary, N.; Saad, E.; Handoussa, H.; el Nashar, R.M. Isolation of sinapic acid from broccoli using molecularly imprinted polymers. J. Sep. Sci. 2018, 41, 1164–1172. [Google Scholar] [CrossRef]
- Nicolescu, T.-V.; Sarbu, A.; Dima, Ş.-O.; Nicolae, C.-A.; Donescu, D. Molecularly imprinted “bulk” copolymers as selective sorbents for gallic acid. J. Appl. Polym. Sci. 2013, 127, 366–374. [Google Scholar] [CrossRef]
- Michailof, C.; Manesiotis, P.; Panayiotou, C. Synthesis of caffeic acid and p-hydroxybenzoic acid molecularly imprinted polymers and their application for the selective extraction of polyphenols from olive mill waste waters. J. Chromatogr. A 2008, 1182, 25–33. [Google Scholar] [CrossRef]
- Li, X.; Shian, Z.; Chen, L.; Whittaker, A. Computer simulation and preparation of molecularly imprinted polymer membranes with chlorogenic acid as template. Polym. Int. 2011, 60, 592–598. [Google Scholar] [CrossRef]
- Liu, Z.; Xu, Z.; Wang, D.; Yang, Y.; Duan, Y.; Ma, L.; Lin, T.; Liu, H. A Review on Molecularly Imprinted Polymers Preparation by Computational Simulation-Aided Methods. Polymers 2021, 13, 2657. [Google Scholar] [CrossRef]
- Roy, E.; Patra, S.; Madhuri, R.; Sharma, P.K. Gold nanoparticle mediated designing of non-hydrolytic sol-gel cross-linked metformin imprinted polymer network: A theoretical and experimental study. Talanta 2014, 120, 198–207. [Google Scholar] [CrossRef]
- Mennucci, B. Polarizable Continuum Model. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2012, 2, 386–404. [Google Scholar] [CrossRef]
- Arshady, R.; Mosbach, K. Synthesis of substrate-selective polymers by host-guest polymerization. Die Makromol. Chem. 1981, 182, 687–692. [Google Scholar] [CrossRef]
- Li, N.; Ng, T.B.; Wong, J.H.; Qiao, J.X.; Zhang, Y.N.; Zhou, R.; Chen, R.R.; Liu, F. Separation and purification of the antioxidant compounds, caffeic acid phenethyl ester and caffeic acid from mushrooms by molecularly imprinted polymer. Food Chem. 2013, 139, 1161–1167. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.S.; McKay, G. Pseudo-second order model for sorption processes. Process Biochem. 1999, 34, 451–465. [Google Scholar] [CrossRef]
- Langmuir, I. The constitution and fundamental properties of solids and liquids. II. liquids.1. J. Am. Chem. Soc. 1917, 39, 1848–1906. [Google Scholar] [CrossRef]
- Ghani, N.T.A.; el Nashar, R.M.; Abdel-Haleem, F.M.; Madbouly, A. Computational Design, Synthesis and Application of a New Selective Molecularly Imprinted Polymer for Electrochemical Detection. Electroanalysis 2016, 28, 1530–1538. [Google Scholar] [CrossRef]
- El Gohary, N.A.; Madbouly, A.; el Nashar, R.M.; Mizaikoff, B. Synthesis and application of a molecularly imprinted polymer for the voltammetric determination of famciclovir. Biosens. Bioelectron. 2015, 65, 108–114. [Google Scholar] [CrossRef]
- Bakas, I.; Oujji, N.B.; Istamboulié, G.; Piletsky, S.; Piletska, E.; Ait-Addi, E.; Ait-Ichou, I.; Noguer, T.; Rouillon, R. Molecularly imprinted polymer cartridges coupled to high performance liquid chromatography (HPLC-UV) for simple and rapid analysis of fenthion in olive oil. Talanta 2014, 125, 313–318. [Google Scholar] [CrossRef]
- Chrzanowska, A.M.; Poliwoda, A.; Wieczorek, P.P. Characterization of particle morphology of biochanin A molecularly imprinted polymers and their properties as a potential sorbent for solid-phase extraction. Mater. Sci. Eng. C Mater. Biol. Appl. 2015, 49, 793–798. [Google Scholar] [CrossRef]
- Li, Y.; Li, X.; Dong, C.; Li, Y.; Jin, P.; Qi, J. Selective recognition and removal of chlorophenols from aqueous solution using molecularly imprinted polymer prepared by reversible addition-fragmentation chain transfer polymerization. Biosens. Bioelectron. 2009, 25, 306–312. [Google Scholar] [CrossRef]
- Le Noir, M.; Plieva, F.; Hey, T.; Guieysse, B.; Mattiasson, B. Macroporous molecularly imprinted polymer/cryogel composite systems for the removal of endocrine disrupting trace contaminants. J. Chromatogr. A 2007, 1154, 158–164. [Google Scholar] [CrossRef]
- Huang, Q.; Hao, X.; Qiao, L.; Wu, M.; Shen, G.; Ma, S. Measurement and thermodynamic functions of solid–liquid phase equilibrium of d-(−)-quinic acid in H2O, methanol, ethanol and (H2O+methanol), (H2O+ethanol) binary solvent mixtures. J. Chem. Thermodyn. 2016, 100, 140–147. [Google Scholar] [CrossRef]
- Pratiwi, R.; Megantara, S.; Rahayu, D.; Pitaloka, I.; Hasanah, A.N. Comparison of Bulk and Precipitation Polymerization Method of Synthesis Molecular Imprinted Solid Phase Extraction for Atenolol using Methacrylic Acid. J. Young Pharm. 2018, 11, 12–16. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, F.; Wang, J.; Li, N.; Li, K. Molecular recognition properties of salicylic acid-imprinted polymers. Chromatographia 2002, 55, 447–451. [Google Scholar] [CrossRef]
- Song, X.; Turiel, E.; He, L.; Martín-Esteban, A. Synthesis of Molecularly Imprinted Polymers for the Selective Extraction of Polymyxins from Environmental Water Samples. Polymers 2020, 12, 131. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Zhao, L.; Chu, B.; Feng, Q.; Yan, W.; Lin, J.M. Molecularly imprinted solid-phase extraction for the selective determination of 17beta-estradiol in fishery samples with high performance liquid chromatography. Talanta 2009, 78, 442–447. [Google Scholar] [CrossRef]
- Boulanouar, S.; Combes, A.; Mezzache, S.; Pichon, V. Synthesis and application of molecularly imprinted polymers for the selective extraction of organophosphorus pesticides from vegetable oils. J. Chromatogr. A 2017, 1513, 59–68. [Google Scholar] [CrossRef]
- Karasova, G.; Lehotay, J.; Sadecka, J.; Skacani, I.; Lachova, M. Selective extraction of derivates of p-hydroxy-benzoic acid from plant material by using a molecularly imprinted polymer. J. Sep. Sci. 2005, 28, 2468–2476. [Google Scholar] [CrossRef]
- Wang, G.F.; Shi, L.P.; Ren, Y.D.; Liu, Q.F.; Liu, H.F.; Zhang, R.J.; Li, Z.; Zhu, F.H.; He, P.L.; Tang, W.; et al. Anti-hepatitis B virus activity of chlorogenic acid, quinic acid and caffeic acid in vivo and in vitro. Antivir. Res. 2009, 83, 186–190. [Google Scholar] [CrossRef]










| Polymer | Type of Polymerization | Template (T) | Functional Monomer (FM) | Cross-Linker (CL) | T:FM:CL Molar Ratio |
|---|---|---|---|---|---|
| A | Bulk | QA | Allylamine | EGDMA | 1:6:20 |
| B | Bulk | QA | MAA | EGDMA | 1:4:20 |
| C | Bulk | QA | 4-VP | EGDMA | 1:5:20 |
| Polymers | A | B | C | ||||
|---|---|---|---|---|---|---|---|
| MIP | NIP | MIP | NIP | MIP | NIP | ||
| BET surface area (m2/g) | 21.51 | 37.13 | 23.80 | 39.04 | 31.41 | 40.90 | |
| BJH Pore volume (cc/g) | Adsorption | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 |
| Desorption | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | 0.02 | |
| BJH Pore radius (nm) | Adsorption | 1.77 | 1.64 | 1.77 | 1.64 | 1.77 | 1.64 |
| Desorption | 1.67 | 1.67 | 1.67 | 1.67 | 1.67 | 1.67 | |
| Polymer | FM | Ratio | Water | ACN:Water (4:1) | Methanol | |||
|---|---|---|---|---|---|---|---|---|
| B (µmol/g) ± SD | IF | B (µmol/g) ± SD | IF | B (µmol/g) ± SD | IF | |||
| MIP A | Allylamine | 1:6:20 | 2.63 ± 0.48 | 2.48 | 2.81 ± 0.57 | 0.82 | 7.52 ± 0.64 | 1.60 |
| NIP A | Allylamine | 0:6:20 | 1.06 ± 0.25 | 3.45 ± 0.49 | 4.70 ± 0.29 | |||
| MIP B | MAA | 1:4:20 | 2.68 ± 0.39 | 2.15 | 0.93 ± 0.27 | 1.69 | 3.14 ± 0.36 | 1.87 |
| NIP B | MAA | 0:4:20 | 1.24 ± 0.22 | 0.55 ± 0.15 | 1.68 ± 0.32 | |||
| MIP C | 4-VP | 1:5:20 | 4.88 ± 0.32 | 2.14 | 9.97 ± 0.66 | 1.60 | 9.05 ± 0.75 | 1.62 |
| NIP C | 4-VP | 0:5:20 | 2.28 ± 0.38 | 6.25 ± 0.36 | 5.58 ± 0.53 | |||
| Trial | Loaded Amount (µmol/g) | Loading Solvent | Washing Solvent | Elution Volume (mL) | MISPE C % Recovery ± SD | NISPE C % Recovery ± SD |
|---|---|---|---|---|---|---|
| 1 | 5 | MeOH | ACN | 2 | 72.53 ± 1.68 | 56.55 ± 2.77 |
| 2 | 10 | MeOH | ACN | 2 | 70.48 ± 2.48 | 55.01 ± 2.89 |
| 3 | 15 | MeOH | ACN | 2 | 51.41 ± 2.04 | 40.59 ± 1.10 |
| 4 | 20 | MeOH | ACN | 2 | 43.31 ± 2.64 | 38.62 ± 3.73 |
| 5 | 5 | H2O | ACN | 2 | 50.21 ± 3.24 | 43.58 ± 4.68 |
| 6 | 5 | EtOH | ACN | 2 | 101.76 ± 1.96 | 63.49 ± 5.84 |
| 7 | 5 | EtOH: H2O (1:1) | ACN | 2 | 50.21 ± 2.97 | 33.87 ± 2.03 |
| 8 | 5 | ACN: H2O (4:1) | ACN | 2 | 54.15 ± 2.03 | 40.24 ± 1.58 |
| 9 | 5 | EtOH | H2O | 2 | 44.46 ± 6.55 | 21.57 ± 4.05 |
| 10 | 5 | EtOH | EtOH | 2 | 56.99 ± 3.28 | 44.06 ± 5.28 |
| 11 | 5 | EtOH | EtOAc | 2 | 68.38 ± 3.98 | 49.24 ± 7.00 |
| 12 | 5 | EtOH | ACN | 1 | 74.77 ± 3.47 | 47.19 ± 2.18 |
| 13 | 5 | EtOH | ACN | 3 | 95.77 ± 5.60 | 60.76 ± 4.93 |
| 14 | 5 | EtOH | ACN | 4 | 98.79 ± 3.10 | 77.10 ± 5.16 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Megahed, S.H.; Abdel-Halim, M.; Hefnawy, A.; Handoussa, H.; Mizaikoff, B.; El Gohary, N.A. Molecularly Imprinted Solid Phase Extraction Strategy for Quinic Acid. Polymers 2022, 14, 3339. https://doi.org/10.3390/polym14163339
Megahed SH, Abdel-Halim M, Hefnawy A, Handoussa H, Mizaikoff B, El Gohary NA. Molecularly Imprinted Solid Phase Extraction Strategy for Quinic Acid. Polymers. 2022; 14(16):3339. https://doi.org/10.3390/polym14163339
Chicago/Turabian StyleMegahed, Sarah H., Mohammad Abdel-Halim, Amr Hefnawy, Heba Handoussa, Boris Mizaikoff, and Nesrine A. El Gohary. 2022. "Molecularly Imprinted Solid Phase Extraction Strategy for Quinic Acid" Polymers 14, no. 16: 3339. https://doi.org/10.3390/polym14163339