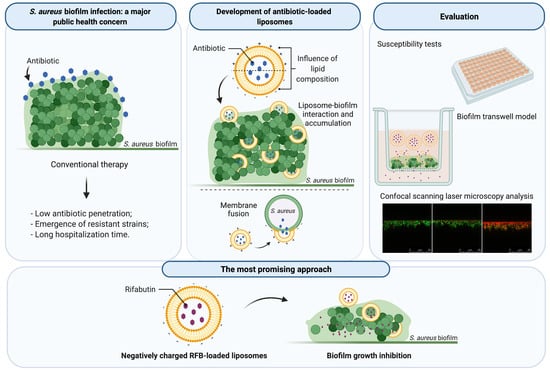
Liposomes as a Nanoplatform to Improve the Delivery of Antibiotics into Staphylococcus aureus Biofilms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Preparation of VCM- and LEV-Loaded Liposomes
2.3. Preparation of RFB-Loaded Liposomes
2.4. Liposomes Characterization
2.5. Bacterial Strain and Culture Conditions
2.6. Planktonic S. aureus Susceptibility to Antibiotics
2.7. S. aureus Biofilm Susceptibility to Antibiotics
2.8. Influence of Lipid Composition on S. aureus Biofilm Interaction
2.9. In Vitro Evaluation of RFB Formulations in a Biofilm Transwell Model
2.10. In Vitro Evaluation of RFB Formulations by Confocal Scanning Laser Microscopy
2.11. Cell Lines Culture Conditions
2.12. Preliminary In Vitro Safety Assessment of Liposomes
2.13. Statistical Analysis
3. Results and Discussion
3.1. Planktonic and Biofilm S. aureus Susceptibility to Antibiotics
3.2. Physicochemical Characterization of Antibiotics-Loaded Liposomes
3.3. Planktonic and Biofilm S. aureus Susceptibility to RFB-Loaded Liposomes
3.4. Influence of Lipid Composition on S. aureus Biofilm Interaction
3.5. In Vitro Evaluation of RFB Formulations in a Biofilm Transwell Model
3.6. Preliminary In Vitro Safety Assessment of RFB Liposomes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zimmerli, W.; Sendi, P. Orthopaedic biofilm infections. APMIS 2017, 125, 353–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parastan, R.; Kargar, M.; Solhjoo, K.; Kafilzadeh, F. Staphylococcus aureus biofilms: Structures, antibiotic resistance, inhibition, and vaccines. Gene Rep. 2020, 20, 100739. [Google Scholar] [CrossRef]
- Bui, L.M.G.; Conlon, B.P.; Kidd, S.P. Antibiotic tolerance and the alternative lifestyles of Staphylococcus aureus. Essays Biochem. 2017, 61, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Conlon, B.P. Staphylococcus aureus chronic and relapsing infections: Evidence of a role for persister cells. BioEssays 2014, 36, 991–996. [Google Scholar] [CrossRef]
- Grassi, L.; Maisetta, G.; Esin, S.; Batoni, G. Combination Strategies to Enhance the Efficacy of Antimicrobial Peptides against Bacterial Biofilms. Front. Microbiol. 2017, 8, 2409. [Google Scholar] [CrossRef]
- Sahukhal, G.S.; Pandey, S.; Elasri, M.O. msaABCR operon is involved in persister cell formation in Staphylococcus aureus. BMC Microbiol. 2017, 17, 218. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, M.; Rzhepishevska, O.; Grenho, L.; Malheiros, D.; Gonçalves, L.; Almeida, A.J.; Jordão, L.; Ribeiro, I.A.; Ramstedt, M.; Gomes, P.; et al. Levofloxacin-loaded bone cement delivery system: Highly effective against intracellular bacteria and Staphylococcus aureus biofilms. Int. J. Pharm. 2017, 532, 241–248. [Google Scholar] [CrossRef]
- Kavanagh, N.; Ryan, E.J.; Widaa, A.; Sexton, G.; Fennell, J.; O’Rourke, S.; Cahill, K.C.; Kearney, C.J.; O’Brien, F.J.; Kerrigan, S.W. Staphylococcal osteomyelitis: Disease progression, treatment challenges, and future directions. Clin. Microbiol. Rev. 2018, 31, 1–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donlan, R.M. Biofilms: Microbial Life on Surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.-Y.; van der Mei, H.C.; Ren, Y.; Busscher, H.J.; Shi, L. Lipid-Based Antimicrobial Delivery-Systems for the Treatment of Bacterial Infections. Front. Chem. 2020, 7, 1–15. [Google Scholar] [CrossRef]
- Ferreira, M.; Aguiar, S.; Bettencourt, A.; Gaspar, M.M. Lipid-based nanosystems for targeting bone implant-associated infections: Current approaches and future endeavors. Drug Deliv. Transl. Res. 2020, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Scriboni, A.B.; Couto, V.M.; Ribeiro, L.N.D.M.; Freires, I.A.; Groppo, F.C.; de Paula, E.; Franz-Montan, M.; Cogo-Müller, K. Fusogenic Liposomes Increase the Antimicrobial Activity of Vancomycin Against Staphylococcus aureus Biofilm. Front. Pharmacol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Konreddy, A.K.; Rani, G.U.; Lee, K.; Choi, Y. Recent Drug-Repurposing-Driven Advances in the Discovery of Novel Antibiotics. Curr. Med. Chem. 2019, 26, 5363–5388. [Google Scholar] [CrossRef] [PubMed]
- Czech, T.; Lalani, R.; Oyewumi, M.O. Delivery Systems as Vital Tools in Drug Repurposing. AAPS PharmSciTech 2019, 20, 116. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, M.; Cruz, A.; Fraga, A.; Castro, A.; Cruz, M.; Pedrosa, J. Developments on Drug Delivery Systems for the Treatment of Mycobacterial Infections. Curr. Top. Med. Chem. 2008, 8, 579–591. [Google Scholar] [CrossRef]
- Gaspar, M.M.; Calado, S.; Pereira, J.; Ferronha, H.; Correia, I.; Castro, H.; Tomás, A.M.; Cruz, M.E.M. Targeted delivery of paromomycin in murine infectious diseases through association to nano lipid systems. Nanomedicine 2015, 11, 1851–1860. [Google Scholar] [CrossRef]
- Nave, M.; Castro, R.E.; Rodrigues, C.M.; Casini, A.; Soveral, G.; Gaspar, M.M. Nanoformulations of a potent copper-based aquaporin inhibitor with cytotoxic effect against cancer cells. Nanomedicine 2016, 11, 1817–1830. [Google Scholar] [CrossRef] [Green Version]
- Gaspar, M.M.; Cruz, A.; Penha, A.F.; Reymão, J.; Sousa, A.C.; Eleutério, C.V.; Domingues, S.A.; Fraga, A.G.; Filho, A.L.; Cruz, M.E.M.; et al. Rifabutin encapsulated in liposomes exhibits increased therapeutic activity in a model of disseminated tuberculosis. Int. J. Antimicrob. Agents 2008, 31, 37–45. [Google Scholar] [CrossRef] [Green Version]
- Pinho, J.O.; Amaral, J.D.; Castro, R.E.; Rodrigues, C.M.P.; Casini, A.; Soveral, G.; Gaspar, M.M. Copper complex nanoformulations featuring highly promising therapeutic potential in murine melanoma models. Nanomedicine 2019, 14, 835–850. [Google Scholar] [CrossRef]
- Elron-Gross, I.; Glucksam, Y.; Margalit, R. Liposomal dexamethasone–diclofenac combinations for local osteoarthritis treatment. Int. J. Pharm. 2009, 376, 84–91. [Google Scholar] [CrossRef]
- Van den Hoven, J.M.; Van Tomme, S.R.; Metselaar, J.M.; Nuijen, B.; Beijnen, J.H.; Storm, G. Liposomal Drug Formulations in the Treatment of Rheumatoid Arthritis. Mol. Pharm. 2011, 8, 1002–1015. [Google Scholar] [CrossRef] [PubMed]
- Forier, K.; Raemdonck, K.; De Smedt, S.C.; Demeester, J.; Coenye, T.; Braeckmans, K. Lipid and polymer nanoparticles for drug delivery to bacterial biofilms. J. Control. Release 2014, 190, 607–623. [Google Scholar] [CrossRef] [Green Version]
- Rukavina, Z.; Vanić, Ž. Current Trends in Development of Liposomes for Targeting Bacterial Biofilms. Pharmaceutics 2016, 8, 18. [Google Scholar] [CrossRef] [Green Version]
- Asseray, N.; Bourigault, C.; Boutoille, D.; Happi, L.; Touchais, S.; Corvec, S.; Bemer, P.; Navas, D. Levofloxacin at the usual dosage to treat bone and joint infections: A cohort analysis. Int. J. Antimicrob. Agents 2016, 47, 478–481. [Google Scholar] [CrossRef] [PubMed]
- Blaskovich, M.A.T.; Hansford, K.A.; Butler, M.S.; Jia, Z.; Mark, A.E.; Cooper, M.A. Developments in Glycopeptide Antibiotics. ACS Infect. Dis. 2018, 4, 715–735. [Google Scholar] [CrossRef] [Green Version]
- Albano, M.; Karau, M.J.; Greenwood-Quaintance, K.E.; Osmon, D.R.; Oravec, C.P.; Berry, D.J.; Abdel, M.P.; Patel, R. In Vitro Activity of Rifampin, Rifabutin, Rifapentine, and Rifaximin against Planktonic and Biofilm States of Staphylococci Isolated from Periprosthetic Joint Infection. Antimicrob. Agents Chemother. 2019, 63, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Kirby, C.; Gregoriadis, G. Dehydration-rehydration vesicles: A simple method for high yield drug entrapment in liposomes. Bio/Technology 1984, 2, 979–984. [Google Scholar] [CrossRef]
- Bhattacharjee, S.; Gaspar, M.M.; Scholz, D.; Almeida, A.J.; Brayden, D.J. Track analysis of the passage of rhodamine-labeled liposomes across porcine jejunal mucus in a microchannel device. Ther. Deliv. 2018, 9, 419–433. [Google Scholar] [CrossRef] [PubMed]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Wang, C.-S.; Smith, R.L. Lowry determination of protein in the presence of Triton X-100. Anal. Biochem. 1975, 63, 414–417. [Google Scholar] [CrossRef]
- Gaspar, M.M.; Perez-Soler, R.; Cruz, M.E.M. Biological characterization of L-asparginase liposomal formulations. Cancer Chemother. Pharmacol. 1996, 38, 373–377. [Google Scholar] [CrossRef] [PubMed]
- Rouser, G.; Fleischer, S.; Akira, Y. Two Dimensional Thin Layer Chromatographic Separation of Polar Lipids and Determination of Phospholipids by Phosphorus Analysis of Spots. Lipids 1970, 5, 494–496. [Google Scholar] [CrossRef]
- Clinical Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; Supplement M100; CLSI: Wayne, PA, USA, 2014; M100-S24. [Google Scholar]
- Brambilla, L.Z.S.; Endo, E.H.; Cortez, D.A.G.; Dias Filho, B.P. Anti-biofilm activity against Staphylococcus aureus MRSA and MSSA of neolignans and extract of Piper regnellii. Rev. Bras. Farmacogn. 2017, 27, 112–117. [Google Scholar] [CrossRef] [Green Version]
- Pontes, C.; Alves, M.; Santos, C.; Ribeiro, M.H.; Gonçalves, L.; Bettencourt, A.F.; Ribeiro, I.A.C. Can Sophorolipids prevent biofilm formation on silicone catheter tubes? Int. J. Pharm. 2016, 513, 697–708. [Google Scholar] [CrossRef]
- Cruz, C.D.; Shah, S.; Tammela, P. Defining conditions for biofilm inhibition and eradication assays for Gram-positive clinical reference strains. BMC Microbiol. 2018, 18, 173. [Google Scholar] [CrossRef]
- Harriott, M.M.; Noverr, M.C. Candida albicans and Staphylococcus aureus Form Polymicrobial Biofilms: Effects on Antimicrobial Resistance. Antimicrob. Agents Chemother. 2009, 53, 3914–3922. [Google Scholar] [CrossRef] [Green Version]
- Pinto, S.N.; Dias, S.A.; Cruz, A.F.; Mil-Homens, D.; Fernandes, F.; Valle, J.; Andreu, D.; Prieto, M.; Castanho, M.A.R.B.; Coutinho, A.; et al. The mechanism of action of pepR, a viral-derived peptide, against Staphylococcus aureus biofilms. J. Antimicrob. Chemother. 2019, 74, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Santos-Rebelo, A.; Garcia, C.; Eleutério, C.; Bastos, A.; Coelho, S.C.; Coelho, M.A.N.; Molpeceres, J.; Viana, A.S.; Ascensão, L.; Pinto, J.F.; et al. Development of parvifloron D-loaded smart nanoparticles to target pancreatic cancer. Pharmaceutics 2018, 10, 216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos-Ferreira, I.; Bettencourt, A.; Almeida, A.J. Nanoparticulate platforms for targeting bone infections: Meeting a major therapeutic challenge. Nanomedicine 2015, 10, 3131–3145. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.; Allan, R.N.; Howlin, R.P.; Stoodley, P.; Hall-Stoodley, L. Targeting microbial biofilms: Current and prospective therapeutic strategies. Nat. Rev. Microbiol. 2017, 15, 740–755. [Google Scholar] [CrossRef]
- Portilla, S.; Fernández, L.; Gutiérrez, D.; Rodríguez, A.; García, P. Encapsulation of the Antistaphylococcal Endolysin LysRODI in pH-Sensitive Liposomes. Antibiotics 2020, 9, 242. [Google Scholar] [CrossRef]
- Bhatia, E.; Sharma, S.; Jadhav, K.; Banerjee, R. Combinatorial liposomes of berberine and curcumin inhibit biofilm formation and intracellular methicillin resistant Staphylococcus aureus infections and associated inflammation. J. Mater. Chem. B 2021, 9, 864–875. [Google Scholar] [CrossRef]
- Dong, D.; Thomas, N.; Thierry, B.; Vreugde, S.; Prestidge, C.A.; Wormald, P.-J. Distribution and Inhibition of Liposomes on Staphylococcus aureus and Pseudomonas aeruginosa Biofilm. PLoS ONE 2015, 10, e0131806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattacharya, M.; Wozniak, D.J.; Stoodley, P.; Hall-Stoodley, L. Prevention and treatment of Staphylococcus aureus biofilms. Expert Rev. Anti-Infect. Ther. 2015, 13, 1499–1516. [Google Scholar] [CrossRef] [Green Version]
- Doub, J.B.; Heil, E.L.; Ntem-Mensah, A.; Neeley, R.; Ching, P.R. Rifabutin use in Staphylococcus biofilm infections: A case series. Antibiotics 2020, 9, 326. [Google Scholar] [CrossRef]
- Levofloxacin | C18H20FN3O4—PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Levofloxacin#section=LogP (accessed on 9 November 2020).
- Levofloxacin | DrugBank Online. Available online: https://go.drugbank.com/drugs/DB01137 (accessed on 9 November 2020).
- Vancomycin Hydrochloride | C66H76Cl3N9O24—PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/6420023#section=Drug-Classes (accessed on 9 November 2020).
- Vancomycin | DrugBank Online. Available online: https://go.drugbank.com/drugs/DB00512 (accessed on 9 November 2020).
- Rifabutin | C46H62N4O11—PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/135398743 (accessed on 9 November 2020).
- Rifabutin | DrugBank Online. Available online: https://go.drugbank.com/drugs/DB00615 (accessed on 9 November 2020).
- Grohs, P.; Kitzis, M.D.; Gutmann, L. In vitro bactericidal activities of linezolid in combination with vancomycin, gentamicin, ciprofloxacin, fusidic acid, and rifampin against Staphylococcus aureus. Antimicrob. Agents Chemother. 2003, 47, 418–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemaire, S.; Kosowska-Shick, K.; Julian, K.; Tulkens, P.M.; Van Bambeke, F.; Appelbaum, P.C. Activities of antistaphylococcal antibiotics towards the extracellular and intraphagocytic forms of Staphylococcus aureus isolates from a patient with persistent bacteraemia and endocarditis. Clin. Microbiol. Infect. 2008, 14, 766–777. [Google Scholar] [CrossRef] [Green Version]
- Chartpitak, T.; Tulakarnwong, S.; Riansuwan, K.; Kiratisin, P.; Nasongkla, N. Vancomycin-impregnated polymer on Schanz pin for prolonged release and antibacterial application. J. Drug Deliv. Sci. Technol. 2018, 47, 223–229. [Google Scholar] [CrossRef]
- Croom, K.F.; Goa, K.L. Levofloxacin: A review of its use in the treatment of bacterial infections in the United States. Drugs 2003, 63, 2769–2802. [Google Scholar] [CrossRef]
- Fish, D.N.; Chow, A.T. The clinical pharmacokinetics of levofloxacin. Clin. Pharmacokinet. 1997, 32, 101–119. [Google Scholar] [CrossRef] [PubMed]
- Mandell, J.B.; Orr, S.; Koch, J.; Nourie, B.; Ma, D.; Bonar, D.D.; Shah, N.; Urish, K.L. Large variations in clinical antibiotic activity against Staphylococcus aureus biofilms of periprosthetic joint infection isolates. J. Orthop. Res. 2019, 37, 1604–1609. [Google Scholar] [CrossRef] [PubMed]
- Macià, M.D.; Rojo-Molinero, E.; Oliver, A. Antimicrobial susceptibility testing in biofilm-growing bacteria. Clin. Microbiol. Infect. 2014, 20, 981–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Jong, W.H.; Borm, P.J.A. Drug delivery and nanoparticles:applications and hazards. Int. J. Nanomed. 2008, 3, 133–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibaraki, H.; Kanazawa, T.; Chien, W.Y.; Nakaminami, H.; Aoki, M.; Ozawa, K.; Kaneko, H.; Takashima, Y.; Noguchi, N.; Seta, Y. The effects of surface properties of liposomes on their activity against Pseudomonas aeruginosa PAO-1 biofilm. J. Drug Deliv. Sci. Technol. 2020, 57. [Google Scholar] [CrossRef]
- Nicolosi, D.; Scalia, M.; Nicolosi, V.M.; Pignatello, R. Encapsulation in fusogenic liposomes broadens the spectrum of action of vancomycin against Gram-negative bacteria. Int. J. Antimicrob. Agents 2010, 35, 553–558. [Google Scholar] [CrossRef] [Green Version]
- Oliveira Pinho, J.; Matias, M.; Gaspar, M.M. Emergent Nanotechnological Strategies for Systemic Chemotherapy against Melanoma. Nanomaterials 2019, 9, 1455. [Google Scholar] [CrossRef] [Green Version]
- Gaspar, M.M.; Neves, S.; Portaels, F.; Pedrosa, J.; Silva, M.T.; Cruz, M.E. Therapeutic efficacy of liposomal rifabutin in a Mycobacterium avium model of infection. Antimicrob. Agents Chemother. 2000, 44, 2424–2430. [Google Scholar] [CrossRef] [Green Version]
- Gomes, M.; Gaspar, M.; Carvalheiro, M.; Cruz, M.; Vale, F. Fusogenic Liposomes As Promising Delivery Systems against Helicobacter Pylori. Helicobacter 2014, 19, 137–138. [Google Scholar]
- Gomes, M.; Lehours, P.; Gaspar, M.; Carvalheiro, M.; Vale, F. Liposomes As Drug Delivery Systems Against Helicobacter Pylori.: Abstract no.: P11. 19. Helicobacter 2013, 18, 30. [Google Scholar]
- Bolotin, E.M.; Cohen, R.; Bar, L.K.; Emanuel, N.; Ninio, S.; Barenholz, Y.; Lasic, D.D. Ammonium Sulfate Gradients for Efficient and Stable Remote Loading of Amphipathic Weak Bases into Liposomes and Ligandoliposomes. J. Liposome Res. 1994, 4, 455–479. [Google Scholar] [CrossRef]
- Bozzuto, G.; Molinari, A. Liposomes as nanomedical devices. Int. J. Nanomed. 2015, 10, 975–999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Sun, P.; Bi, R.; Wang, J.; Zhang, N.; Huang, G. Targeted delivery of levofloxacin-liposomes for the treatment of pulmonary inflammation. J. Drug Target. 2009, 17, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Bhise, K.; Sau, S.; Kebriaei, R.; Rice, S.A.; Stamper, K.C.; Alsaab, H.O.; Rybak, M.J.; Iyer, A.K. Combination of Vancomycin and Cefazolin Lipid Nanoparticles for Overcoming Antibiotic Resistance of MRSA. Materials 2018, 11, 1245. [Google Scholar] [CrossRef] [Green Version]
- Ranjan, A.; Pothayee, N.; Seleem, M.N.; Tyler, R.D.; Brenseke, B.; Sriranganathan, N.; Riffle, J.S.; Kasimanickam, R. Antibacterial efficacy of core-shell nanostructures encapsulating gentamicin against an in vivo intracellular Salmonella model. Int. J. Nanomed. 2009, 4, 289–297. [Google Scholar] [CrossRef] [Green Version]
- Ranjan, A.; Pothayee, N.; Vadala, T.P.; Seleem, M.N.; Restis, E.; Sriranganathan, N.; Riffle, J.S.; Kasimanickam, R. Efficacy of Amphiphilic Core-Shell Nanostructures Encapsulating Gentamicin in an In Vitro Salmonella and Listeria Intracellular Infection Model. Antimicrob. Agents Chemother. 2010, 54, 3524–3526. [Google Scholar] [CrossRef] [Green Version]
- Pinheiro, M.; Magalhães, J.; Reis, S. Antibiotic interactions using liposomes as model lipid membranes. Chem. Phys. Lipids 2019, 222, 36–46. [Google Scholar] [CrossRef]
- Kadry, A.A.; Al-Suwayeh, S.A.; Abd-Allah, A.R.A.; Bayomi, M.A. Treatment of experimental osteomyelitis by liposomal antibiotics. J. Antimicrob. Chemother. 2004, 54, 1103–1108. [Google Scholar] [CrossRef] [Green Version]
- Messiaen, A.-S.; Forier, K.; Nelis, H.; Braeckmans, K.; Coenye, T. Transport of nanoparticles and tobramycin-loaded liposomes in Burkholderia cepacia complex biofilms. PLoS ONE 2013, 8, e79220. [Google Scholar] [CrossRef] [Green Version]
- Köse, N. Biological Response to Orthopedic Implants and Biomaterials. In Musculoskeletal Research and Basic Science; Korkusuz, F., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 3–14. ISBN 978-3-319-20776-6. [Google Scholar]
- Alhariri, M.; Azghani, A.; Omri, A. Liposomal antibiotics for the treatment of infectious diseases. Expert Opin. Drug Deliv. 2013, 10, 1515–1532. [Google Scholar] [CrossRef] [PubMed]
- Arisaka, M.; Nakamura, T.; Yamada, A.; Negishi, Y.; Aramaki, Y. Involvement of protein kinase Cdelta in induction of apoptosis by cationic liposomes in macrophage-like RAW264.7 cells. FEBS Lett. 2010, 584, 1016–1020. [Google Scholar] [CrossRef] [Green Version]
- Iwaoka, S.; Nakamura, T.; Takano, S.; Tsuchiya, S.; Aramaki, Y. Cationic liposomes induce apoptosis through p38 MAP kinase-caspase-8-Bid pathway in macrophage-like RAW264.7 cells. J. Leukoc. Biol. 2006, 79, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Aramaki, Y.; Takano, S.; Tsuchiya, S. Cationic liposomes induce macrophage apoptosis through mitochondrial pathway. Arch. Biochem. Biophys. 2001, 392, 245–250. [Google Scholar] [CrossRef] [PubMed]






| Antibiotics | MIC 1 (µg/mL) | MBIC50 2 (µg/mL) |
|---|---|---|
| RFB | 0.006 ± 0.000 | 0.005 ± 0.002 |
| VCM | 1.562 ± 0.033 | >200.000 |
| LEV | 0.125 ± 0.068 | 9.468 ± 0.672 |
| Antibiotics | Lipid Composition (Molar Ratio) | Loading Capacity (µg/µmol) | I.E. (%) | Ø (µm) (P.I.) | Zeta Potential (mV) |
|---|---|---|---|---|---|
| RFB | DMPC:DOPE:CHEMS (4:4:2) | 57 ± 9 | 87 ± 5 | 0.11 (<0.10) | −22 ± 3 |
| DMPC:DMPG (8:2) | 36 ± 5 | 51 ± 7 | 0.15 (<0.10) | −21 ± 3 | |
| DMPC:SA (9:1) | 24 ± 4 | 32 ± 3 | 0.12 (<0.10) | +13 ± 2 | |
| VCM | DPPC:DOPE:CHEMS (4:4:2) | 45 ± 3 | 19 ± 4 | 0.17 (<0.10) | −30 ± 1 |
| DPPC:DPPG (8:2) | 23 ± 2 | 32 ± 8 | 0.15 (<0.15) | −23 ± 1 | |
| LEV | DMPC:DOPE:CHEMS (4:4:2) | <2 | <3 | 0.12 (<0.10) | −21 ± 2 |
| DMPC:DMPG (8:2) | <2 | <3 | 0.11 (<0.10) | −24 ± 1 | |
| DMPC:SA (9:1) | <2 | <3 | 0.13 (<0.10) | +18 ± 1 |
| Formulation | MIC 1 (µg/mL) | MBIC50 2 (µg/mL) |
|---|---|---|
| Free RFB | 0.006 ± 0.000 | 0.005 ± 0.0032 |
| LIP1 | 0.006 ± 0.004 | 0.002 ± 0.0001 |
| LIP2 | 0.006 ± 0.000 | 0.002 ± 0.0002 |
| LIP3 | 0.006 ± 0.004 | 0.006 ± 0.0030 |
| Lipid Composition | BEL (%) | Biofilm Biomass Reduction (%) 1 |
|---|---|---|
| RFB LIP1 | 17 ± 9 | 72 ± 5 |
| RFB LIP2 | 23 ± 6 | 64 ± 9 |
| RFB LIP3 | 40 ± 8 | 32 ± 14 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferreira, M.; Pinto, S.N.; Aires-da-Silva, F.; Bettencourt, A.; Aguiar, S.I.; Gaspar, M.M. Liposomes as a Nanoplatform to Improve the Delivery of Antibiotics into Staphylococcus aureus Biofilms. Pharmaceutics 2021, 13, 321. https://doi.org/10.3390/pharmaceutics13030321
Ferreira M, Pinto SN, Aires-da-Silva F, Bettencourt A, Aguiar SI, Gaspar MM. Liposomes as a Nanoplatform to Improve the Delivery of Antibiotics into Staphylococcus aureus Biofilms. Pharmaceutics. 2021; 13(3):321. https://doi.org/10.3390/pharmaceutics13030321
Chicago/Turabian StyleFerreira, Magda, Sandra N. Pinto, Frederico Aires-da-Silva, Ana Bettencourt, Sandra I. Aguiar, and Maria Manuela Gaspar. 2021. "Liposomes as a Nanoplatform to Improve the Delivery of Antibiotics into Staphylococcus aureus Biofilms" Pharmaceutics 13, no. 3: 321. https://doi.org/10.3390/pharmaceutics13030321