Layered Double Hydroxide as a Potent Non-viral Vector for Nucleic Acid Delivery Using Gene-Activated Scaffolds for Tissue Regeneration Applications
Abstract
:1. Introduction
2. Materials and Methods
2.1. Synthesis of MgAl-NO3 LDH Nanoparticles
2.2. Complexation of LDH with Nucleic Acid (pDNA, siRNA, miRNA)
2.2.1. Plasmid DNA (pDNA) Propagation
2.2.2. Physicochemical Nanoparticle Characterization
2.2.3. Nucleic Acid Encapsulation
2.3. Mesenchymal Stromal Cell Uptake and Cytocompatibility of LDH–Nucleic Acid Complexes in 2D Monolayer
2.3.1. Cellular Uptake
2.3.2. LDH–Nucleic Acid Cytocompatibility
2.3.3. Assessment of LDH–Nucleic Acid Nanoparticle Efficacy in 2D
2.4. LDH–Nanoparticle Activated Collagen-Nanohydroxyapatite Scaffold Systems
2.5. Confocal Imaging
2.6. Scanning Electron Microscopy
2.7. Statistical Analysis
3. Results
3.1. Efficiency of LDH–pDNA Nanoparticle Complexation
3.2. Efficiency of LDH–siRNA Complexation and Encapsulation
3.3. Efficiency of LDH–miRNA Complexation and Encapsulation
3.4. Uptake and Cytocompatibility of LDH–pDNA Nanoparticles in Monolayer Transfection
3.5. Uptake and Cytocompatibility of LDH–SiRNA Nanoparticles in Monolayer Transfection
3.6. Uptake and Cytocompatibility of LDH–MiRNA Mimic and LDH–MiRNA Inhibitor Nanoparticles in Monolayer Transfection
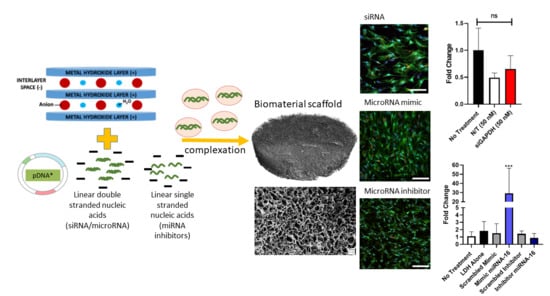
3.7. SiRNA Delivery from Gene-Activated Collagen-Nanohydroxyapatite Scaffolds
3.8. MicroRNA Delivery from Gene-Activated Collagen-Nanohydroxyapatite Scaffolds
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A




References
- Naldini, L. Gene therapy returns to centre stage. Nature 2015, 526, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Curtin, C.M.; Tierney, E.G.; McSorley, K.; Cryan, S.A.; Duffy, G.P.; O’Brien, F.J. Combinatorial gene therapy accelerates bone regeneration: Non-viral dual delivery of VEGF and BMP2 in a collagen-nanohydroxyapatite scaffold. Adv. Healthc. Mater. 2015, 4, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Kelly, D.C.; Raftery, R.M.; Curtin, C.M.; O’Driscoll, C.M.; O’Brien, F.J. Scaffold-Based Delivery of Nucleic Acid Therapeutics for Enhanced Bone and Cartilage Repair. J. Orthop. Res. Off. Publ. Orthop. Res. Soc. 2019, 37, 1671–1680. [Google Scholar] [CrossRef] [PubMed]
- Mencía Castaño, I.; Curtin, C.M.; Shaw, G.; Murphy, J.M.; Duffy, G.P.; O’Brien, F.J. A novel collagen-nanohydroxyapatite microRNA-activated scaffold for tissue engineering applications capable of efficient delivery of both miR-mimics and antagomiRs to human mesenchymal stem cells. J. Control. Release Off. J. Control. Release Soc. 2015, 200, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Castaño, I.M.; Raftery, R.M.; Chen, G.; Cavanagh, B.; Quinn, B.; Duffy, G.P.; O’Brien, F.J.; Curtin, C.M. Rapid bone repair with the recruitment of CD206(+)M2-like macrophages using non-viral scaffold-mediated miR-133a inhibition of host cells. Acta Biomater. 2020, 109, 267–279. [Google Scholar] [CrossRef] [PubMed]
- Curtin, C.M.; Cunniffe, G.M.; Lyons, F.G.; Bessho, K.; Dickson, G.R.; Duffy, G.P.; O’Brien, F.J. Innovative collagen nano-hydroxyapatite scaffolds offer a highly efficient non-viral gene delivery platform for stem cell-mediated bone formation. Adv. Mater. 2012, 24, 749–754. [Google Scholar] [CrossRef]
- Mencía Castaño, I.; Curtin, C.M.; Duffy, G.P.; O’Brien, F.J. Next generation bone tissue engineering: Non-viral miR-133a inhibition using collagen-nanohydroxyapatite scaffolds rapidly enhances osteogenesis. Sci. Rep. 2016, 6, 27941. [Google Scholar] [CrossRef]
- Raftery, R.M.; Gonzalez Vazquez, A.G.; Chen, G.; O’Brien, F.J. Activation of the SOX-5, SOX-6, and SOX-9 Trio of Transcription Factors Using a Gene-Activated Scaffold Stimulates Mesenchymal Stromal Cell Chondrogenesis and Inhibits Endochondral Ossification. Adv. Healthc. Mater. 2020, 9, e1901827. [Google Scholar] [CrossRef]
- Yan, L.P.; Castaño, I.M.; Sridharan, R.; Kelly, D.; Lemoine, M.; Cavanagh, B.L.; Dunne, N.J.; McCarthy, H.O.; O’Brien, F.J. Collagen/GAG scaffolds activated by RALA-siMMP-9 complexes with potential for improved diabetic foot ulcer healing. Mater. Sci. Eng. C Mater. Biol. Appl. 2020, 114, 111022. [Google Scholar] [CrossRef]
- Laiva, A.L.; O’Brien, F.J.; Keogh, M.B. SDF-1α gene-activated collagen scaffold drives functional differentiation of human Schwann cells for wound healing applications. Biotechnol. Bioeng. 2020. [Google Scholar] [CrossRef]
- Liu, H.; Yang, Z.; Xun, Z.; Gao, Z.; Sun, Y.; Yu, J.; Yang, T.; Zhao, X.; Cai, C.; Ding, P. Nuclear delivery of plasmid DNA determines the efficiency of gene expression. Cell Biol. Int. 2019, 43, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leng, Q.; Chen, L.; Lv, Y. RNA-based scaffolds for bone regeneration: Application and mechanisms of mRNA, miRNA and siRNA. Theranostics 2020, 10, 3190–3205. [Google Scholar] [CrossRef] [PubMed]
- Sadakierska-Chudy, A. MicroRNAs: Diverse Mechanisms of Action and Their Potential Applications as Cancer Epi-Therapeutics. Biomolecules 2020, 10, 1285. [Google Scholar] [CrossRef] [PubMed]
- Arabian, M.; Mirzadeh Azad, F.; Maleki, M.; Malakootian, M. Insights into role of microRNAs in cardiac development, cardiac diseases, and developing novel therapies. Iran. J. Basic Med. Sci. 2020, 23, 961–969. [Google Scholar] [CrossRef] [PubMed]
- Bajan, S.; Hutvagner, G. RNA-Based Therapeutics: From Antisense Oligonucleotides to miRNAs. Cells 2020, 9, 137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Urits, I.; Swanson, D.; Swett, M.C.; Patel, A.; Berardino, K.; Amgalan, A.; Berger, A.A.; Kassem, H.; Kaye, A.; Viswanath, O. A Review of Patisiran (ONPATTRO®) for the Treatment of Polyneuropathy in People with Hereditary Transthyretin Amyloidosis. Neurol. Ther. 2020. [Google Scholar] [CrossRef]
- Helal, N.A.; Osami, A.; Helmy, A.; McDonald, T.; Shaaban, L.A.; Nounou, M.I. Non-viral gene delivery systems: Hurdles for bench-to-bedside transformation. Pharmazie 2017, 72, 627–693. [Google Scholar] [CrossRef]
- Benskey, M.J.; Sandoval, I.M.; Miller, K.; Sellnow, R.L.; Gezer, A.; Kuhn, N.C.; Vashon, R.; Manfredsson, F.P. Basic Concepts in Viral Vector-Mediated Gene Therapy. Methods Mol. Biol. 2019, 1937, 3–26. [Google Scholar] [CrossRef]
- Heyde, M.; Partridge, K.A.; Oreffo, R.O.C.; Howdle, S.M.; Shakesheff, K.M.; Garnett, M.C. Gene therapy used for tissue engineering applications. J. Pharm. Pharmacol. 2007, 59, 329–350. [Google Scholar] [CrossRef] [PubMed]
- Bono, N.; Ponti, F.; Mantovani, D.; Candiani, G. Non-Viral in Vitro Gene Delivery: It is Now Time to Set the Bar! Pharmaceutics 2020, 12, 183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, X.; Huang, L. Recent advances in nonviral vectors for gene delivery. Acc. Chem. Res. 2012, 45, 971–979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patil, S.; Gao, Y.G.; Lin, X.; Li, Y.; Dang, K.; Tian, Y.; Zhang, W.J.; Jiang, S.F.; Qadir, A.; Qian, A.R. The Development of Functional Non-Viral Vectors for Gene Delivery. Int. J. Mol. Sci. 2019, 20, 5491. [Google Scholar] [CrossRef] [Green Version]
- Cullis, P.R.; Hope, M.J. Lipid Nanoparticle Systems for Enabling Gene Therapies. Mol. Ther. J. Am. Soc. Gene Ther. 2017, 25, 1467–1475. [Google Scholar] [CrossRef] [Green Version]
- Kulkarni, J.A.; Cullis, P.R.; van der Meel, R. Lipid Nanoparticles Enabling Gene Therapies: From Concepts to Clinical Utility. Nucleic Acid Ther. 2018, 28, 146–157. [Google Scholar] [CrossRef] [Green Version]
- Raftery, R.M.; Tierney, E.G.; Curtin, C.M.; Cryan, S.A.; O’Brien, F.J. Development of a gene-activated scaffold platform for tissue engineering applications using chitosan-pDNA nanoparticles on collagen-based scaffolds. J. Control. Release Off. J. Control. Release Soc. 2015, 210, 84–94. [Google Scholar] [CrossRef]
- Dixon, J.E.; Osman, G.; Morris, G.E.; Markides, H.; Rotherham, M.; Bayoussef, Z.; El Haj, A.J.; Denning, C.; Shakesheff, K.M. Highly efficient delivery of functional cargoes by the synergistic effect of GAG binding motifs and cell-penetrating peptides. Proc. Natl. Acad. Sci. USA 2016, 113, E291–E299. [Google Scholar] [CrossRef] [Green Version]
- Cryan, S.A.; Holohan, A.; Donohue, R.; Darcy, R.; O’Driscoll, C.M. Cell transfection with polycationic cyclodextrin vectors. Eur. J. Pharm. Sci. Off. J. Eur. Fed. Pharm. Sci. 2004, 21, 625–633. [Google Scholar] [CrossRef]
- Elsabahy, M.; Nazarali, A.; Foldvari, M. Non-viral nucleic acid delivery: Key challenges and future directions. Curr. Drug Deliv. 2011, 8, 235–244. [Google Scholar] [CrossRef]
- Bai, H.; Lester, G.M.S.; Petishnok, L.C.; Dean, D.A. Cytoplasmic transport and nuclear import of plasmid DNA. Biosci. Rep. 2017, 37. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Xiu, K.M.; Xu, S.L.; Yang, W.T.; Xu, F.J. Functionalized layered double hydroxide nanoparticles conjugated with disulfide-linked polycation brushes for advanced gene delivery. Bioconjug. Chem. 2013, 24, 968–978. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, C.; Jaskaniec, S.; McCarthy, E.K.; Downing, C.; Opelt, K.; Güth, K.; Shmeliov, A.; Mourad, M.C.D.; Mandel, K.; Nicolosi, V. Structural transformation of layered double hydroxides: An in situ TEM analysis. NPJ 2D Mater. Appl. 2018, 2, 4. [Google Scholar] [CrossRef]
- Xu, Z.P.; Niebert, M.; Porazik, K.; Walker, T.L.; Cooper, H.M.; Middelberg, A.P.; Gray, P.P.; Bartlett, P.F.; Lu, G.Q. Subcellular compartment targeting of layered double hydroxide nanoparticles. J. Control. Release Off. J. Control. Release Soc. 2008, 130, 86–94. [Google Scholar] [CrossRef]
- Choi, S.J.; Oh, J.M.; Choy, J.H. Biocompatible nanoparticles intercalated with anticancer drug for target delivery: Pharmacokinetic and biodistribution study. J. Nanosci. Nanotechnol. 2010, 10, 2913–2916. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Choi, S.J.; Oh, J.M.; Park, T.; Choy, J.H. Anticancer drug-inorganic nanohybrid and its cellular interaction. J. Nanosci. Nanotechnol. 2007, 7, 3700–3705. [Google Scholar] [CrossRef]
- Choy, J.H.; Jung, J.S.; Oh, J.M.; Park, M.; Jeong, J.; Kang, Y.K.; Han, O.J. Layered double hydroxide as an efficient drug reservoir for folate derivatives. Biomaterials 2004, 25, 3059–3064. [Google Scholar] [CrossRef]
- Oh, J.M.; Choi, S.J.; Kim, S.T.; Choy, J.H. Cellular uptake mechanism of an inorganic nanovehicle and its drug conjugates: Enhanced efficacy due to clathrin-mediated endocytosis. Bioconjug. Chem. 2006, 17, 1411–1417. [Google Scholar] [CrossRef]
- Kriven, W.; Kwak, S.Y.; Wallig, M.; Choy, J.H. Bio-Resorbable Nanoceramics for Gene and Drug Delivery. MRS Bull. Mater. Res. Soc. 2004, 29, 33–37. [Google Scholar] [CrossRef]
- Ladewig, K.; Niebert, M.; Xu, Z.P.; Gray, P.P.; Lu, G.Q. Efficient siRNA delivery to mammalian cells using layered double hydroxide nanoparticles. Biomaterials 2010, 31, 1821–1829. [Google Scholar] [CrossRef]
- Desigaux, L.; Belkacem, M.B.; Richard, P.; Cellier, J.; Léone, P.; Cario, L.; Leroux, F.; Taviot-Guého, C.; Pitard, B. Self-assembly and characterization of layered double hydroxide/DNA hybrids. Nano Lett. 2006, 6, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.P.; Walker, T.L.; Liu, K.L.; Cooper, H.M.; Lu, G.Q.; Bartlett, P.F. Layered double hydroxide nanoparticles as cellular delivery vectors of supercoiled plasmid DNA. Int. J. Nanomed. 2007, 2, 163–174. [Google Scholar]
- Choi, S.J.; Oh, J.M.; Choy, J.H. Safety Aspect of Inorganic Layered Nanoparticles: Size-Dependency In Vitro and In Vivo. J. Nanosci. Nanotechnol. 2008, 8, 5297–5301. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.M.; Lim, S.K. Role of miRNAs in bone and their potential as therapeutic targets. Curr. Opin. Pharmacol. 2014, 16, 133–141. [Google Scholar] [CrossRef]
- Cunniffe, G.M.; Dickson, G.R.; Partap, S.; Stanton, K.T.; O’Brien, F.J. Development and characterisation of a collagen nano-hydroxyapatite composite scaffold for bone tissue engineering. J. Mater. Sci. Mater. Med. 2010, 21, 2293–2298. [Google Scholar] [CrossRef] [PubMed]
- Fayyazbakhsh, F.; Solati-Hashjin, M.; Keshtkar, A.; Shokrgozar, M.A.; Dehghan, M.M.; Larijani, B. Novel layered double hydroxides-hydroxyapatite/gelatin bone tissue engineering scaffolds: Fabrication, characterization, and in vivo study. Mater. Sci. Eng. C Mater. Biol. Appl. 2017, 76, 701–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fayyazbakhsh, F.; Solati-Hashjin, M.; Keshtkar, A.; Shokrgozar, M.A.; Dehghan, M.M.; Larijani, B. Release behavior and signaling effect of vitamin D3 in layered double hydroxides-hydroxyapatite/gelatin bone tissue engineering scaffold: An in vitro evaluation. Colloids Surf. B Biointerfaces 2017, 158, 697–708. [Google Scholar] [CrossRef]
- Li, L.; Zhang, R.; Gu, W.; Xu, Z.P. Mannose-conjugated layered double hydroxide nanocomposite for targeted siRNA delivery to enhance cancer therapy. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 2355–2364. [Google Scholar] [CrossRef]
- Park, D.H.; Cho, J.; Kwon, O.J.; Yun, C.O.; Choy, J.H. Biodegradable Inorganic Nanovector: Passive versus Active Tumor Targeting in siRNA Transportation. Angew. Chem. 2016, 55, 4582–4586. [Google Scholar] [CrossRef]
- Li, L.; Gu, W.; Chen, J.; Chen, W.; Xu, Z.P. Co-delivery of siRNAs and anti-cancer drugs using layered double hydroxide nanoparticles. Biomaterials 2014, 35, 3331–3339. [Google Scholar] [CrossRef] [Green Version]
- Zheng, W.; Yin, T.; Chen, Q.; Qin, X.; Huang, X.; Zhao, S.; Xu, T.; Chen, L.; Liu, J. Co-delivery of Se nanoparticles and pooled SiRNAs for overcoming drug resistance mediated by P-glycoprotein and class III β-tubulin in drug-resistant breast cancers. Acta Biomater. 2016, 31, 197–210. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Bao, W.; Umar, A.; Wang, Q.; O’Hare, D.; Wan, Y. Delaminated Layered Double Hydroxide Nanosheets as an Efficient Vector for DNA Delivery. J. Biomed. Nanotechnol. 2016, 12, 922–933. [Google Scholar] [CrossRef]
- Senapati, S.; Sarkar, T.; Das, P.; Maiti, P. Layered Double Hydroxide Nanoparticles for Efficient Gene Delivery for Cancer Treatment. Bioconjug. Chem. 2019, 30, 2544–2554. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, J.; Wang, C.J.; Wang, Q.; Cader, M.Z.; Lu, J.; Evans, D.G.; Duan, X.; O’Hare, D. Cellular uptake and gene delivery using layered double hydroxide nanoparticles. J. Mater. Chem. B 2013, 1, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Wong, Y.; Markham, K.; Xu, Z.P.; Chen, M.; Max Lu, G.Q.; Bartlett, P.F.; Cooper, H.M. Efficient delivery of siRNA to cortical neurons using layered double hydroxide nanoparticles. Biomaterials 2010, 31, 8770–8779. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.Y.; van Ee, R.J.; Timmer, K.; Craenmehr, E.G.M.; Huang, J.H.; Öner, F.C.; Dhert, W.J.A.; Kragten, A.H.M.; Willems, N.; Grinwis, G.C.M.; et al. A novel injectable thermoresponsive and cytocompatible gel of poly(N-isopropylacrylamide) with layered double hydroxides facilitates siRNA delivery into chondrocytes in 3D culture. Acta Biomater. 2015, 23, 214–228. [Google Scholar] [CrossRef] [PubMed]
- González-Vázquez, A.; Planell, J.A.; Engel, E. Extracellular calcium and CaSR drive osteoinduction in mesenchymal stromal cells. Acta Biomater. 2014, 10, 2824–2833. [Google Scholar] [CrossRef]
- Tierney, E.G.; Duffy, G.P.; Hibbitts, A.J.; Cryan, S.A.; O’Brien, F.J. The development of non-viral gene-activated matrices for bone regeneration using polyethyleneimine (PEI) and collagen-based scaffolds. J. Control. Release Off. J. Control. Release Soc. 2012, 158, 304–311. [Google Scholar] [CrossRef]
- Cunniffe, G.M.; Curtin, C.M.; Thompson, E.M.; Dickson, G.R.; O’Brien, F.J. Content-Dependent Osteogenic Response of Nanohydroxyapatite: An in Vitro and in Vivo Assessment within Collagen-Based Scaffolds. ACS Appl. Mater. Interfaces 2016, 8, 23477–23488. [Google Scholar] [CrossRef]
- Curtin, C.M.; Castaño, I.M.; O’Brien, F.J. Scaffold-Based microRNA Therapies in Regenerative Medicine and Cancer. Adv. Healthc. Mater. 2018, 7. [Google Scholar] [CrossRef]
- Wu, Y.; Gu, W.; Chen, C.; Do, S.T.; Xu, Z.P. Optimization of Formulations Consisting of Layered Double Hydroxide Nanoparticles and Small Interfering RNA for Efficient Knockdown of the Target Gene. ACS Omega 2018, 3, 4871–4877. [Google Scholar] [CrossRef] [PubMed]
- Faraji, A.H.; Wipf, P. Nanoparticles in cellular drug delivery. Bioorg. Med. Chem. 2009, 17, 2950–2962. [Google Scholar] [CrossRef]
- Yan, L.; Gonca, S.; Zhu, G.; Zhang, W.; Chen, X. Layered double hydroxide nanostructures and nanocomposites for biomedical applications. J. Mater. Chem. B 2019, 7, 5583–5601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrea, K.A.; Wang, L.; Carrier, A.J.; Campbell, M.; Buhariwalla, M.; Mutch, M.; MacQuarrie, S.L.; Bennett, C.; Mkandawire, M.; Oakes, K.; et al. Adsorption of Oligo-DNA on Magnesium Aluminum-Layered Double-Hydroxide Nanoparticle Surfaces: Mechanistic Implication in Gene Delivery. Langmuir ACS J. Surf. Colloids 2017, 33, 3926–3933. [Google Scholar] [CrossRef]
- Bao, W.; Wang, J.; Wang, Q.; O’Hare, D.; Wan, Y. Layered Double Hydroxide Nanotransporter for Molecule Delivery to Intact Plant Cells. Sci. Rep. 2016, 6, 26738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanderson, B.A.; Sowersby, D.S.; Crosby, S.; Goss, M.; Lewis, L.K.; Beall, G.W. Charge density and particle size effects on oligonucleotide and plasmid DNA binding to nanosized hydrotalcite. Biointerphases 2013, 8, 8. [Google Scholar] [CrossRef] [Green Version]
- Yazdani, P.; Mansouri, E.; Eyvazi, S.; Yousefi, V.; Kahroba, H.; Hejazi, M.S.; Mesbahi, A.; Tarhriz, V.; Abolghasemi, M.M. Layered double hydroxide nanoparticles as an appealing nanoparticle in gene/plasmid and drug delivery system in C2C12 myoblast cells. Artif. Cells Nanomed. Biotechnol. 2019, 47, 436–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fitzgerald, K.A.; Guo, J.; Raftery, R.M.; Castaño, I.M.; Curtin, C.M.; Gooding, M.; Darcy, R.; FJ, O.B.; CM, O.D. Nanoparticle-mediated siRNA delivery assessed in a 3D co-culture model simulating prostate cancer bone metastasis. Int. J. Pharm. 2016, 511, 1058–1069. [Google Scholar] [CrossRef] [PubMed]
- Ladewig, K.; Xu, Z.P.; Lu, G.Q. Layered double hydroxide nanoparticles in gene and drug delivery. Expert Opin. Drug Deliv. 2009, 6, 907–922. [Google Scholar] [CrossRef]
- Chugh, P.; Dittmer, D.P. Potential pitfalls in microRNA profiling. Wiley Interdiscip. Rev. RNA 2012, 3, 601–616. [Google Scholar] [CrossRef] [Green Version]
- Huang, M.; Fong, C.W.; Khor, E.; Lim, L.Y. Transfection efficiency of chitosan vectors: Effect of polymer molecular weight and degree of deacetylation. J. Control. Release Off. J. Control. Release Soc. 2005, 106, 391–406. [Google Scholar] [CrossRef]
- Ladewig, K.; Niebert, M.; Xu, Z.P.; Gray, P.P.; Lu, G.Q. Controlled preparation of layered double hydroxide nanoparticles and their application as gene delivery vehicles. Appl. Clay Sci. 2010, 48, 280–289. [Google Scholar] [CrossRef]
- Choi, S.J.; Oh, J.M.; Choy, J.H. Toxicological effects of inorganic nanoparticles on human lung cancer A549 cells. J. Inorg. Biochem. 2009, 103, 463–471. [Google Scholar] [CrossRef]
- Chen, M.; Cooper, H.M.; Zhou, J.Z.; Bartlett, P.F.; Xu, Z.P. Reduction in the size of layered double hydroxide nanoparticles enhances the efficiency of siRNA delivery. J. Colloid Interface Sci. 2013, 390, 275–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acharya, R.; Chakraborty, M.; Chakraborty, J. Prospective treatment of Parkinson’s disease by a siRNA-LDH nanoconjugate. MedChemComm 2019, 10, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Wong, Y.; Cooper, H.M.; Zhang, K.; Chen, M.; Bartlett, P.; Xu, Z.P. Efficiency of layered double hydroxide nanoparticle-mediated delivery of siRNA is determined by nucleotide sequence. J. Colloid Interface Sci. 2012, 369, 453–459. [Google Scholar] [CrossRef]
- Yang, L.; Sun, J.; Liu, Q.; Zhu, R.; Yang, Q.; Hua, J.; Zheng, L.; Li, K.; Wang, S.; Li, A. Synergetic Functional Nanocomposites Enhance Immunotherapy in Solid Tumors by Remodeling the Immunoenvironment. Adv. Sci. 2019, 6, 1802012. [Google Scholar] [CrossRef] [Green Version]
- Yoo, S.S.; Razzak, R.; Bédard, E.; Guo, L.; Shaw, A.R.; Moore, R.B.; Roa, W.H. Layered gadolinium-based nanoparticle as a novel delivery platform for microRNA therapeutics. Nanotechnology 2014, 25, 425102. [Google Scholar] [CrossRef]
- Mencía Castaño, I.; Curtin, C.M.; Duffy, G.P.; O’Brien, F.J. Harnessing an Inhibitory Role of miR-16 in Osteogenesis by Human Mesenchymal Stem Cells for Advanced Scaffold-Based Bone Tissue Engineering. Tissue Eng. Part A 2019, 25, 24–33. [Google Scholar] [CrossRef]
- Chen, Y.X.; Zhu, R.; Ke, Q.F.; Gao, Y.S.; Zhang, C.Q.; Guo, Y.P. MgAl layered double hydroxide/chitosan porous scaffolds loaded with PFTalpha to promote bone regeneration. Nanoscale 2017, 9, 6765–6776. [Google Scholar] [CrossRef]
- Cao, D.; Xu, Z.; Chen, Y.; Ke, Q.; Zhang, C.; Guo, Y. Ag-loaded MgSrFe-layered double hydroxide/chitosan composite scaffold with enhanced osteogenic and antibacterial property for bone engineering tissue. J. Biomed. Mater. Res. Part B Appl. Biomater. 2018, 106, 863–873. [Google Scholar] [CrossRef] [PubMed]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costard, L.S.; Kelly, D.C.; Power, R.N.; Hobbs, C.; Jaskaniec, S.; Nicolosi, V.; Cavanagh, B.L.; Curtin, C.M.; O’Brien, F.J. Layered Double Hydroxide as a Potent Non-viral Vector for Nucleic Acid Delivery Using Gene-Activated Scaffolds for Tissue Regeneration Applications. Pharmaceutics 2020, 12, 1219. https://doi.org/10.3390/pharmaceutics12121219
Costard LS, Kelly DC, Power RN, Hobbs C, Jaskaniec S, Nicolosi V, Cavanagh BL, Curtin CM, O’Brien FJ. Layered Double Hydroxide as a Potent Non-viral Vector for Nucleic Acid Delivery Using Gene-Activated Scaffolds for Tissue Regeneration Applications. Pharmaceutics. 2020; 12(12):1219. https://doi.org/10.3390/pharmaceutics12121219
Chicago/Turabian StyleCostard, Lara S., Domhnall C. Kelly, Rachael N. Power, Christopher Hobbs, Sonia Jaskaniec, Valeria Nicolosi, Brenton L. Cavanagh, Caroline M. Curtin, and Fergal J. O’Brien. 2020. "Layered Double Hydroxide as a Potent Non-viral Vector for Nucleic Acid Delivery Using Gene-Activated Scaffolds for Tissue Regeneration Applications" Pharmaceutics 12, no. 12: 1219. https://doi.org/10.3390/pharmaceutics12121219