Temporal Dynamics of the Intestinal Microbiome Following Short-Term Dietary Restriction
Abstract
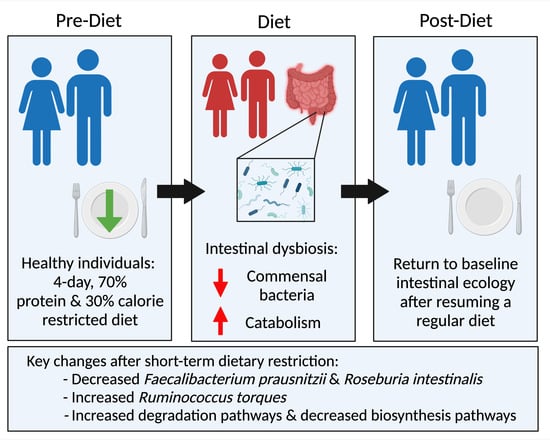
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Subjects and Dietary Intervention
2.3. Sample Collection, DNA Extraction, and Shotgun Metagenomic Sequencing
2.4. Taxonomic and Functional Pathway Analysis
3. Results
3.1. Subject Characteristics and Specimen Collection
3.2. Compositional Analysis
3.3. Functional Pathway Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Brandhorst, S.; Harputlugil, E.; Mitchell, J.R.; Longo, V.D. Protective effects of short-term dietary restriction in surgical stress and chemotherapy. Ageing Res. Rev. 2017, 39, 68–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Longchamp, A.; Harputlugil, E.; Corpataux, J.M.; Ozaki, C.K.; Mitchell, J.R. Is Overnight Fasting before Surgery Too Much or Not Enough? How Basic Aging Research Can Guide Preoperative Nutritional Recommendations to Improve Surgical Outcomes: A Mini-Review. Gerontology 2017, 63, 228–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, J.R.; Beckman, J.A.; Nguyen, L.L.; Ozaki, C.K. Reducing elective vascular surgery perioperative risk with brief preoperative dietary restriction. Surgery 2013, 153, 594–598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gredilla, R.; Barja, G. Minireview: The role of oxidative stress in relation to caloric restriction and longevity. Endocrinology 2005, 146, 3713–3717. [Google Scholar] [CrossRef] [PubMed]
- Fontana, L.; Klein, S. Aging, adiposity, and calorie restriction. JAMA 2007, 297, 986–994. [Google Scholar] [CrossRef] [PubMed]
- Fontana, L.; Partridge, L. Promoting health and longevity through diet: From model organisms to humans. Cell 2015, 161, 106–118. [Google Scholar] [CrossRef] [Green Version]
- Di Francesco, A.; Di Germanio, C.; Bernier, M.; de Cabo, R. A time to fast. Science 2018, 362, 770–775. [Google Scholar] [CrossRef] [Green Version]
- Robertson, L.T.; Mitchell, J.R. Benefits of short-term dietary restriction in mammals. Exp. Gerontol. 2013, 48, 1043–1048. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, J.R.; Verweij, M.; Brand, K.; van de Ven, M.; Goemaere, N.; van den Engel, S.; Chu, T.; Forrer, F.; Müller, C.; de Jong, M.; et al. Short-term dietary restriction and fasting precondition against ischemia reperfusion injury in mice. Aging Cell 2010, 9, 40–53. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, S.E.; Delville, C.; Konstantopedos, P.; Hurst, J.; Derous, D.; Green, C.; Chen, L.; Han, J.J.; Wang, Y.; Promislow, D.E.; et al. The effects of graded levels of calorie restriction: II. Impact of short term calorie and protein restriction on circulating hormone levels, glucose homeostasis and oxidative stress in male C57BL/6 mice. Oncotarget 2015, 6, 23213–23237. [Google Scholar] [CrossRef] [Green Version]
- Trocha, K.; Kip, P.; MacArthur, M.R.; Mitchell, S.J.; Longchamp, A.; Treviño-Villarreal, J.H.; Tao, M.; Bredella, M.A.; De Amorim Bernstein, K.; Mitchell, J.R.; et al. Preoperative Protein or Methionine Restriction Preserves Wound Healing and Reduces Hyperglycemia. J. Surg. Res. 2019, 235, 216–222. [Google Scholar] [CrossRef] [PubMed]
- Robertson, L.T.; Treviño-Villarreal, J.H.; Mejia, P.; Grondin, Y.; Harputlugil, E.; Hine, C.; Vargas, D.; Zheng, H.; Ozaki, C.K.; Kristal, B.S.; et al. Protein and Calorie Restriction Contribute Additively to Protection from Renal Ischemia Reperfusion Injury Partly via Leptin Reduction in Male Mice. J. Nutr. 2015, 145, 1717–1727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verweij, M.; van Ginhoven, T.M.; Mitchell, J.R.; Sluiter, W.; van den Engel, S.; Roest, H.P.; Torabi, E.; Ijzermans, J.N.; Hoeijmakers, J.H.; de Bruin, R.W. Preoperative fasting protects mice against hepatic ischemia/reperfusion injury: Mechanisms and effects on liver regeneration. Liver Transpl. 2011, 17, 695–704. [Google Scholar] [CrossRef] [PubMed]
- Mauro, C.R.; Tao, M.; Yu, P.; Treviño-Villerreal, J.H.; Longchamp, A.; Kristal, B.S.; Ozaki, C.K.; Mitchell, J.R. Preoperative dietary restriction reduces intimal hyperplasia and protects from ischemia-reperfusion injury. J. Vasc. Surg. 2016, 63, 500–509.e501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trocha, K.M.; Kip, P.; Tao, M.; MacArthur, M.R.; Treviño-Villarreal, J.H.; Longchamp, A.; Toussaint, W.; Lambrecht, B.N.; de Vries, M.R.; Quax, P.H.A.; et al. Short-term preoperative protein restriction attenuates vein graft disease via induction of cystathionine γ-lyase. Cardiovasc. Res. 2020, 116, 416–428. [Google Scholar] [CrossRef]
- Nguyen, B.; Tao, M.; Yu, P.; Mauro, C.; Seidman, M.A.; Wang, Y.E.; Mitchell, J.; Ozaki, C.K. Preoperative diet impacts the adipose tissue response to surgical trauma. Surgery 2013, 153, 584–593. [Google Scholar] [CrossRef] [Green Version]
- Peng, W.; Robertson, L.; Gallinetti, J.; Mejia, P.; Vose, S.; Charlip, A.; Chu, T.; Mitchell, J.R. Surgical stress resistance induced by single amino acid deprivation requires Gcn2 in mice. Sci. Transl. Med. 2012, 4, 118ra111. [Google Scholar] [CrossRef] [Green Version]
- Hew, J.J.; Parungao, R.J.; Mooney, C.P.; Smyth, J.K.; Kim, S.; Tsai, K.H.; Shi, H.; Chong, C.; Chan, R.C.F.; Attia, B.; et al. Low-protein diet accelerates wound healing in mice post-acute injury. Burns Trauma 2021, 9, tkab010. [Google Scholar] [CrossRef]
- Kip, P.; Sluiter, T.J.; Moore, J.K.; Hart, A.; Ruske, J.; O’Leary, J.; Jung, J.; Tao, M.; MacArthur, M.R.; Heindel, P.; et al. Short-Term Pre-Operative Protein Caloric Restriction in Elective Vascular Surgery Patients: A Randomized Clinical Trial. Nutrients 2021, 13, 4024. [Google Scholar] [CrossRef]
- Jongbloed, F.; de Bruin, R.W.; Klaassen, R.A.; Beekhof, P.; van Steeg, H.; Dor, F.J.; van der Harst, E.; Dollé, M.E.; JN, I.J. Short-Term Preoperative Calorie and Protein Restriction Is Feasible in Healthy Kidney Donors and Morbidly Obese Patients Scheduled for Surgery. Nutrients 2016, 8, 306. [Google Scholar] [CrossRef] [Green Version]
- van Ginhoven, T.M.; de Bruin, R.W.; Timmermans, M.; Mitchell, J.R.; Hoeijmakers, J.H.; Ijzermans, J.N. Pre-operative dietary restriction is feasible in live-kidney donors. Clin. Transplant. 2011, 25, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Grundmann, F.; Müller, R.U.; Reppenhorst, A.; Hülswitt, L.; Späth, M.R.; Kubacki, T.; Scherner, M.; Faust, M.; Becker, I.; Wahlers, T.; et al. Preoperative Short-Term Calorie Restriction for Prevention of Acute Kidney Injury After Cardiac Surgery: A Randomized, Controlled, Open-Label, Pilot Trial. J. Am. Heart Assoc. 2018, 7, e008181. [Google Scholar] [CrossRef] [PubMed]
- Gallinetti, J.; Harputlugil, E.; Mitchell, J.R. Amino acid sensing in dietary-restriction-mediated longevity: Roles of signal-transducing kinases GCN2 and TOR. Biochem. J. 2013, 449, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harputlugil, E.; Hine, C.; Vargas, D.; Robertson, L.; Manning, B.D.; Mitchell, J.R. The TSC complex is required for the benefits of dietary protein restriction on stress resistance in vivo. Cell Rep. 2014, 8, 1160–1170. [Google Scholar] [CrossRef] [Green Version]
- Hill, C.M.; Berthoud, H.R.; Münzberg, H.; Morrison, C.D. Homeostatic sensing of dietary protein restriction: A case for FGF21. Front. Neuroendocrinol. 2018, 51, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Jongbloed, F.; Saat, T.C.; Verweij, M.; Payan-Gomez, C.; Hoeijmakers, J.H.; van den Engel, S.; van Oostrom, C.T.; Ambagtsheer, G.; Imholz, S.; Pennings, J.L.; et al. A signature of renal stress resistance induced by short-term dietary restriction, fasting, and protein restriction. Sci. Rep. 2017, 7, 40901. [Google Scholar] [CrossRef]
- Soultoukis, G.A.; Partridge, L. Dietary Protein, Metabolism, and Aging. Annu. Rev. Biochem. 2016, 85, 5–34. [Google Scholar] [CrossRef] [Green Version]
- Yap, Y.W.; Rusu, P.M.; Chan, A.Y.; Fam, B.C.; Jungmann, A.; Solon-Biet, S.M.; Barlow, C.K.; Creek, D.J.; Huang, C.; Schittenhelm, R.B.; et al. Restriction of essential amino acids dictates the systemic metabolic response to dietary protein dilution. Nat. Commun. 2020, 11, 2894. [Google Scholar] [CrossRef]
- Solon-Biet, S.M.; Mitchell, S.J.; de Cabo, R.; Raubenheimer, D.; Le Couteur, D.G.; Simpson, S.J. Macronutrients and caloric intake in health and longevity. J. Endocrinol. 2015, 226, R17–R28. [Google Scholar] [CrossRef]
- Ignacio, A.; Morales, C.I.; Câmara, N.O.; Almeida, R.R. Innate Sensing of the Gut Microbiota: Modulation of Inflammatory and Autoimmune Diseases. Front. Immunol. 2016, 7, 54. [Google Scholar] [CrossRef] [Green Version]
- Hansen, T.H.; Gøbel, R.J.; Hansen, T.; Pedersen, O. The gut microbiome in cardio-metabolic health. Genome Med. 2015, 7, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richards, E.M.; Li, J.; Stevens, B.R.; Pepine, C.J.; Raizada, M.K. Gut Microbiome and Neuroinflammation in Hypertension. Circ. Res. 2022, 130, 401–417. [Google Scholar] [CrossRef] [PubMed]
- Nakai, M.; Ribeiro, R.V.; Stevens, B.R.; Gill, P.; Muralitharan, R.R.; Yiallourou, S.; Muir, J.; Carrington, M.; Head, G.A.; Kaye, D.M.; et al. Essential Hypertension Is Associated With Changes in Gut Microbial Metabolic Pathways: A Multisite Analysis of Ambulatory Blood Pressure. Hypertension 2021, 78, 804–815. [Google Scholar] [CrossRef] [PubMed]
- Stevens, B.R.; Goel, R.; Seungbum, K.; Richards, E.M.; Holbert, R.C.; Pepine, C.J.; Raizada, M.K. Increased human intestinal barrier permeability plasma biomarkers zonulin and FABP2 correlated with plasma LPS and altered gut microbiome in anxiety or depression. Gut 2018, 67, 1555–1557. [Google Scholar] [CrossRef] [PubMed]
- Prasad, R.; Patton, M.J.; Floyd, J.L.; Vieira, C.P.; Fortmann, S.; DuPont, M.; Harbour, A.; Jeremy, C.S.; Wright, J.; Lamendella, R.; et al. Plasma microbiome in COVID-19 subjects: An indicator of gut barrier defects and dysbiosis. bioRxiv 2021. [Google Scholar] [CrossRef]
- Leeming, E.R.; Johnson, A.J.; Spector, T.D.; Le Roy, C.I. Effect of Diet on the Gut Microbiota: Rethinking Intervention Duration. Nutrients 2019, 11, 2862. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.K.; Chang, H.W.; Yan, D.; Lee, K.M.; Ucmak, D.; Wong, K.; Abrouk, M.; Farahnik, B.; Nakamura, M.; Zhu, T.H.; et al. Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 2017, 15, 73. [Google Scholar] [CrossRef] [Green Version]
- Rinninella, E.; Cintoni, M.; Raoul, P.; Ianiro, G.; Laterza, L.; Lopetuso, L.R.; Ponziani, F.R.; Gasbarrini, A.; Mele, M.C. Gut Microbiota during Dietary Restrictions: New Insights in Non-Communicable Diseases. Microorganisms 2020, 8, 1140. [Google Scholar] [CrossRef]
- Zheng, X.; Wang, S.; Jia, W. Calorie restriction and its impact on gut microbial composition and global metabolism. Front. Med. 2018, 12, 634–644. [Google Scholar] [CrossRef] [Green Version]
- Paffenbarger, R.S., Jr.; Wing, A.L.; Hyde, R.T. Physical activity as an index of heart attack risk in college alumni. Am. J. Epidemiol. 1978, 108, 161–175. [Google Scholar] [CrossRef]
- Ainsworth, B.E.; Leon, A.S.; Richardson, M.T.; Jacobs, D.R.; Paffenbarger, R.S., Jr. Accuracy of the College Alumnus Physical Activity Questionnaire. J. Clin. Epidemiol. 1993, 46, 1403–1411. [Google Scholar] [CrossRef]
- Mifflin, M.D.; St Jeor, S.T.; Hill, L.A.; Scott, B.J.; Daugherty, S.A.; Koh, Y.O. A new predictive equation for resting energy expenditure in healthy individuals. Am. J. Clin. Nutr. 1990, 51, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Song, L.; Breitwieser, F.P.; Salzberg, S.L. Centrifuge: Rapid and sensitive classification of metagenomic sequences. Genome Res. 2016, 26, 1721–1729. [Google Scholar] [CrossRef] [Green Version]
- Franzosa, E.A.; McIver, L.J.; Rahnavard, G.; Thompson, L.R.; Schirmer, M.; Weingart, G.; Lipson, K.S.; Knight, R.; Caporaso, J.G.; Segata, N.; et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat. Methods 2018, 15, 962–968. [Google Scholar] [CrossRef]
- Suzek, B.E.; Wang, Y.; Huang, H.; McGarvey, P.B.; Wu, C.H. UniRef clusters: A comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 2015, 31, 926–932. [Google Scholar] [CrossRef] [Green Version]
- Caspi, R.; Billington, R.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Midford, P.E.; Ong, W.K.; Paley, S.; Subhraveti, P.; Karp, P.D. The MetaCyc database of metabolic pathways and enzymes—A 2019 update. Nucleic Acids Res. 2020, 48, D445–D453. [Google Scholar] [CrossRef] [Green Version]
- Stevens, B.R.; Roesch, L.; Thiago, P.; Russell, J.T.; Pepine, C.J.; Holbert, R.C.; Raizada, M.K.; Triplett, E.W. Depression phenotype identified by using single nucleotide exact amplicon sequence variants of the human gut microbiome. Mol. Psychiatry 2020, 26, 4277–4287. [Google Scholar] [CrossRef]
- Roesch, L.F.W.; Dobbler, P.T.; Pylro, V.S.; Kolaczkowski, B.; Drew, J.C.; Triplett, E.W. pime: A package for discovery of novel differences among microbial communities. Mol. Ecol. Resour. 2020, 20, 415–428. [Google Scholar] [CrossRef]
- Nearing, J.T.; Douglas, G.M.; Hayes, M.G.; MacDonald, J.; Desai, D.K.; Allward, N.; Jones, C.M.A.; Wright, R.J.; Dhanani, A.S.; Comeau, A.M.; et al. Microbiome differential abundance methods produce different results across 38 datasets. Nat. Commun. 2022, 13, 342. [Google Scholar] [CrossRef]
- Fernandes, A.D.; Reid, J.N.; Macklaim, J.M.; McMurrough, T.A.; Edgell, D.R.; Gloor, G.B. Unifying the analysis of high-throughput sequencing datasets: Characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome 2014, 2, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chong, J.; Liu, P.; Zhou, G.; Xia, J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 2020, 15, 799–821. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Foster, Z.S.; Sharpton, T.J.; Grünwald, N.J. Metacoder: An R package for visualization and manipulation of community taxonomic diversity data. PLoS Comput. Biol. 2017, 13, e1005404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salonen, A.; Lahti, L.; Salojärvi, J.; Holtrop, G.; Korpela, K.; Duncan, S.H.; Date, P.; Farquharson, F.; Johnstone, A.M.; Lobley, G.E.; et al. Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. ISME J. 2014, 8, 2218–2230. [Google Scholar] [CrossRef]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [Green Version]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef]
- Lopez-Siles, M.; Duncan, S.H.; Garcia-Gil, L.J.; Martinez-Medina, M. Faecalibacterium prausnitzii: From microbiology to diagnostics and prognostics. ISME J. 2017, 11, 841–852. [Google Scholar] [CrossRef]
- Nie, K.; Ma, K.; Luo, W.; Shen, Z.; Yang, Z.; Xiao, M.; Tong, T.; Yang, Y.; Wang, X. Roseburia intestinalis: A Beneficial Gut Organism from the Discoveries in Genus and Species. Front. Cell. Infect. Microbiol. 2021, 11, 757718. [Google Scholar] [CrossRef]
- Zhu, C.; Song, K.; Shen, Z.; Quan, Y.; Tan, B.; Luo, W.; Wu, S.; Tang, K.; Yang, Z.; Wang, X. Roseburia intestinalis inhibits interleukin--17 excretion and promotes regulatory T cells differentiation in colitis. Mol. Med. Rep. 2018, 17, 7567–7574. [Google Scholar] [CrossRef] [Green Version]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martín, R.; Miquel, S.; Benevides, L.; Bridonneau, C.; Robert, V.; Hudault, S.; Chain, F.; Berteau, O.; Azevedo, V.; Chatel, J.M.; et al. Functional Characterization of Novel Faecalibacterium prausnitzii Strains Isolated from Healthy Volunteers: A Step Forward in the Use of F. prausnitzii as a Next-Generation Probiotic. Front. Microbiol. 2017, 8, 1226. [Google Scholar] [CrossRef] [PubMed]
- Sokol, H.; Seksik, P.; Furet, J.P.; Firmesse, O.; Nion-Larmurier, I.; Beaugerie, L.; Cosnes, J.; Corthier, G.; Marteau, P.; Doré, J. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm. Bowel. Dis 2009, 15, 1183–1189. [Google Scholar] [CrossRef]
- Chen, X.; Devaraj, S. Gut Microbiome in Obesity, Metabolic Syndrome, and Diabetes. Curr. Diab. Rep. 2018, 18, 129. [Google Scholar] [CrossRef] [PubMed]
- Jie, Z.; Xia, H.; Zhong, S.L.; Feng, Q.; Li, S.; Liang, S.; Zhong, H.; Liu, Z.; Gao, Y.; Zhao, H.; et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat. Commun. 2017, 8, 845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benítez-Páez, A.; Gómez Del Pugar, E.M.; López-Almela, I.; Moya-Pérez, Á.; Codoñer-Franch, P.; Sanz, Y. Depletion of Blautia Species in the Microbiota of Obese Children Relates to Intestinal Inflammation and Metabolic Phenotype Worsening. mSystems 2020, 5, e00587-19. [Google Scholar] [CrossRef] [Green Version]
- Kasai, C.; Sugimoto, K.; Moritani, I.; Tanaka, J.; Oya, Y.; Inoue, H.; Tameda, M.; Shiraki, K.; Ito, M.; Takei, Y.; et al. Comparison of the gut microbiota composition between obese and non-obese individuals in a Japanese population, as analyzed by terminal restriction fragment length polymorphism and next-generation sequencing. BMC Gastroenterol. 2015, 15, 100. [Google Scholar] [CrossRef] [Green Version]
- Le Roy, T.; Moens de Hase, E.; Van Hul, M.; Paquot, A.; Pelicaen, R.; Régnier, M.; Depommier, C.; Druart, C.; Everard, A.; Maiter, D.; et al. Dysosmobacter welbionis is a newly isolated human commensal bacterium preventing diet-induced obesity and metabolic disorders in mice. Gut 2022, 71, 534–543. [Google Scholar] [CrossRef]
- Kasahara, K.; Krautkramer, K.A.; Org, E.; Romano, K.A.; Kerby, R.L.; Vivas, E.I.; Mehrabian, M.; Denu, J.M.; Bäckhed, F.; Lusis, A.J.; et al. Interactions between Roseburia intestinalis and diet modulate atherogenesis in a murine model. Nat. Microbiol. 2018, 3, 1461–1471. [Google Scholar] [CrossRef]
- Wexler, H.M. Bacteroides: The good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 2007, 20, 593–621. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, N.; Emoto, T.; Yamashita, T.; Watanabe, H.; Hayashi, T.; Tabata, T.; Hoshi, N.; Hatano, N.; Ozawa, G.; Sasaki, N.; et al. Bacteroides vulgatus and Bacteroides dorei Reduce Gut Microbial Lipopolysaccharide Production and Inhibit Atherosclerosis. Circulation 2018, 138, 2486–2498. [Google Scholar] [CrossRef] [PubMed]
- Davis-Richardson, A.G.; Ardissone, A.N.; Dias, R.; Simell, V.; Leonard, M.T.; Kemppainen, K.M.; Drew, J.C.; Schatz, D.; Atkinson, M.A.; Kolaczkowski, B.; et al. Bacteroides dorei dominates gut microbiome prior to autoimmunity in Finnish children at high risk for type 1 diabetes. Front. Microbiol. 2014, 5, 678. [Google Scholar] [CrossRef] [PubMed]
- Gurung, M.; Li, Z.; You, H.; Rodrigues, R.; Jump, D.B.; Morgun, A.; Shulzhenko, N. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine 2020, 51, 102590. [Google Scholar] [CrossRef] [Green Version]
- Murphy, E.A.; Velazquez, K.T.; Herbert, K.M. Influence of high-fat diet on gut microbiota: A driving force for chronic disease risk. Curr. Opin. Clin. Nutr. Metab. Care 2015, 18, 515–520. [Google Scholar] [CrossRef]
- Tan, H.; Yu, Z.; Wang, C.; Zhang, Q.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. Pilot Safety Evaluation of a Novel Strain of Bacteroides ovatus. Front. Genet. 2018, 9, 539. [Google Scholar] [CrossRef]
- Deaver, J.A.; Eum, S.Y.; Toborek, M. Circadian Disruption Changes Gut Microbiome Taxa and Functional Gene Composition. Front. Microbiol. 2018, 9, 737. [Google Scholar] [CrossRef]
- Tailford, L.E.; Crost, E.H.; Kavanaugh, D.; Juge, N. Mucin glycan foraging in the human gut microbiome. Front. Genet. 2015, 6, 81. [Google Scholar] [CrossRef] [Green Version]
- Png, C.W.; Lindén, S.K.; Gilshenan, K.S.; Zoetendal, E.G.; McSweeney, C.S.; Sly, L.I.; McGuckin, M.A.; Florin, T.H. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 2010, 105, 2420–2428. [Google Scholar] [CrossRef] [PubMed]
- Malinen, E.; Krogius-Kurikka, L.; Lyra, A.; Nikkilä, J.; Jääskeläinen, A.; Rinttilä, T.; Vilpponen-Salmela, T.; von Wright, A.J.; Palva, A. Association of symptoms with gastrointestinal microbiota in irritable bowel syndrome. World J. Gastroenterol. 2010, 16, 4532–4540. [Google Scholar] [CrossRef]
- Parker, B.J.; Wearsch, P.A.; Veloo, A.C.M.; Rodriguez-Palacios, A. The Genus Alistipes: Gut Bacteria with Emerging Implications to Inflammation, Cancer, and Mental Health. Front. Immunol. 2020, 11, 906. [Google Scholar] [CrossRef]
- Dziarski, R.; Park, S.Y.; Kashyap, D.R.; Dowd, S.E.; Gupta, D. Pglyrp-Regulated Gut Microflora Prevotella falsenii, Parabacteroides distasonis and Bacteroides eggerthii Enhance and Alistipes finegoldii Attenuates Colitis in Mice. PLoS ONE 2016, 11, e0146162. [Google Scholar] [CrossRef] [PubMed]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.M.; Bitterman, K.J.; Wood, J.G.; Medvedik, O.; Cohen, H.; Lin, S.S.; Manchester, J.K.; Gordon, J.I.; Sinclair, D.A. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J. Biol. Chem. 2002, 277, 18881–18890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S.J.; Kaeberlein, M.; Andalis, A.A.; Sturtz, L.A.; Defossez, P.A.; Culotta, V.C.; Fink, G.R.; Guarente, L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature 2002, 418, 344–348. [Google Scholar] [CrossRef] [PubMed]
- von Schwartzenberg, R.J.; Bisanz, J.E.; Lyalina, S.; Spanogiannopoulos, P.; Ang, Q.Y.; Cai, J.; Dickmann, S.; Friedrich, M.; Liu, S.Y.; Collins, S.L.; et al. Caloric restriction disrupts the microbiota and colonization resistance. Nature 2021, 595, 272–277. [Google Scholar] [CrossRef]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012, 13, R79. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Li, S.; Yang, L.; Huang, P.; Li, W.; Wang, S.; Zhao, G.; Zhang, M.; Pang, X.; Yan, Z.; et al. Structural modulation of gut microbiota in life-long calorie-restricted mice. Nat. Commun. 2013, 4, 2163. [Google Scholar] [CrossRef] [Green Version]
- Duszka, K.; Ellero-Simatos, S.; Ow, G.S.; Defernez, M.; Paramalingam, E.; Tett, A.; Ying, S.; König, J.; Narbad, A.; Kuznetsov, V.A.; et al. Complementary intestinal mucosa and microbiota responses to caloric restriction. Sci. Rep. 2018, 8, 11338. [Google Scholar] [CrossRef]
- Wang, S.; Huang, M.; You, X.; Zhao, J.; Chen, L.; Wang, L.; Luo, Y.; Chen, Y. Gut microbiota mediates the anti-obesity effect of calorie restriction in mice. Sci. Rep. 2018, 8, 13037. [Google Scholar] [CrossRef] [Green Version]
- Pan, F.; Zhang, L.; Li, M.; Hu, Y.; Zeng, B.; Yuan, H.; Zhao, L.; Zhang, C. Predominant gut Lactobacillus murinus strain mediates anti-inflammaging effects in calorie-restricted mice. Microbiome 2018, 6, 54. [Google Scholar] [CrossRef] [PubMed]
- Fabersani, E.; Russo, M.; Marquez, A.; Abeijón-Mukdsi, C.; Medina, R.; Gauffin-Cano, P. Modulation of intestinal microbiota and immunometabolic parameters by caloric restriction and lactic acid bacteria. Food Res. Int. 2019, 124, 188–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, H.J.; Yu, J.G.; Lee, I.A.; Liu, M.J.; Shen, Y.F.; Sharma, S.P.; Jamal, M.A.; Yoo, J.H.; Kim, H.J.; Hong, S.T. Intestinal removal of free fatty acids from hosts by Lactobacilli for the treatment of obesity. FEBS Open Bio 2016, 6, 64–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabbiano, S.; Suárez-Zamorano, N.; Trajkovski, M. Host-Microbiota Mutualism in Metabolic Diseases. Front. Endocrinol. 2017, 8, 267. [Google Scholar] [CrossRef] [PubMed]
- Russo, M.; Fabersani, E.; Abeijón-Mukdsi, M.C.; Ross, R.; Fontana, C.; Benítez-Páez, A.; Gauffin-Cano, P.; Medina, R.B. Lactobacillus fermentum CRL1446 Ameliorates Oxidative and Metabolic Parameters by Increasing Intestinal Feruloyl Esterase Activity and Modulating Microbiota in Caloric-Restricted Mice. Nutrients 2016, 8, 415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, D.; Stasse-Wolthuis, M. The bifidogenic effect of inulin and oligofructose and its consequences for gut health. Eur. J. Clin. Nutr. 2009, 63, 1277–1289. [Google Scholar] [CrossRef] [PubMed]
- Ott, B.; Skurk, T.; Hastreiter, L.; Lagkouvardos, I.; Fischer, S.; Büttner, J.; Kellerer, T.; Clavel, T.; Rychlik, M.; Haller, D.; et al. Effect of caloric restriction on gut permeability, inflammation markers, and fecal microbiota in obese women. Sci. Rep. 2017, 7, 11955. [Google Scholar] [CrossRef]
- Simões, C.D.; Maukonen, J.; Scott, K.P.; Virtanen, K.A.; Pietiläinen, K.H.; Saarela, M. Impact of a very low-energy diet on the fecal microbiota of obese individuals. Eur. J. Nutr. 2014, 53, 1421–1429. [Google Scholar] [CrossRef]
- Cani, P.D.; Bibiloni, R.; Knauf, C.; Waget, A.; Neyrinck, A.M.; Delzenne, N.M.; Burcelin, R. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 2008, 57, 1470–1481. [Google Scholar] [CrossRef] [Green Version]
- Carmody, R.N.; Gerber, G.K.; Luevano, J.M., Jr.; Gatti, D.M.; Somes, L.; Svenson, K.L.; Turnbaugh, P.J. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 2015, 17, 72–84. [Google Scholar] [CrossRef] [Green Version]
- Wali, J.A.; Milner, A.J.; Luk, A.W.S.; Pulpitel, T.J.; Dodgson, T.; Facey, H.J.W.; Wahl, D.; Kebede, M.A.; Senior, A.M.; Sullivan, M.A.; et al. Impact of dietary carbohydrate type and protein-carbohydrate interaction on metabolic health. Nat. Metab. 2021, 3, 810–828. [Google Scholar] [CrossRef] [PubMed]
- Solon-Biet, S.M.; McMahon, A.C.; Ballard, J.W.; Ruohonen, K.; Wu, L.E.; Cogger, V.C.; Warren, A.; Huang, X.; Pichaud, N.; Melvin, R.G.; et al. The ratio of macronutrients, not caloric intake, dictates cardiometabolic health, aging, and longevity in ad libitum-fed mice. Cell Metab. 2014, 19, 418–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solon-Biet, S.M.; Mitchell, S.J.; Coogan, S.C.; Cogger, V.C.; Gokarn, R.; McMahon, A.C.; Raubenheimer, D.; de Cabo, R.; Simpson, S.J.; Le Couteur, D.G. Dietary Protein to Carbohydrate Ratio and Caloric Restriction: Comparing Metabolic Outcomes in Mice. Cell Rep. 2015, 11, 1529–1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lasker, D.A.; Evans, E.M.; Layman, D.K. Moderate carbohydrate, moderate protein weight loss diet reduces cardiovascular disease risk compared to high carbohydrate, low protein diet in obese adults: A randomized clinical trial. Nutr. Metab. 2008, 5, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fontana, L.; Cummings, N.E.; Arriola Apelo, S.I.; Neuman, J.C.; Kasza, I.; Schmidt, B.A.; Cava, E.; Spelta, F.; Tosti, V.; Syed, F.A.; et al. Decreased Consumption of Branched-Chain Amino Acids Improves Metabolic Health. Cell Rep. 2016, 16, 520–530. [Google Scholar] [CrossRef] [Green Version]








| Participant | Gender | Race/ Ethnicity | Age | Height (cm) | Weight (kg) | BMI | Tobacco Use | Medications/ Supplements | Daily Energy Requirement (kcal) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | F | Asian | 22 | 160 | 60 | 23.4 | No | Oral contraceptive | 1940 |
| 2 | M | White | 30 | 188 | 102 | 28.9 | No | - | 3267 |
| 3 | M | White | 26 | 191 | 98 | 26.9 | No | Magnesium, krill oil | 2983 |
| 4 | M | Pacific Islander | 51 | 188 | 88 | 24.9 | Yes | - | 3078 |
| 5 | F | White | 21 | 185 | 70 | 20.5 | No | - | 3177 |
| 6 | M | White | 28 | 188 | 79 | 22.4 | No | - | 2884 |
| 7 | F | Hispanic | 23 | 168 | 91 | 32.2 | No | Oral contraceptive | 2116 |
| 8 | F | Asian | 33 | 152 | 50 | 21.6 | No | - | 1738 |
| 9 | F | White | 19 | 175 | 58 | 18.9 | No | Oral contraceptive | 2221 |
| 10 | M | White | 29 | 178 | 86 | 27.1 | Yes | - | 3118 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anderson, E.M.; Rozowsky, J.M.; Fazzone, B.J.; Schmidt, E.A.; Stevens, B.R.; O’Malley, K.A.; Scali, S.T.; Berceli, S.A. Temporal Dynamics of the Intestinal Microbiome Following Short-Term Dietary Restriction. Nutrients 2022, 14, 2785. https://doi.org/10.3390/nu14142785
Anderson EM, Rozowsky JM, Fazzone BJ, Schmidt EA, Stevens BR, O’Malley KA, Scali ST, Berceli SA. Temporal Dynamics of the Intestinal Microbiome Following Short-Term Dietary Restriction. Nutrients. 2022; 14(14):2785. https://doi.org/10.3390/nu14142785
Chicago/Turabian StyleAnderson, Erik M., Jared M. Rozowsky, Brian J. Fazzone, Emilie A. Schmidt, Bruce R. Stevens, Kerri A. O’Malley, Salvatore T. Scali, and Scott A. Berceli. 2022. "Temporal Dynamics of the Intestinal Microbiome Following Short-Term Dietary Restriction" Nutrients 14, no. 14: 2785. https://doi.org/10.3390/nu14142785