Facile Cellulase Immobilisation on Bioinspired Silica
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis Bio-Inspired Silica Supports
2.3. Materials Characterization
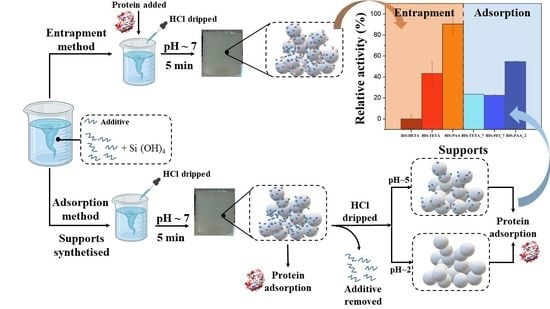
2.4. Protein Adsorption Method
2.5. Protein Entrapment Method
2.6. Protein Quantification
2.7. Cellulase Activity Assay
3. Results and Discussion
3.1. Analysis of the Synthesized BIS Supports
3.2. Confinement of the Proteins
3.2.1. Protein Adsorption
3.2.2. Protein Entrapment
3.3. Catalytic Activity of Immobilised Cellulase
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Tripathi, N.; Hills, C.D.; Singh, R.S.; Atkinson, C.J. Biomass waste utilisation in low-carbon products: Harnessing a major potential resource. Npj Clim. Atmos. Sci. 2019, 2, 35. [Google Scholar] [CrossRef] [Green Version]
- Sherwood, J. The significance of biomass in a circular economy. Bioresour. Technol. 2020, 300, 122–755. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Sun, J.; Shi, Y.; Wang, Q.; Wu, J.; Liu, J. Nanocellulose from various biomass wastes: Its preparation and potential usages towards the high value-added products. Environ. Sci. Ecotechnol. 2021, 5, 100077. [Google Scholar] [CrossRef]
- Wilson, D.B. Cellulases and biofuels. Curr. Opin. Biotechnol. 2009, 20, 295–299. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Tan, L.; Kida, K.; Morimura, S.; Sun, Z.-Y.; Tang, Y.-Q. Potential for reduced water consumption in biorefining of lignocellulosic biomass to bioethanol and biogas. J. Biosci. Bioeng. 2021, 131, 461–468. [Google Scholar] [CrossRef]
- Duque, A.; Álvarez, C.; Doménech, P.; Manzanares, P.; Moreno, A.D. Advanced bioethanol production: From novel raw materials to integrated biorefineries. Processes 2021, 9, 206. [Google Scholar] [CrossRef]
- Koehler, N.; Mccaherty, J.; Wilson, C.; Cooper, G.; Schwarck, R.; Kemmet, N.; Baker, R.; Mcafee, E.; Drook, R.; Markham, S.; et al. Focus Forward. 2020 RFA’s Ethanol Industry Outlook; Renewable Fuels Association: Ellisvile, MO, USA, 2020; pp. 1–40. [Google Scholar]
- Lamed, R.; Bayer, E.A.; Shoham, Y.; Chanzy, H. Cellulose, cellulases and cellulosomes. Curr. Opin. Struct. Biol. 1998, 8, 548–557. [Google Scholar]
- El-Said, A.H.M.; Saleem, A.; Maghraby, T.A.; Hussein, M.A. Cellulase activity of some phytopathogenic fungi isolated from diseased leaves of broad bean. Arch. Phytopathol. Plant Prot. 2014, 47, 2078–2094. [Google Scholar] [CrossRef]
- Siqueira, J.G.W.; Rodrigues, C.; Vandenberghe, L.P.d.S.; Woiciechowski, A.L.; Soccol, C.R. Current advances in on-site cellulase production and application on lignocellulosic biomass conversion to biofuels: A review. Biomass Bioenergy 2020, 132, 105419. [Google Scholar] [CrossRef]
- Arantes, V.; Saddler, J.N. Access to cellulose limits the efficiency of enzymatic hydrolysis: The role of amorphogenesis. Biotechnol. Biofuels 2010, 3, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Liu, W.; Li, Y.; Lai, H.L.; Zheng, Y.; Huang, J.W.; Chen, C.C.; Chen, Y.; Jin, J.; Li, H.; et al. Functional and structural analysis of Pichia pastoris-expressed Aspergillus Niger 1,4-β-endoglucanase. Biochem. Biophys. Res. Commun. 2016, 475, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Kassim, E.A. Cellulase Enzyme from Aspergillus niger. Microbiol. Immunol. 1982, 26, 449–454. [Google Scholar] [CrossRef] [PubMed]
- Okada, G. Cellulase of Aspergillus niger. Methods Enzymol. 1988, 160, 259–264. [Google Scholar]
- Chang, R.H.-Y.; Jang, J.; Wu, K.C.W. Cellulase immobilized mesoporous silica nanocatalysts for efficient cellulose-to-glucose conversion. Green Chem. 2011, 13, 2844. [Google Scholar] [CrossRef]
- Vasić, K.; Knez, Ž.; Leitgeb, M. Bioethanol Production by Enzymatic Hydrolysis from Different Lignocellulosic Sources. Molecules 2021, 26, 753. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Tiwari, M.K.; Singh, R.; Lee, J.K. From protein engineering to immobilization: Promising strategies for the upgrade of industrial enzymes. Int. J. Mol. Sci. 2013, 14, 1232–1277. [Google Scholar] [CrossRef]
- Zhou, H.; Pu, S.; Huo, J.; Cao, W.; Wang, B.; Li, J. Facile corrosion synthesis and sintering of disperse pure tetragonal zirconia nanoparticles. Ceram. Int. 2016, 42, 15005–15011. [Google Scholar] [CrossRef]
- Walker, J.M. Enzyme Stabilization and Immobilization; Minteer, S.D., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2017; Volume 1504, ISBN 978-1-4939-6497-0. [Google Scholar]
- Ansari, S.A.; Husain, Q. Potential applications of enzymes immobilized on/in nano materials: A review. Biotechnol. Adv. 2012, 30, 512–523. [Google Scholar] [CrossRef]
- Tran, D.N.; Balkus, K.J. Perspective of recent progress in immobilization of enzymes. ACS Catal. 2011, 1, 956–968. [Google Scholar] [CrossRef]
- Campbell, A.S.; Dong, C.; Meng, F.; Hardinger, J.; Perhinschi, G.; Wu, N.; Dinu, C.Z. Enzyme catalytic efficiency: A function of bio-nano interface reactions. ACS Appl. Mater. Interfaces 2014, 6, 5393–5403. [Google Scholar] [CrossRef]
- Abaházi, E.; Lestál, D.; Boros, Z.; Poppe, L. Tailoring the Spacer Arm for Covalent Immobilization of Candida antarctica Lipase B—Thermal Stabilization by Bisepoxide-Activated Aminoalkyl Resins in Continuous-Flow Reactors. Molecules 2016, 21, 767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Shora, H.M.; El-Sharkawy, R.M.; Khateb, A.M.; Darwish, D.B. Production and immobilization of β-glucanase from Aspergillus niger with its applications in bioethanol production and biocontrol of phytopathogenic fungi. Sci. Rep. 2021, 11, 21000. [Google Scholar] [CrossRef] [PubMed]
- Bourkaib, M.C.; Guiavarc’h, Y.; Chevalot, I.; Delaunay, S.; Gleize, J.; Ghanbaja, J.; Valsaque, F.; Berrada, N.; Desforges, A.; Vigolo, B. Non-Covalent and covalent immobilization of Candida antarctica lipase B on chemically modified multiwalled carbon nanotubes for a green acylation process in supercritical CO2. Catal. Today 2020, 348, 26–36. [Google Scholar] [CrossRef]
- Zhao, F.; Li, H.; Jiang, Y.; Wang, X.; Mu, X. Co-Immobilization of multi-enzyme on control-reduced graphene oxide by non-covalent bonds: An artificial biocatalytic system for the one-pot production of gluconic acid from starch. Green Chem. 2014, 16, 2558–2565. [Google Scholar] [CrossRef] [Green Version]
- Califano, V.; Sannino, F.; Costantini, A.; Avossa, J.; Cimino, S.; Aronne, A. Wrinkled Silica Nanoparticles: Efficient Matrix for β-Glucosidase Immobilization. J. Phys. Chem. C 2018, 122, 8373–8379. [Google Scholar] [CrossRef]
- Machado, N.B.; Miguez, J.P.; Bolina, I.C.A.; Salviano, A.B.; Gomes, R.A.B.; Tavano, O.L.; Luiz, J.H.H.; Tardioli, P.W.; Cren, É.C.; Mendes, A.A. Preparation, functionalization and characterization of rice husk silica for lipase immobilization via adsorption. Enzym. Microb. Technol. 2019, 128, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, I.N.; Yang, X.L.; Dubale, A.A.; Li, R.F.; Ma, Y.M.; Wang, L.M.; Hou, G.H.; Guan, R.F.; Xie, M.H. Hydrolysis of cellulose using cellulase physically immobilized on highly stable zirconium based metal-organic frameworks. Bioresour. Technol. 2018, 270, 377–382. [Google Scholar] [CrossRef]
- Ungurean, M.; Paul, C.; Peter, F. Cellulase immobilized by sol-gel entrapment for efficient hydrolysis of cellulose. Bioprocess Biosyst. Eng. 2013, 36, 1327–1338. [Google Scholar] [CrossRef]
- Takimoto, A.; Shiomi, T.; Ino, K.; Tsunoda, T.; Kawai, A.; Mizukami, F.; Sakaguchi, K. Encapsulation of cellulase with mesoporous silica (SBA-15). Microporous Mesoporous Mater. 2008, 116, 601–606. [Google Scholar] [CrossRef]
- Aznar, E.; Villalonga, R.; Giménez, C.; Sancenón, F.; Marcos, M.D.; Martínez-Máñez, R.; Díez, P.; Pingarrón, J.M.; Amorós, P. Glucose-triggered release using enzyme-gated mesoporous silica nanoparticles. Chem. Commun. 2013, 49, 6391–6393. [Google Scholar] [CrossRef]
- Afsahi, B.; Alomoum, A.K.; Nejati, S.; Kazemi, A. Immobilization of Cellulase on non-porous ultrafine silica particles. Sci. Iran. 2007, 14, 379–383. [Google Scholar]
- Verma, M.L.; Chaudhary, R.; Tsuzuki, T.; Barrow, C.J.; Puri, M. Immobilization of β-glucosidase on a magnetic nanoparticle improves thermostability: Application in cellobiose hydrolysis. Bioresour. Technol. 2013, 135, 2–6. [Google Scholar] [CrossRef] [PubMed]
- Hartono, S.B.; Qiao, S.Z.; Liu, J.; Jack, K.; Ladewig, B.P.; Hao, Z.; Lu, G.Q.M. Functionalized mesoporous silica with very large pores for cellulase immobilization. J. Phys. Chem. C 2010, 114, 8353–8362. [Google Scholar] [CrossRef]
- Alahakoon, T.; Koh, J.W.; Chong, X.W.C.; Lim, W.T.L. Immobilization of cellulases on amine and aldehyde functionalized Fe2O3 magnetic nanoparticles. Prep. Biochem. Biotechnol. 2012, 42, 234–248. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Singh, S.; Tiwari, R.; Goel, R.; Nain, L. Immobilization of indigenous holocellulase on iron oxide (Fe2O3) nanoparticles enhanced hydrolysis of alkali pretreated paddy straw. Int. J. Biol. Macromol. 2017, 96, 538–549. [Google Scholar] [CrossRef]
- Bohara, R.A.; Thorat, N.D.; Pawar, S.H. Immobilization of cellulase on functionalized cobalt ferrite nanoparticles. Korean J. Chem. Eng. 2016, 33, 216–222. [Google Scholar] [CrossRef]
- Liang, W.; Cao, X. Preparation of a pH-sensitive polyacrylate amphiphilic copolymer and its application in cellulase immobilization. Bioresour. Technol. 2012, 116, 140–146. [Google Scholar] [CrossRef]
- Zaccariello, G.; Back, M.; Benedetti, A.; Canton, P.; Cattaruzza, E.; Onoda, H.; Glisenti, A.; Alimonti, A.; Bocca, B.; Riello, P. Bismuth titanate-based UV filters embedded mesoporous silica nanoparticles: Role of bismuth concentration in the self-sealing process. J. Colloid Interface Sci. 2019, 549, 1–8. [Google Scholar] [CrossRef]
- Yang, S.J.; Song, W.J.; Dingwell, D.B.; He, J.; Guo, H.B. Surface roughness affects metastable non-wetting behavior of silicate melts on thermal barrier coatings. Rare Met. 2022, 41, 469–481. [Google Scholar] [CrossRef]
- Zhang, W.; Qiu, J.; Feng, H.; Zang, L.; Sakai, E. Increase in stability of cellulase immobilized on functionalized magnetic nanospheres. J. Magn. Magn. Mater. 2015, 375, 117–123. [Google Scholar] [CrossRef]
- Jordan, J.; Kumar, C.S.S.R.; Theegala, C. Preparation and characterization of cellulase-bound magnetite nanoparticles. J. Mol. Catal. B Enzym. 2011, 68, 139–146. [Google Scholar] [CrossRef]
- Qi, H.; Duan, H.; Wang, X.; Meng, X.; Yin, X.; Ma, L. Preparation of magnetic porous terpolymer and its application in cellulase immobilization. Polym. Eng. Sci. 2015, 55, 1039–1045. [Google Scholar] [CrossRef]
- Li, Y.; Wang, X.-Y.; Jiang, X.-P.; Ye, J.-J.; Zhang, Y.-W.; Zhang, X.-Y. Fabrication of graphene oxide decorated with Fe3O4@SiO2 for immobilization of cellulase. J. Nanopart. Res. 2015, 17, 8. [Google Scholar] [CrossRef]
- Salem, K.; Jabalera, Y.; Puentes-Pardo, J.D.; Vilchez-Garcia, J.; Sayari, A.; Hmida-Sayari, A.; Jimenez-Lopez, C.; Perduca, M. Enzyme Storage and Recycling: Nanoassemblies of α-Amylase and Xylanase Immobilized on Biomimetic Magnetic Nanoparticles. ACS Sustain. Chem. Eng. 2021, 9, 4054–4063. [Google Scholar] [CrossRef]
- Zang, L.; Qiu, J.; Wu, X.; Zhang, W.; Sakai, E.; Wei, Y. Preparation of magnetic chitosan nanoparticles as support for cellulase immobilization. Ind. Eng. Chem. Res. 2014, 53, 3448–3454. [Google Scholar] [CrossRef]
- Zhang, D.; Hegab, H.E.; Lvov, Y.; Dale Snow, L.; Palmer, J. Immobilization of cellulase on a silica gel substrate modified using a 3-APTES self-assembled monolayer. Springerplus 2016, 5, 48. [Google Scholar] [CrossRef] [Green Version]
- Matisons, J. Bio-Inspired Silicon-Based Materials; Springer: Cham, Switzerland, 2014; ISBN 978-1-4020-8171-2. [Google Scholar]
- Forsyth, C.; Patwardhan, S.V. Bioinspired Silica for Enzyme Immobilisation: A Comparison with Traditional Methods; Springer: Dordrecht, The Netherlands, 2014; pp. 39–62. [Google Scholar]
- Forsyth, C.; Patwardhan, S.V. Controlling performance of lipase immobilised on bioinspired silica. J. Mater. Chem. B 2013, 1, 1164–1174. [Google Scholar] [CrossRef]
- Cazaban, D.; Illanes, A.; Wilson, L.; Betancor, L. Bio-inspired silica lipase nanobiocatalysts for the synthesis of fatty acid methyl esters. Process Biochem. 2018, 74, 86–93. [Google Scholar] [CrossRef]
- Edwards, J.S.; Roberts, A.; Hemmert, A.C.; Edwards, C.C.; Potter, P.M.; Redinbo, M.R. Immobilization of Active Human Carboxylesterase 1 in Biomimetic Silica Nanoparticles. Biotechnol. Prog. 2011, 27, 863–869. [Google Scholar] [CrossRef]
- Zamora, P.; Narváez, A.; Domínguez, E. Enzyme-modified nanoparticles using biomimetically synthesized silica. Bioelectrochemistry 2009, 76, 100–106. [Google Scholar] [CrossRef]
- Ryu, Y.H.; Yeo, K.B.; Ki, M.R.; Kim, Y.J.; Pack, S.P. Improved stability and reusability of endoglucanase from Clostridium thermocellum by a biosilica-based auto-encapsulation method. Biochem. Eng. J. 2016, 105, 144–149. [Google Scholar] [CrossRef]
- Patwardhan, S.V.; Manning, J.R.H.; Chiacchia, M. Bioinspired synthesis as a potential green method for the preparation of nanomaterials: Opportunities and challenges. Curr. Opin. Green Sustain. Chem. 2018, 12, 110–116. [Google Scholar] [CrossRef]
- Patwardhan, S.V.; Staniland, S.S. Green Nanomaterials: From Bioinspired Synthesis to Sustainable Manufacturing of Inorganic Nanomaterials; IOP Publishing: Bristol, UK, 2019; ISBN 978-0-7503-1221-9. [Google Scholar]
- Forsyth, C.; Yip, T.W.S.; Patwardhan, S.V. CO2 sequestration by enzyme immobilized onto bioinspired silica. Chem. Commun. 2013, 49, 3191–3193. [Google Scholar] [CrossRef]
- Koutsopoulos, S.; Van Der Oost, J.; Norde, W. Adsorption of an endoglucanase from the hyperthermophilic Pyrococcus furiosus on hydrophobic (polystyrene) and hydrophilic (silica) surfaces increases protein heat stability. Langmuir 2004, 20, 6401–6406. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, Y.; Parashar, A.; Bressler, D.C. Highly retained enzymatic activities of two different cellulases immobilized on non-porous and porous silica particles. Biotechnol. Bioprocess Eng. 2014, 19, 621–628. [Google Scholar] [CrossRef]
- Chen, B.; Qiu, J.; Mo, H.; Yu, Y.; Ito, K.; Sakai, E.; Feng, H. Synthesis of mesoporous silica with different pore sizes for cellulase immobilization: Pure physical adsorption. New J. Chem. 2017, 41, 9338–9345. [Google Scholar] [CrossRef]
- Manning, J.R.H.; Yip, T.W.S.; Centi, A.; Jorge, M.; Patwardhan, S.V. An Eco-Friendly, Tunable and Scalable Method for Producing Porous Functional Nanomaterials Designed Using Molecular Interactions. ChemSusChem 2017, 10, 1683–1691. [Google Scholar] [CrossRef]
- Bradford Protocol; Sigma-Aldrich: St. Louis, MO, USA, 2011; pp. 3–8.
- Ghose, T.K. Measurement of Cellulase Activities. Pure Appl. Chem. 1987, 59, 257–268. [Google Scholar] [CrossRef]
- Rahman, M.S.; Fernando, S.; Ross, B.; Wu, J.; Qin, W. Endoglucanase (eg) activity assays. Methods Mol. Biol. 2018, 1796, 169–183. [Google Scholar]
- Sotomayor, F.J.; Cychosz, K.A.; Thommes, M. Characterization of Micro/Mesoporous Materials by Physisorption: Concepts and Case Studies. Acc. Mater. Surf. Res. 2018, 3, 34–50. [Google Scholar]
- Davidson, S.; Lamprou, D.A.; Urquhart, A.J.; Grant, M.H.; Patwardhan, S.V. Bioinspired Silica Offers a Novel, Green, and Biocompatible Alternative to Traditional Drug Delivery Systems. ACS Biomater. Sci. Eng. 2016, 2, 1493–1503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ball, V.; Winterhalter, M.; Schwinte, P.; Lavalle, P.; Voegel, J.C.; Schaaf, P. Complexation mechanism of bovine serum albumin and poly(allylamine hydrochloride). J. Phys. Chem. B 2002, 106, 2357–2364. [Google Scholar] [CrossRef]
- Vander Straeten, A.; Lefèvre, D.; Demoustier-Champagne, S.; Dupont-Gillain, C. Protein-based polyelectrolyte multilayers. Adv. Colloid Interface Sci. 2020, 280, 102161. [Google Scholar] [CrossRef]
- Shi, J.; Yang, C.; Zhang, S.; Wang, X.; Jiang, Z.; Zhang, W.; Song, X.; Ai, Q.; Tian, C. Polydopamine microcapsules with different wall structures prepared by a template-mediated method for enzyme immobilization. ACS Appl. Mater. Interfaces 2013, 5, 9991–9997. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Xu, Q.; Zou, C.; He, Z.; Tang, Y.; Gao, T.; Han, M.; Dai, Z. A boronate-modified renewable nanointerface for ultrasensitive electrochemical assay of cellulase activity. Chin. Chem. Lett. 2020, 32, 1470–1474. [Google Scholar] [CrossRef]
- Ahmad, R.; Sardar, M. Immobilization of cellulase on TiO2 nanoparticles by physical and covalent methods: A comparative study. Indian J. Biochem. Biophys. 2014, 51, 314–320. [Google Scholar] [PubMed]
- Taipakova, S.M.; Smekenov, I.T.; Saparbaev, M.K.; Bissenbaev, A.K. Characterization of Aspergillus niger endo-1,4-β-glucanase ENG1 secreted from Saccharomyces cerevisiae using different expression vectors. Genet. Mol. Res. 2015, 14, 6439–6452. [Google Scholar] [CrossRef]
- Lübeck, M. Cellulases: Method and Protocols; Springer Humana Press: New York, NY, USA, 2018. [Google Scholar]







| Additives | Final pH | ||
|---|---|---|---|
| 7 | 5 | 2 | |
| DETA | BIS-DETA_7 | BIS-DETA_5 | BIS-DETA_2 |
| TETA | BIS-TETA_7 | BIS-TETA_5 | BIS-TETA_2 |
| PEHA | BIS-PEHA_7 | BIS-PEHA_5 | BIS-PEHA_2 |
| PEI | BIS-PEI_7 | BIS-PEI_5 | BIS-PEI_2 |
| PAA | BIS-PAA_7 | BIS-PAA_5 | BIS-PAA_2 |
| Enzyme | Support Material | Reaction Condition | Immobilisation Techniques | Activity Enzyme after Immobilisation (%) | Reference |
|---|---|---|---|---|---|
| Cellulase from Robillarda sp. and Cellulase from Trichoderma reesei | Silica fumed (commercial support) | n.a | Adsorption | 42–48 | [59] |
| Covalent bond (R-NH2+Glu) | 24 | ||||
| Covalent bond (R-NH2+Carbodiimide) | 18.8 | ||||
| Cellulase from Trichoderma reesei | Silica particles (14 nm mean size) | n.a | Adsorption | 35 | [33] |
| Covalent bond (R-NH2+Glu) | 25 | ||||
| Cellulase | SBA-15 (Particle size ~200–250 nm, pore size = 8.9 nm, >700 m2g) | Acid condition 35–60–80 °C for 20 h + calcination | Encapsulation | 65 | [31] |
| Endoglucanase | FDU-12 | Acid condition 160 °C for 72 h for hydrothermal treatment + acid purification | Adsorption | 75.3 | [35] |
| FDU-12@APTES | 15.6 | ||||
| FDU-12@VTMS (three-dimensional mesoporous material with pore size ~10 nm) | 80.3 | ||||
| Cellulase 1 from Trichoderma reesei Cellulases 2 which originated from Aspergillus niger | SiO2 non-porous (Fumed silica)- S1 | n.a | Adsorption | >90 | [60] |
| SiO2-porous (Davisil chromatographic silica 633N)-S2 | ~60 | ||||
| Cellulase from Aspergillus niger | MSN-3.8 nm | 80–90 °C for 48 h in water solution + calcination | Adsorption | 63.3 | [61] |
| MSN-17.6 nm | 26.6 | ||||
| MSN-25 nm | 35.8 | ||||
| MSN-200 nm | 13.5 | ||||
| Cellulase from Aspergillus niger | Bio-inspired silica (BIS) Particle size~150 nm | Neutral pH, room temperature for 5 min of the reaction | Entrapment | 90 | This work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lombardi, V.; Trande, M.; Back, M.; Patwardhan, S.V.; Benedetti, A. Facile Cellulase Immobilisation on Bioinspired Silica. Nanomaterials 2022, 12, 626. https://doi.org/10.3390/nano12040626
Lombardi V, Trande M, Back M, Patwardhan SV, Benedetti A. Facile Cellulase Immobilisation on Bioinspired Silica. Nanomaterials. 2022; 12(4):626. https://doi.org/10.3390/nano12040626
Chicago/Turabian StyleLombardi, Vincenzo, Matteo Trande, Michele Back, Siddharth V. Patwardhan, and Alvise Benedetti. 2022. "Facile Cellulase Immobilisation on Bioinspired Silica" Nanomaterials 12, no. 4: 626. https://doi.org/10.3390/nano12040626