1. Introduction
Type I interferons (IFNs) represent a class of cytokines that are naturally secreted in response to viral infections. They are produced by a variety of cells during the inflammatory response and can, upon activation, induce numerous molecular changes that affect several cellular processes including cell growth and differentiation. These proteins have garnered biomedical interest thanks to their therapeutical action against viral infections, neuro-inflammatory diseases, and tumors including melanoma, hairy cell leukemia, and lymphoma [
1,
2,
3]. Type I IFNs were the first of their type to be produced by recombinant DNA technology as well as the first to be used therapeutically. The first report on their anti-tumoral activity in mice was published almost half a century ago [
4]. Although, in recent years, more targeted therapies have been favored in the treatment of tumors, type I IFNs are still highly considered thanks to their ability to induce the expression of more than 200 proteins that are implicated in therapeutic processes, although some are still of unknown activity [
5,
6]. Consequently, research studies are focusing on unraveling the wide variety of molecular pathways prompted by IFNs to characterize and target them for the treatment of different pathologies. Previously, it was reported that type I IFNs can directly impair the survival of neuroblastoma SH-SY5Y cells by promoting intrinsic apoptosis [
5]. Moreover, in different types of cultured malignant cells, type I IFNs were successful in inducing cell death by promoting intracellular events leading to apoptosis [
6,
7].
PPARα is a member of the peroxisome-proliferator activated receptors (PPARs) family, which includes the PPARα, PPARγ, and PPARβ/δ subtypes that are characterized by distinct tissue distribution. These nuclear receptors promote ligand-dependent transcription of target genes that regulate energy production, lipid metabolism, and inflammation. PPARα is highly expressed in the muscles, heart, kidneys, liver, and small and large intestines [
8]. Notably, PPARα agonists are known for their significant role in the treatment of dyslipidemia or metabolic syndromes by reducing plasma triglyceride levels [
9] together with the modulation of glucose homeostasis and insulin resistance [
10]. The expression of PPARα in peripheral tissue underlies its crucial implication in metabolic pathways that are linked to several conditions including inflammation, cancer, and neurodegeneration [
11,
12].
Oleoylethanolamide (OEA) and palmitoylethanolamide (PEA) are a class of naturally occurring bioactive lipids derived from saturated and unsaturated fatty acid precursors that display high affinity for the nuclear PPARα receptors [
13]. OEA is an endogenous lipid derived from oleic acid, a monounsaturated fatty acid synthesized from membrane glycerophospholipids, while PEA is an endogenous compound belonging to the family of N-acylethanolamines isolated for the first time from purified lipid fractions of soybeans, egg yolk, and peanut meal [
13,
14,
15]. Lipids are a group of complex biomolecules that not only form the structural basis of biological membranes but also function as signaling molecules and as a source of energy. In recent years, lipids have emerged as central players in a complex network that modulates cellular and molecular actions associated with different physiopathological states [
14,
15]. While the effect of PEA on B16 melanoma, MCF-7 breast, colon HCT116, and astrocytoma cells has been documented, little is known about the impact of OEA and PEA exposure on neuroblastoma [
12,
16,
17]. The focus of our research is mainly directed towards elucidating any possible overlapping signaling pathways that may be activated by these two lipids in the human neuroblastoma SH-SY5Y cell line when present simultaneously with IFNβ in a pharmacological combination, thereby suggesting a possible contributing role of OEA and PEA in the development of alternative therapeutic strategies.
3. Discussion
In this study, we investigated the effects of OEA and PEA, in association with IFNβ, on human neuroblastoma SH-SY5Y cells. We also focused on elucidating the intracellular mechanisms that underlie the observed impairments in cell viability and proliferation to provide key proof of the detrimental effects mediated by these two lipids in SH-SY5Y neuroblastoma cells. The enhanced synthesis and absorption of lipids are known to contribute to the rapid growth of cells, the onset of inflammation, and tumorigenesis [
14]. As a bioactive lipid, PEA’s involvement has been previously documented in different cancer cell lines of different origins, such as B16 melanoma, MCF-7 breast, and HCT116 colon cells [
12,
16,
17,
26]. So far, little is known about how OEA and PEA may affect human neuroblastoma SH-SY5Y cells. Initially, the toxicity of OEA and PEA in SH-SY5Y cells was evaluated using a cell viability MTT assay. After 24 h of exposure to OEA and PEA at concentrations ranging from 0.3 to 30 µM, SH-SY5Y cells showed no signs of cellular suffering. In contrast, Pagano and colleagues reported a reduction in the proliferation of colon cancer cells after exposure to PEA at concentrations ranging from 3 μM to 30 μM, while treatment at 1 μM showed no significant effect [
12]. In addition, the exposure of N1E-115 mouse neuroblastoma cells to OEA and PEA at 1 μM and 5 μM, respectively, was able to reduce cell viability [
18]. These findings demonstrate how the sensitivity and the response to OEA and PEA treatment can change depending on the concentration, the treatment duration, and the type of the cancer cell line itself (i.e., colon cancer cells, mouse neuroblastoma cells) [
12,
18] as well as the cellular microenvironment, which can also be very different from what could potentially be observed in vivo as the data reported above all refer to in vitro investigations. The molecular and phenotypic heterogeneity of tumors and, consequently, their susceptibility to various forms of treatment must also be considered along with the nature of the patient’s physiological state dictated by age, inflammation, immune system response, pathologies, and so on.
Interestingly, our results show that OEA and PEA can drastically lower cell viability when co-exposed with IFNβ in SH-SY5Y cells. The same effect was also observed through the wound healing assay in which the proliferation was negatively affected. Furthermore, the co-treatment with either OEA or PEA and IFNβ for 10 days was able to decrease colony formation in the clonogenic test, an approach that is regularly used in in vitro settings to assess clonogenicity. These results are in line with previous findings demonstrating that PEA inhibited tumor cell proliferation and migration through the PPARα receptor in colon cancer cells [
12]. Moreover, our results also show that treatment with OEA and PEA alone can hinder cell proliferation and colony formation in comparison to the control group, showing that these two lipids are not harmful to SH-SY5Y cells after a 24 h treatment but do, however, exert detrimental effects at longer periods of exposure.
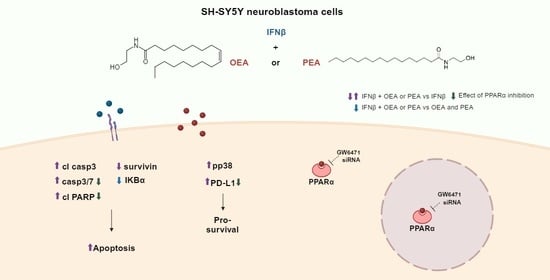
To determine the intracellular events that underlie the enhanced impairment in both cell viability and proliferation, we investigated the intrinsic apoptotic pathway, a mechanism that has been previously recognized to prompt IFNβ’s detrimental effect in SH-SY5Y cells [
5]. Our findings showed that the co-administration of the two lipids with IFNβ is more effective than IFNβ alone at promoting the cleavage of caspase 3 and PARP, therefore indicating the potentiation of intrinsic apoptotic cell death without the amplification of the main IFNβ signaling cascade. Future research is necessary to determine how these compounds can affect other neuroblastoma cell lines. A study reported that an engraftment of human IMR32 neuroblastoma cells in vivo was responsive and restricted by type I IFN treatment [
27]. Nevertheless, more investigations should be carried out while taking into account neuroblastoma cell lines that display differences in genetic amplification, p53 mutation or overexpression, and ALK receptor constitutive active involvement. Previous research has shown that IFNβ signaling through STAT1 affects survivin, an inhibitor of apoptosis (IAP) protein, whose levels may be modulated by the JAK-STAT system [
28,
29,
30]. Our investigation found that IFNβ increased survivin levels, but co-exposure to OEA and PEA impaired this effect. This result might explain the lipid-mediated enhancement of IFNβ-induced apoptosis, as the decrease in survivin levels may represent another mechanism that contributes to the increase in cell death, along with the enhancement of the cleavage of caspase 3 and PARP. A study by Wang and colleagues reported that the stimulation of PPARγ induces cell death through the downregulation of survivin expression and the increase in caspase 3 activity in colorectal cancer cells [
19]. These findings are in line with our results that demonstrate both a decrease in survivin and an increase in caspase 3 cleavage and caspase 3/7 activity upon co-treatment with OEA or PEA. Furthermore, OEA and PEA actions are inherently STAT1-independent as demonstrated by the fact that these two lipids are unable to potentiate the JAK-STAT pathway. IFNβ, OEA, and PEA modulate survivin through distinct mechanisms, explaining their different effects when used alone or in combination. However, it is crucial to emphasize that survivin expression undergoes regulation through several other pathways, and many variables, including alternative signaling cascades, might have a significant impact on this protein. The intricate processes by which STAT1 as well as PPARα regulate survivin expression require more in-depth investigations.
Studies have also reported that prolonged exposure to OEA and PEA alone elevates IKBα levels in SH-SY5Y cells [
20,
21]. Nevertheless, our findings suggest that co-treatment with IFNβ disrupts this enhancement, indicating that IKBα failed at counteracting the cellular damage elicited by this pro-inflammatory cytokine. Mcl-1 is another anti-apoptotic protein from the Bcl-2 family that regulates cell survival, affecting the balance between cell viability and death. Similarly, IFNγ has been reported to downregulate Mcl-1 expression, leading to the promotion of apoptosis [
31]. In addition, the suggested association of IFNs with apoptosis can be potentially traced to the downregulation of anti-apoptotic proteins such as Mcl-1 [
32]. In our experiments, IFNβ alone decreased Mcl-1 levels. This decrease was not further modulated by co-exposure with either OEA or PEA. The totality of these findings suggests a decrease in the anti-apoptotic cell response, which may lead to increased cell death by IFNβ in combination with OEA or PEA. Most likely, the amalgamation between the reduction in Mcl-1 by IFNβ along with the downregulating action of OEA and PEA on survivin expression and the reduction in IKBα levels by IFNs compromise cellular homeostasis and render cells more susceptible to apoptosis.
IFNs can also induce PD-L1, a ligand of the PD-1 receptor that works as an immune-inhibiting checkpoint, leading to immune evasion and inhibiting antitumoral immune responses through the JAK-STAT pathway [
22,
23]. Our results demonstrate, for the first time, an increase in the levels of PD-L1 in both cell lysate and cell membranes of SH-SY5Y cells co-treated with OEA or PEA and IFNβ. PPARγ agonists have been shown to increase PD-L1 protein expression in human gastrointestinal and colorectal cancer cell lines [
33]. In our investigations, OEA and PEA alone were unable to induce any variation in PD-L1 levels. We also formerly reported that during the induction of apoptosis in SH-SY5Y neuroblastoma cells, IFNβ triggers a collateral signaling pathway mediated by p38 MAPK that opposes the activity of programmed cell death to counteract the cell damage induced by this cytokine [
24]. Our current work reports a slight but significant increase in p38 MAPK phosphorylation in samples treated with IFNβ and OEA or PEA concurrently. Although the contribution of these two distinct proteins, p38 MAPK phosphorylation, and PD-L1 might point to a possible synergism attempting to block the apoptotic damage elicited by IFNβ, our results prove that this effect is insufficient in shielding SH-SY5Y cells from the damage brought on by concomitant treatment as suggested by the decrease in cell survival and proliferation as well as survivin, Mcl-1, and IKBα levels as mentioned above. On the other hand, studies have shown that OEA and oleic acid decrease PD-L1 expression induced by IFNγ in human lung carcinoma cells [
34]. The varied results obtained by Yamagata and colleagues may be due to the different cellular lines and the different kinds of IFNs employed in their study. Previous reports demonstrate that PD-L1 likewise increases in other neuroblastoma cells apart from SH-SY5Y by IFNγ [
35], indicating that different human neuroblastoma cell lines are responsive to IFNs when it comes to PD-L1 rise; however, future studies are necessary to determine if the OEA- and PEA-mediated potentiation of IFNβ-induced PD-L1 levels is present in other neuroblastoma cell lines. Increased expression of PD-L1 can also represent an important target for cancer therapy. Indeed, several treatments that target PD-L1/PD-1 interaction have been recently approved for cancer therapy, including monoclonal antibodies and immune checkpoint inhibitors, since tumors displaying a higher expression of PD-L1 appear to be more sensitive to such treatments [
36].
To assess the involvement of the PPARα receptor in the mediation of the observed effects elicited by treatment with OEA, PEA, and IFNβ, we first investigated the expression of the latter after IFNβ exposure and we found no significant difference. Furthermore, to address the direct role of this receptor in OEA- and PEA-induced apoptosis as well as PD-L1 potentiation, we used a specific PPARα receptor antagonist, the compound GW6471. This inhibitor partially blocked PD-L1 potentiation and reduced the PARP cleavage elicited by both IFNβ alone and by the co-treatment with OEA and PEA, while no effect in vehicle-pretreated cells or GW6471 was detected. These findings are further corroborated by the measurement of caspase 3/7 activity which shows the implication of PPARα in the mediation of the effects prompted by the different treatments. Additionally, the fact that GW6471 did not affect the signaling pathway of IFNβ reveals the PPARα inhibitor’s selectivity.
In favor of gaining more insight into PPARα involvement, this receptor was silenced by cell transfection with a specific PPARα siRNA. The experiment demonstrated that in PPARα siRNA-treated samples, IFNβ alone or in combination with OEA or PEA reduced PD-L1 and, to a lesser extent, the potentiation of PARP cleavage. The results suggest that the PPARα receptor is an active contributing element to the increase in apoptosis and PD-L1 by the co-treatment of OEA, PEA, and IFNβ. However, current data leave the possibility for other cellular mechanisms that may participate in OEA- and PEA-mediated apoptosis with IFNβ. Further research is needed to evaluate other collateral pathways that can intervene in OEA and PEA signaling in the SH-SY5Y neuroblastoma cell line.
4. Materials and Methods
4.1. Materials
OEA and GW6471 were purchased from Tocris (Abingdon, UK) and PEA was obtained from Abcam (Cambridge, UK). rh IFN-beta 1b was from ImmunoTolls (Friesoythe, Germany). Muse™ reagents were obtained from Luminex Corporation (Austin, TX, USA). Caspase-Glo® 3/7 and the RealTime-Glo™ MT Cell Viability assay kit were purchased from Promega (Madison, WI, USA).
4.2. Cell Culture
Human neuroblastoma cell line SH-SY5Y was obtained from the European Collection of Authenticated Cell Cultures (ECACC) (Salisbury, UK). The cell line was authenticated by the vendors. SH-SY5Y cells were grown in Ham’s F12/MEM medium (1:1) (Sigma-Aldrich, St. Louis, MO, USA) containing 2 mM L-glutamine (Sigma-Aldrich) and 1% non-essential amino acids (NEAA) (Sigma-Aldrich). The cells were maintained at 37 °C in a humidified atmosphere of 5% CO2 in air. Cells were split every 72 h using 0.25% trypsin/EDTA (Sigma-Aldrich). After resuscitation, the cells were used for no more than 15 passages. The cells were checked for mycoplasma by using the MycoFluor Mycoplasma Detection kit (Invitrogen-Life Technologies, Monza, Italy).
4.3. Cell Treatment and Cell Lysate Preparation
Unless otherwise specified, neuroblastoma cells were washed and incubated in a medium containing no FCS. The cells were treated with the test agents as indicated in the text and were maintained at constant temperature and humidity conditions as mentioned earlier. To prepare cell lysates, cells were first washed with PBS and then scraped into an ice-cold lysis buffer (RIPA buffer), supplemented with 1 mM phenylmethylsulphonyl fluoride (PMSF), 0.5% phosphatase inhibitor cocktail 3, and 1% protease inhibitor cocktail (Sigma-Aldrich). The samples were sonicated for 5 s and cell extract aliquots were taken for protein analysis by using the Bio-Rad protein assay (Bio-Rad Lab, Hercules, CA, USA).
4.4. Biotinylation of Surface Proteins
Surface biotinylation of cell proteins was performed as previously described [
37,
38]. Briefly, SH-SY5Y cells treated with either vehicle or IFNβ for 24 h were incubated for 1 h at 4 °C with the cell-impermeable biotinylating agent sulfosuccinimidyl-6-(biotin-amido) hexanoate (sulpho-NHS-LC-biotin) (Pierce, Rockford, IL, USA). After that, the cells were washed with PBS containing 20 mM glycine and solubilized by incubation in RIPA buffer supplemented with 1% Triton X 100. Cell extracts were centrifuged at 10,000×
g for 5 min at 4 °C and the supernatants were incubated overnight at 4 °C with streptavidin-conjugated agarose beads. The beads were mixed with sample buffer and incubated for 4 min at 100 °C. The proteins were then analyzed by Western blot.
4.5. Westen Blotting
Cell proteins were separated by SDS-polyacrylamide gel electrophoresis and were transferred to polyvinylidene difluoride membranes (Millipore). Membranes were blocked, washed, and incubated overnight at 4 °C with one of the following primary antibodies: PD-L1 (cat. no. 13684, Cell Signaling Technology, Danvers, MA, USA) (1:1000); IKBα (cat. no. 4814, Cell Signaling Technology) (1:1000); cleaved caspase 3 (Asp175) (cat. no. 9664, Cell Signaling Technology) (1:1000); caspase 3 (cat no. 9665, Cell Signaling Technology) (1:1000); cleaved-poly (ADP-ribose) polymerase (PARP) (Asp214) (cat. no. 5625, Cell Signaling Technology) (1:1000); PARP (cat. no. 9542, Cell Signaling Technology) (1:1000); phospho-Tyr701-STAT1 (1:1000) (cat no. ST1P-11A5, Thermo Fisher Scientific, Rockford, IL, USA); anti-STAT1 (1:500) (cat no. sc-592, Santa Cruz Biotechnology, Paso Robles, CA, USA); PKR (1:1000) (cat no. 3072, Cell Signaling Technology); survivin (cat. no. 2808, Cell Signaling Technology); Mcl-1 (1:1000) (sc-819, Santa Cruz Biotechnology); pan cadherin (cat. no. 4073, Cell Signaling Technology) (1:2000); actin (1:3000) (cat no. A2066, Sigma-Aldrich); GAPDH (1:5000) (cat no. 247-002, Synaptic Systems, Gottingen, Germany). Thereafter, the membranes were washed and incubated with an appropriate horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology). Immunoreactive bands were detected by Clarity Western ECL substrate (Bio-Rad Laboratory, Hercules, CA, USA) and were visualized using an ImageQuant LAS-4000 (GE Healthcare, Little Chalfont, UK). The size of immunoreactive bands was determined by using molecular weight standards detected with an ECL suitable antibody (1:1000) (sc-2035, Santa Cruz Biotechnology). Band densities were determined using NIH ImageJ software (US National Institutes of Health, Bethesda, MA, USA,
https://imagej.net/ij/ accessed on 24 March 2024). The optical density of the phosphorylated protein bands was normalized to the density of the corresponding total protein in the same sample. For analysis of caspases and PARP, the formation of the cleaved protein was normalized to the level of the corresponding procaspase or non-cleaved PARP measured in the same sample. For the remaining proteins, the densitometric values were normalized to the levels of either actin or a subcellular fraction marker, as indicated.
4.6. Isolation of Cell Nuclei
SH-SY5Y nuclei isolation was performed as previously described [
39]. Cells were grown in Petri dishes. After drug treatment, cells were washed with ice-cold PBS (pH 7.4) and scraped in an ice-cold lysis buffer. Cell lysates were subjected to centrifugation at 3000×
g for 10 min at 4 °C while the supernatant was collected and centrifuged at 24,000×
g for 20 min (cytosolic fraction). The pellets were washed three times in ice-cold washing buffer and layered over a cushion of 1 mL of 1 M sucrose buffer. Following centrifugation at 3000×
g for 10 min at 4 °C, the nuclei were washed, and the proteins were extracted by incubating the nuclei for 30 min in an extraction buffer. Following centrifugation at 24,000×
g for 10 min at 4 °C, the nuclear extracts were heated at 100 °C with sample buffer.
4.7. Transfection of Small Interfering RNA (siRNA)
SH-SY5Y cells were transfected with either Trilencer-27 Universal scrambled negative control siRNA duplex (SR30004) or PPARA (Human)-3 unique 27mer siRNA duplexes (SR303653) using Lipofectamine 2000 (Invitrogen-Thermo Fisher Scientific, Rockford, IL, USA) as a transfection reagent. Cells grown in 6-well plates were incubated in an antibiotic-free medium for 24 h. The medium was renewed, and the cells were incubated with siRNA duplexes for 4–5 h at 37 °C. Thereafter, the medium was replaced by the growth medium, and the cells were analyzed 48 h post-transfection.
4.8. Determination of Cell Count and Viability
A Muse™ Cell Analyzer was used for determining the count and viability of cellular samples using the Muse™ Viability assay kit as instructed by the manufacturer (Millipore Corporation, Merck Life Sciences, Darmstadt, Germany). Briefly, SH-SY5Y cells were treated with OEA or PEA in the presence or absence of IFNβ and were incubated at 37 °C for 24 h. Cells were detached and centrifuged at 300× g for 5 min. Finally, the obtained cell pellet was suspended in a complete medium. An amount of 20 µL of this cell suspension was mixed with 380 µL of Count & Viability reagent. The suspension was then kept for 5 min at room temperature and thereafter examined for cell count and viability by the Muse™ Cell Analyzer.
4.9. MTT Assay
Cell viability was assessed by an MTT assay in a 96-well plate. SH-SY5Y cells were treated with either the vehicle, OEA, or PEA at different concentrations for 24 h. Cells were incubated with MTT. The blue formazan product was solubilized by the addition of 10% SDS with 10 mM HCl. Absorbance was measured using a Wallach Victor microplate reader (PerkinElmer, Waltham, MA, USA).
4.10. Scratch Wound Healing Assay
SH-SY5Y cells were seeded into 24-well cell culture plates and were allowed to grow to 100% confluence as a monolayer. The monolayer was gently scratched across the center of the well with a sterile pipette tip. After scratching, the medium was removed, and the wells were washed twice in PBS solution. Fresh medium containing no FBS and designated treatments were added to each well. Images were obtained from the same fields immediately after scratching (t0) and 48 h later. The scratch was visualized by phase-contrast light microscopy using an Olympus IX51 inverted microscope (Olympus Optical Co., Hamburg, Germany). The images were acquired in randomly selected fields by using an Olympus digital camera and analyzed by ImageJ software (National Institutes of Health, Bethesda, MD, USA). The percentage of the closure of the scratch was then calculated.
4.11. Clonogenic Assay
Single-cell suspensions of exponentially growing cultures were seeded into six-well plates in a range of 200 cells/well and were allowed to adhere for 24 h. Cells were then incubated at 37 °C for 10 days. The culture medium was changed every 2 days. At the end of the 10 days, cell growth of all six-well plates was stopped simultaneously. Colonies were fixed with 100% ethanol and were stained with 0.5% crystal violet. A cell colony was defined as a group formation of at least 50 cells and was counted using the Image J software.
4.12. RealTime-Glo MT Cell Viability Assay
Luminescence analysis using the RealTime-Glo MT assay kit (Promega, Madison, WI, USA) was used to determine cell viability. Cells grown in 96-well plates (ViewPlate, PerkinElmer) were exposed to the test agents and then incubated with the reagents provided by the kit following the manufacturer’s instructions. The intensity of the luminescent signal generated in viable cells was measured by a Wallac Victor III microplate reader (PerkinElmer, Waltham, MA, USA). Assays were performed in triplicate.
4.13. Caspase-Glo 3/7 Assay
The cells that were grown in 96-well plates (ViewPlate-96) were incubated as specified in the text. The cells were then assayed for caspase activity by using the Caspase-Glo 3/7 assay kit (Promega, Madison, USA), according to the manufacturer’s instructions. Luminescence intensity was measured by using a Wallac Victor III microplate reader (PerkinElmer, Waltham, MA, USA). The assays were performed in triplicate.