miR-370-3p Regulates Adipogenesis through Targeting Mknk1
Abstract
:1. Introduction
2. Results
2.1. miR-370-3p Is Associated with Adipogenesis
2.2. miR-370-3p Promotes Preadipocyte Proliferation
2.3. miR-370-3p Inhibits Preadipocyte Differentiation
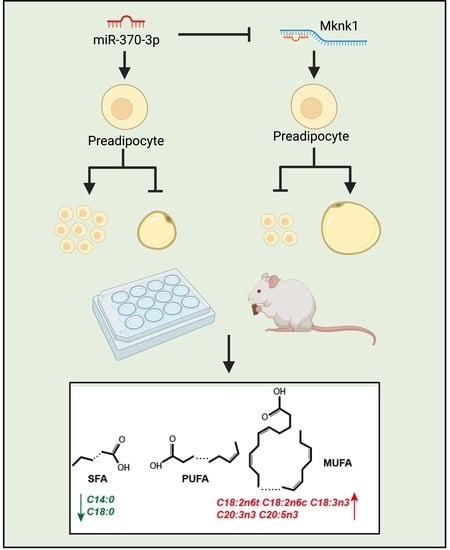
2.4. miR-370-3p Changes Mature Adipocyte Fatty Acid Composition In Vitro and In Vivo, and Inhibit Adipogenesis In Vivo
2.5. Mknk1 Is a Target Gene of miR-370-3p
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Ethics Statement
5.2. Experimental Animals
5.3. Cell Culture and Transfection
5.4. Cell Proliferation Assay
5.5. Isolation of RNA and RT-PCR
5.6. Oil Red O Staining and Triglyceride Assay
5.7. Luciferase Reporter Assay
5.8. Westen Blotting
5.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PPARγ | Peroxisome proliferator activated receptor γ |
| C/EBPα | CCAAT/enhancer binding protein α |
| FABP4 | Adipocyte fatty acid-binding protein 4 |
| CDK2 | Cyclin-dependent kinase 2 |
| CDK4 | Cyclin-dependent kinases 4 |
| CyclinD1 | Cyclin D1 |
| CyclinE | Cyclin E1 |
| P21 | Cyclin-dependent kinase inhibitor 1 |
| Adipoq | Adiponectin, C1Q and collagen domain containing |
| ACOX2 | Acyl-Coenzyme A oxidase 2, branched chain |
| ACSS1 | Acyl-CoA synthetase short-chain family member 1 |
| CD36 | CD36 molecule |
| ACADL | Acyl-Coenzyme A dehydrogenase, long-chain |
| VLDL | CD320 antigen |
| SCD | Stearoyl-Coenzyme A desaturase 1 |
| FAS | Fatty acid synthase |
| DGAT | Diacylglycerol O-acyltransferase 1 |
| PNPLA3 | Patatin-like phospholipase domain containing 3 |
| BMP2 | Bone morphogenetic protein 2 |
| Mknk1 | MAP kinase-interacting serine/threonine kinase 1 |
| miRNA | MicroRNA |
| UTR | 3′untranslated region |
| RISC | RNA-induced silencing complex |
| HFD | high fat diet |
| NCW | normal chow |
| eWAT | epididymal white adipose tissue |
| iWAT | inguinal white adipose tissue |
| TG | triglycerides |
| TC | total cholesterol |
| NC | negative control |
| HFD-OE group | high fat diet-induced obesity mice which overexpressed miR-370-3p |
| WT | wild type |
| MUT | mutant |
| SFAs | saturated fatty acids |
| MUFAs | monounsaturated fatty acids |
| PUFAs | polyunsaturated fatty acids |
| EdU | 5-ethynyl-20-deoxyuridine |
| qRT-PCR | Quantitative real-time PCR |
| WT-Mknk1 | wild-type 3′UTR of Mknk1 |
| MUT-Mknk1 | mutant-type 3’UTR of Mknk1 |
References
- Yamauchi, T.; Kamon, J.; Waki, H.; Terauchi, Y.; Kubota, N.; Hara, K.; Mori, Y.; Ide, T.; Murakami, K.; Tsuboyama-Kasaoka, N.; et al. The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat. Med. 2001, 7, 941–946. [Google Scholar] [CrossRef] [PubMed]
- Jung, R.T. Obesity as a disease. Br. Med Bull. 1997, 53, 307–321. [Google Scholar] [CrossRef]
- Spiegelman, B.M.; Flier, J.S. Obesity and the Regulation of Energy Balance. Cell 2001, 104, 531–543. [Google Scholar] [CrossRef] [Green Version]
- Rosen, E.D.; MacDougald, O. Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Biol. 2006, 7, 885–896. [Google Scholar] [CrossRef] [PubMed]
- Obregon, M.-J. Thyroid Hormone and Adipocyte Differentiation. Thyroid 2008, 18, 185–195. [Google Scholar] [CrossRef] [Green Version]
- Gregoire, F.M.; Smas, C.M.; Sul, H.S. Understanding adipocyte differentiation. Physiol. Rev. 1998, 78, 783–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved Seed Pairing, Often Flanked by Adenosines, Indicates that Thousands of Human Genes are MicroRNA Targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Xu, B.; Shen, J.; Li, D.; Ning, B.; Guo, L.; Bing, H.; Chen, J.; Li, Y. Overexpression of microRNA-9 inhibits 3T3-L1 cell adipogenesis by targeting PNPLA3 via activation of AMPK. Gene 2020, 730, 144260. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, T.; Wang, S.; Wei, J.; Fan, J.; Li, J.; Han, Q.; Liao, L.; Shao, C.; Zhao, R.C. miR-17-5p and miR-106a are involved in the balance between osteogenic and adipogenic differentiation of adipose-derived mesenchymal stem cells. Stem Cell Res. 2013, 10, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Moore, C.E.; Pickford, J.; Cagampang, F.R.; Stead, R.L.; Tian, S.; Tang, X.; Byrne, C.D.; Proud, C.G. MNK1 and MNK2 mediate adverse effects of high-fat feeding in distinct ways. Sci. Rep. 2016, 6, 23476. [Google Scholar] [CrossRef] [Green Version]
- Sandeman, L.Y.; Kang, W.X.; Wang, X.; Jensen, K.B.; Wong, D.; Bo, T.; Gao, L.; Zhao, J.; Byrne, C.D.; Page, A.J.; et al. Disabling MNK protein kinases promotes oxidative metabolism and protects against diet-induced obesity. Mol. Metab. 2020, 42, 101054. [Google Scholar] [CrossRef]
- Xu, N.; Geller, D.H.; Jones, M.R.; Funari, V.A.; Azziz, R.; Goodarzi, M.O. Comprehensive assessment of expression of insulin signaling pathway components in subcutaneous adipose tissue of women with and without polycystic ovary syndrome. J. Clin. Transl. Endocrinol. 2015, 2, 99–104. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.B.; Spiegelman, B.M. ADD1/SREBP1 promotes adipocyte differentiation and gene expression linked to fatty acid metabolism. Genes Dev. 1996, 10, 1096–1107. [Google Scholar] [CrossRef] [Green Version]
- Trayhurn, P.; Wood, I.S. Adipokines: Inflammation and the pleiotropic role of white adipose tissue. Br. J. Nutr. 2004, 92, 347–355. [Google Scholar] [CrossRef] [Green Version]
- Cancello, R.; Henegar, C.; Viguerie, N.; Taleb, S.; Poitou, C.; Rouault, C.; Coupaye, M.; Pelloux, V.; Hugol, D.; Bouillot, J.-L.; et al. Reduction of Macrophage Infiltration and Chemoattractant Gene Expression Changes in White Adipose Tissue of Morbidly Obese Subjects After Surgery-Induced Weight Loss. Diabetes 2005, 54, 2277–2286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, C.-Y.; Zhu, M.-X.; Lu, N.-H.; Liu, J.-Q.; Yang, Y.-W.; Zhang, Y.; Shi, Y.-D.; Feng, Z.-H.; Li, J.-X.; Qi, F.-Z.; et al. Circular RNA circ_0020710 drives tumor progression and immune evasion by regulating the miR-370-3p/CXCL12 axis in melanoma. Mol. Cancer 2020, 19, 1–14. [Google Scholar] [CrossRef]
- Liu, L.; Yan, C.; Tao, S.; Wang, H. Circ_0058124 Aggravates the Progression of Papillary Thyroid Carcinoma by Activating LMO4 Expression via Targeting miR-370-3p. Cancer Manag. Res. 2020, 12, 9459–9470. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Song, G.; Yuan, J.; Qiao, S.; Xu, S.; Si, Z.; Yang, Y.; Xu, X.; Wang, A. RETRACTED ARTICLE: Circular RNA circ_0003204 inhibits proliferation, migration and tube formation of endothelial cell in atherosclerosis via miR-370-3p/TGFβR2/phosph-SMAD3 axis. J. Biomed. Sci. 2020, 27, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, Y.; Zhang, M.; Duan, R.; Yang, J.; Yang, Y.; Wang, J.; Jiang, C.; Yao, B.; Li, L.; Yuan, H.; et al. Long noncoding RNA FGF14-AS2 inhibits breast cancer metastasis by regulating the miR-370-3p/FGF14 axis. Cell Death Discov. 2020, 6, 1–14. [Google Scholar] [CrossRef]
- Mizuno, H.; Hedrick, M.H. Multilineage cells from human adipose tissue; implications for future cell-based tissue engineering in the field of plastic surgery. Tissue Eng. 2001, 7, 211–228. [Google Scholar]
- Tilg, H.; Moschen, A. Adipocytokines: Mediators linking adipose tissue, inflammation and immunity. Nat. Rev. Immunol. 2006, 6, 772–783. [Google Scholar] [CrossRef] [PubMed]
- Zuber, M.X.; Wang, S.M.; Thammavaram, K.V.; Reed, D.K.; Reed, B.C. Elevation of the number of cell-surface insulin receptors and the rate of 2-deoxyglucose uptake by exposure of 3T3-L1 adipocytes to tolbutamide. J. Biol. Chem. 1985, 260, 14045–14052. [Google Scholar] [CrossRef]
- Haraguchi, K.; Shimura, H.; Lin, L.; Saito, T.; Endo, T.; Onaya, T. Functional Expression of Thyrotropin Receptor in Differentiated 3T3-L1 Cells: A Possible Model Cell Line of Extrathyroidal Expression of Thyrotropin Receptor. Biochem. Biophys. Res. Commun. 1996, 223, 193–198. [Google Scholar] [CrossRef]
- Lee, H.; Kim, J.; Park, J.Y.; Kang, K.S.; Park, J.H.; Hwang, G.S. Processed Panax ginseng, sun ginseng, inhibits the differentiation and proliferation of 3T3-L1 preadipocytes and fat accumulation in Caenorhabditis elegans. J. Ginseng Res. 2017, 41, 257–267. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Liu, Z.; Zhang, Z.; Liu, G.; Sun, S.; Sun, C. miR-103 promotes 3T3-L1 cell adipogenesis through AKT/mTOR signal pathway with its target being MEF2D. Biol. Chem. 2015, 396, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Mo, D.; Li, M.; Zhang, Y.; Chen, L.; Zhang, X.; Li, M.; Zhou, X.; Chen, Y. miR-709 inhibits 3T3-L1 cell differentiation by targeting GSK3β of Wnt/β-catenin signaling. Cell. Signal. 2014, 26, 2583–2589. [Google Scholar] [CrossRef]
- Tang, R.; Ma, F.; Li, W.; Ouyang, S.; Liu, Z.; Wu, J. miR-206-3p Inhibits 3T3-L1 Cell Adipogenesis via the c-Met/PI3K/Akt Pathway. Int. J. Mol. Sci. 2017, 18, 1510. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, D.; Xu, L.; Zhang, L.; Guo, D.; Tan, X.; Yu, X.; Yaqiong, Y.; Ye, Y.; Liu, Q.; Ma, Y.; et al. MiR-181a-5p regulates 3T3-L1 cell adipogenesis by targetingSmad7andTcf7l2. Acta Biochim. Biophys. Sin. 2016, 48, 1034–1041. [Google Scholar] [CrossRef] [Green Version]
- Alexandre-Gouabau, M.-C.; Moyon, T.; Cariou, V.; Antignac, J.-P.; Qannari, E.M.; Croyal, M.; Soumah, M.; Guitton, Y.; David-Sochard, A.; Billard, H.; et al. Breast Milk Lipidome Is Associated with Early Growth Trajectory in Preterm Infants. Nutrients 2018, 10, 164. [Google Scholar] [CrossRef] [Green Version]
- Menni, C.; Fauman, E.; Erte, I.; Perry, J.R.; Kastenmüller, G.; Shin, S.-Y.; Petersen, A.-K.; Hyde, C.; Psatha, M.; Ward, K.J.; et al. Biomarkers for Type 2 Diabetes and Impaired Fasting Glucose Using a Nontargeted Metabolomics Approach. Diabetes 2013, 62, 4270–4276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guillou, H.; Zadravec, D.; Martin, P.G.; Jacobsson, A. The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog. Lipid Res. 2010, 49, 186–199. [Google Scholar] [CrossRef] [PubMed]
- Wood, R.; Reiser, R. Cyclopropane fatty acid metabolism: Physical and chemical identification of propane ring metabolic products in the adipose tissue. J. Am. Oil Chem. Soc. 1965, 42, 315–320. [Google Scholar] [CrossRef]
- Jensen, M.D. Adipose tissue and fatty acid metabolism in humans. J. R. Soc. Med. 2002, 95, 3–7. [Google Scholar]
- Jensen, M.D. Adipose tissue as an endocrine organ: Implications of its distribution on free fatty acid metabolism. Eur. Heart J. Suppl. 2006, 8, B13–B19. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Du, J.; Zhang, P.; Zhao, X.; Li, Q.; Jiang, A.; Jiang, D.; Wang, J.; Li, X.; Zhang, S.; et al. MicroRNA-125a-5p Mediates 3T3-L1 Preadipocyte Proliferation and Differentiation. Molecules 2018, 23, 317. [Google Scholar] [CrossRef] [Green Version]
- Xing, K.; Zhao, X.; Liu, Y.; Zhang, F.; Tan, Z.; Qi, X.; Wang, X.; Ni, H.; Guo, Y.; Sheng, X.; et al. Identification of Differentially Expressed MicroRNAs and Their Potential Target Genes in Adipose Tissue from Pigs with Highly Divergent Backfat Thickness. Animals 2020, 10, 624. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Du, J.; Wang, L.; Niu, L.; Zhao, Y.; Tang, G.; Jiang, Y.; Shuai, S.; Bai, L.; Li, X.; et al. MicroRNA-143a-3p modulates preadipocyte proliferation and differentiation by targeting MAPK7. Biomed. Pharmacother. 2018, 108, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Du, J.; Guo, X.; Wu, S.; He, J.; Li, X.; Shen, L.; Chen, L.; Li, B.; Zhang, J.; et al. LncMyoD Promotes Skeletal Myogenesis and Regulates Skeletal Muscle Fiber-Type Composition by Sponging miR-370-3p. Genes 2021, 12, 589. [Google Scholar] [CrossRef]





| Fatty Acid | NC | Mimics | Significance |
|---|---|---|---|
| C6:0 | 0.29 | 0.28 | Increase NS |
| C8:0 | 0.1 | 0.11 | Increase NS |
| C10:0 | 0.16 | 0.16 | Increase NS |
| C11:0 | 0 | 0 | —— |
| C12:0 | 0.26 | 0.28 | Increase NS |
| C13:0 | 0.26 | 0.25 | Increase NS |
| C14:0 | 1.6 | 1.35 | Decrease * |
| C14:1 | 0 | 0 | —— |
| C15:0 | 0.38 | 0.4 | Increase NS |
| C15:1 | 0.87 | 0.95 | Increase * |
| C16:0 | 4.01 | 3.64 | Decrease NS |
| C16:1 | 0.24 | 0.22 | Decrease NS |
| C17:0 | 0.44 | 0.45 | Increase NS |
| C17:1 | 0 | 0 | —— |
| C18:0 | 6.59 | 4.77 | Decrease ** |
| C18:1n9t | 0 | 0 | —— |
| C18:1n9c | 7.72 | 8.63 | Increase NS |
| C18:2n6t | 0 | 0 | —— |
| C18:2n6c | 1.39 | 1.63 | Increase * |
| C18:3n6 | 0 | 0 | —— |
| C18:3n3 | 0.58 | 0.67 | Increase * |
| C20:0 | 0.62 | 0.48 | Decrease NS |
| C20:1 | 0 | 0 | —— |
| C20:2 | 0.59 | 0.61 | Increase NS |
| C21:0 | 0 | 0 | —— |
| C20:3n6 | 1.22 | 1.54 | Increase NS |
| C20:4n6 | 2.18 | 2.45 | Increase * |
| C20:3n3 | 0.57 | 0.63 | Increase ** |
| C20:5n3 | 0.49 | 0.89 | Increase ** |
| C22:0 | 0 | 0 | —— |
| C22:1n9 | 0 | 0 | —— |
| C22:2n6 | 0 | 0 | —— |
| C23:0 | 0.12 | 0.13 | |
| C24:0 | 0 | 0 | —— |
| C24:1 | 0 | 0 | —— |
| C22:6 | 1.42 | 1.97 | Increase * |
| Fatty Acid | HFD-NC | HFD-OE | Significance |
|---|---|---|---|
| C6:0 | 29.4 | 36.17 | Increase NS |
| C8:0 | 10.23 | 13.32 | Increase NS |
| C10:0 | 50.51 | 61.86 | Increase NS |
| C11:0 | 1 | 1.11 | Increase NS |
| C12:0 | 263.84 | 334.95 | Increase NS |
| C13:0 | 6.2 | 6.17 | Increase NS |
| C14:0 | 5937.03 | 5544.6 | Decrease ** |
| C14:1 | 258.6 | 305.88 | Increase ** |
| C15:0 | 329.27 | 355.14 | Increase NS |
| C15:1 | 2.87 | 4.44 | Increase ** |
| C16:0 | 48,897.32 | 47,001.13 | Decrease NS |
| C16:1 | 19,479.61 | 21,348.67 | Increase NS |
| C17:0 | 586.42 | 588.27 | Increase NS |
| C17:1 | 767.72 | 832.1 | Increase NS |
| C18:0 | 21,122.84 | 21,110.67 | Decrease ** |
| C18:1n9t | 0 | 0 | —— |
| C18:1n9c | 92,141.78 | 92,929.21 | Increase ** |
| C18:2n6t | 0 | 0 | —— |
| C18:2n6c | 43,726.38 | 44,619.86 | Increase ** |
| C18:3n6 | 0 | 0 | —— |
| C18:3n3 | 3682.38 | 4043 | Increase NS |
| C20:0 | 469.59 | 446.31 | Decrease NS |
| C20:1 | 3283.17 | 3103.03 | Decrease NS |
| C20:2 | 1369.2 | 1390.44 | Increase NS |
| C21:0 | 0 | 0 | —— |
| C20:3n6 | 507.39 | 539.02 | Increase NS |
| C20:4n6 | 801.83 | 778.48 | Decrease NS |
| C20:3n3 | 1899.48 | 2096.02 | Increase ** |
| C20:5n3 | 173.99 | 184.22 | Increase ** |
| C22:0 | 72.72 | 69.17 | Decrease ** |
| C22:1n9 | 149.16 | 137.78 | Decrease NS |
| C22:2n6 | 11.57 | 12.34 | Increase NS |
| C23:0 | 12.71 | 13.08 | Increase NS |
| C24:0 | 52.09 | 51.81 | Decrease NS |
| C24:1 | 88.6 | 85.59 | Decrease NS |
| C22:6 | 1157.08 | 1157.85 | Increase NS |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, P.; Li, X.; Zhang, S.; Wu, S.; Xiao, Q.; Gu, Y.; Guo, X.; Lin, X.; Chen, L.; Zhao, Y.; et al. miR-370-3p Regulates Adipogenesis through Targeting Mknk1. Molecules 2021, 26, 6926. https://doi.org/10.3390/molecules26226926
Zhang P, Li X, Zhang S, Wu S, Xiao Q, Gu Y, Guo X, Lin X, Chen L, Zhao Y, et al. miR-370-3p Regulates Adipogenesis through Targeting Mknk1. Molecules. 2021; 26(22):6926. https://doi.org/10.3390/molecules26226926
Chicago/Turabian StyleZhang, Peiwen, Xinrong Li, Shunhua Zhang, Shuang Wu, Qian Xiao, Yang Gu, Xinyu Guo, Xutao Lin, Lei Chen, Ye Zhao, and et al. 2021. "miR-370-3p Regulates Adipogenesis through Targeting Mknk1" Molecules 26, no. 22: 6926. https://doi.org/10.3390/molecules26226926