Amperometric Biosensors Based on Direct Electron Transfer Enzymes
Abstract
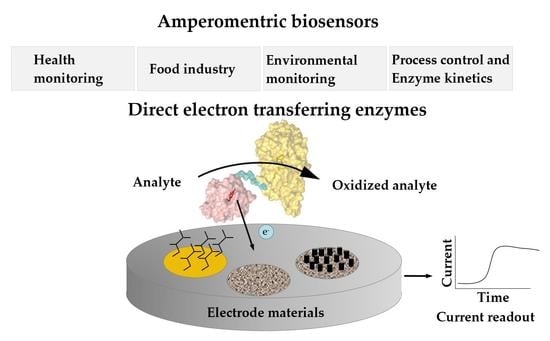
:1. Introduction
1.1. Background
1.2. Amperometric Biosensors
1.3. Applications for DET-Based Biosensors
1.4. Challenges of DET-Based Biosensors
1.5. Scope
2. Enzymes
2.1. Single Cofactor Enzymes
2.1.1. Peroxidases
2.1.2. Theophylline Oxidase Cytochrome P450
2.1.3. Glucose Oxidase
2.1.4. FAD-Dependent Glucose Dehydrogenase
2.1.5. FAD-Domain of Cellobiose Dehydrogenase
2.1.6. FAD-Dependent Pyranose Dehydrogenase
2.1.7. PQQ-Dependent Glucose Dehydrogenase
2.1.8. PQQ-Domain of Pyranose Dehydrogenase
2.1.9. Copper Oxidases
2.2. Multi-Cofactor Direct Electron Transfer Enzymes
2.2.1. Cellobiose Dehydrogenase
2.2.2. Bacterial FAD-Dependent Glucose Dehydrogenase
2.2.3. Bacterial FAD-Dependent Fructose Dehydrogenase
2.2.4. FMN-Dependent L-Lactate Dehydrogenase
2.2.5. Xanthine Oxidase
2.2.6. Sulfite Dehydrogenase and Sulfite Oxidase
2.2.7. PQQ-Dependent Pyranose Dehydrogenase
2.2.8. Bacterial PQQ-Dependent Type II Alcohol Dehydrogenase
2.2.9. Bacterial PQQ-Dependent Lactate Dehydrogenase
2.3. Fusion Enzymes
2.3.1. PQQ-Dependent Glucose Dehydrogenase Fused to a Cytochrome c Domain
2.3.2. FAD-Dependent Glucose Dehydrogenase Fused to a Minimal Cytochrome c Domain
2.3.3. FAD-Dependent Glucose Dehydrogenase Fused to a Cytochrome b Domain
2.3.4. FAD-Dependent Glucose Dehydrogenase Fused to Cytochrome b562
2.4. Performance of DET Enzymes
3. Electrode Materials and Modifications
3.1. Non-Modified Electrodes
3.2. Electrodes Modified with Self-Assembled Monolayers
| SAM Forming Molecule | SAM Terminal Group | Electrode Material | Enzyme | Analyte | Current (Density) or Sensitivity | Ref. |
|---|---|---|---|---|---|---|
| Cysteamine | amino | Au | Cellobiosedehydrogenase | 1 mM cellobiose | est. 4.55 μA cm–2 | [126] |
| Cysteamine | amino | Au-NPs | Glucose dehydrogenase | glucose | 715 µA mM−1 cm−2 | [78] |
| 4,4′-Aldrithiol | hydroxyl | Au | Cellobiose dehydrogenase | 1 mM cellobiose | est. 0.35 μA | [132] |
| 11-Mercapto-1-undecanol | hydroxyl | Au | Cellobiose dehydrogenase | 10 mM lactose | est. 35 µA cm−2 | [127] |
| 4-Mercaptobenzoic acid | carboxyl | Au | Cellobiose dehydrogenase | 10 mM lactose | est. 20 µA cm−2 | [127] |
| 4-Mercaptophenol | hydroxyl | Au | Cellobiose dehydrogenase | 10 mM lactose | est. 6 µA cm−2 | [127] |
| 2-Mercaptoethane | alkyl | Au | Fructose dehydrogenase | 200 mM fructose | est. 40 µA cm−2 | [134] |
| 2-Mercaptoethanol | hydroxyl | Au | Fructosed ehydrogenase | 100 mM fructose | est. 150 µA cm−2 | [133] |
| 4-Mercaptophenol | hydroxyl | Au | Fructose dehydrogenase | fructose | 175 μA mM–1 cm–2 | [85] |
| Dithiobis(succinimidyl hexanoate) | succinimidylester | Au | FAD-Glucose dehydrogenase | 5 mM glucose | est. 2.8 µA cm−2 | [130] |
| 4-Aminothiophenol/ 4-Mercaptobenzoic acid | hydroxyl/ carboxyl | Au | Cellobiose dehydrogenase | glucose | 3.1 µA mM−1 cm−2 | [74] |
| Dithiobis-(succinimidyl octanoate) | succinimidyl ester | Au | FAD-Glucose dehydrogenase | 5 mM glucose | est. 1.2 µA cm−2 | [130] |
| Hexamethylcystamine | amino | Au | Peroxidase | H2O2 | 0.018 µA mM−1 | [34] |
| 3-Carboxy-propyldisulfide | carboxyl | Au | Peroxidase | H2O2 | 0.0025 µA mM−1 | [34] |
| 1,4-Phenylenediamine | amino | SWCNT | Cellobiose dehydrogenase | 5 mM lactose | 500 μA cm−2 | [114] |
3.2.1. Mesoporous or Nanostructured Electrodes
3.2.2. Mesoporous Graphite Electrodes
3.2.3. Mesoporous Gold Electrodes
3.3. Electrodes Modified with Nanomaterials
3.3.1. Carbon Nanomaterials

| Nanomaterials | Material Properties | Comment | Enzyme | Analyte | Current (Density) Sensitivity | Ref. |
|---|---|---|---|---|---|---|
| Ketjen Black | large surface areas | miniaturized electrode | FAD-glucose dehydrogenase | 5 mM glucose | est. 174 μA cm−2 | [145] |
| Ketjen Black | mesoporous | horseradish peroxidase | H2O2 | 4.8 mA mM–1 cm–2 | [27] | |
| carbon cryogel | pore size of 20 nm | PQQ-glucose dehydrogenase | 300 mM glucose | 930 μA cm−2 | [50] | |
| carbon particles | hollow structure, particle size ~40 nm | Fructose dehydrogenase | 200 mM fructose | est. 12 mA | [152] | |
| SWCNT | heme b domain fused | FAD-glucose dehydrogenase | 5 mM glucose | est. 0.011 μA | [148] | |
| SWCNT | Poly(methoxyaniline sulfonic Acid) modified Au-electrode | PQQ-glucose dehydrogenase | 5 mM glucose | 500 μA cm−2 | [51] | |
| SWCNT | sodium cholate assisted dispersion | acetonitrile plasma-polymerized film (2 nm) on Au-electrode | FAD-glucose dehydrogenase | glucose | 110 μA cm−2 mM−1 | [144] |
| SWCNT | oxidatively shortened CNT | cellobiose dehydrogenase | glucose | 0.22 μA cm−2 mM−1 | [155] | |
| graphene | single layer | covalent immobilization of glucose dehydrogenase | FAD-glucose dehydrogenase | 20 mM glucose | est. 0.3 μA cm−2 | [147] |
| graphene/ SWCNT | highly conductive, no stabilizers | final polyethyleneimine layer | FAD-glucose oxidase | glucose | 72 μA μg−1 | [149] |
| Pt- nanoclusters | porous gold electrodes | FAD-glucose dehydrogenase | 100 mM glucose | est. 1000 μA cm−2 | [43] | |
| Pt and Pd NPs | particle sizes Pt 1.3– 2.5 nm, Pd 2.5–5 nm | Nanohybrids with CNT | cellobiose dehydrogenase | lactose | Pt: 3.07 μA mM−1 Pd: 3.28 μA mM−1 | [154] |
| carbon nanofibers | cup-stacked structure controlled O/C ratio | fructose dehydrogenase | 200 mM fructose | est. 1 mA cm−2 | [82] | |
| carbon nanofibers | edge-plane surface, cup-stacked | horseradish peroxidase | H2O2 | est. 0.6 μA cm−2 mM−1 | [150] | |
| Au-NPs | 4-aminothiophenol modification | alcohol dehydrogenase | 50 mM glycerol | 510 μA cm−2 mM−1 | [156] | |
| Au-NPs | 3D printed graphene/polylactic electrode | horseradish peroxidase | H2O2 | est. 65 μA mM−1 | [31] | |
| Au-NPs/graphene/PEI | hybrid | glutaraldehyde crosslinking | FAD-glucose oxidase | glucose | 93 μA mM−1 cm−2 | [157] |
3.3.2. Metal Nanomaterials
3.4. Performance of Electrode Materials and Modifications
4. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Gorton, L.; Lindgren, A.; Larsson, T.; Munteanu, F.D.; Ruzgas, T.; Gazaryan, I. Direct Electron Transfer between Heme-Containing Enzymes and Electrodes as Basis for Third Generation Biosensors. Anal. Chim. Acta 1999, 400, 91–108. [Google Scholar] [CrossRef]
- Cruys-Bagger, N.; Ren, G.; Tatsumi, H.; Baumann, M.J.; Spodsberg, N.; Andersen, H.D.; Gorton, L.; Borch, K.; Westh, P. An Amperometric Enzyme Biosensor for Real-Time Measurements of Cellobiohydrolase Activity on Insoluble Cellulose. Biotechnol. Bioeng. 2012, 109, 3199–3204. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Wohlschlager, L.; Csarman, F.; Ruff, A.; Schuhmann, W.; Scheiblbrandner, S.; Ludwig, R. Real-Time Measurement of Cellobiose and Glucose Formation during Enzymatic Biomass Hydrolysis. Anal. Chem. 2021, 93, 7732–7738. [Google Scholar] [CrossRef] [PubMed]
- Gray, H.B.; Winkler, J.R. Electron Tunneling through Proteins. Q. Rev. Biophys. 2003, 36, 341–372. [Google Scholar] [CrossRef]
- Moser, C.C.; Keske, J.M.; Warncke, K.; Farid, R.S.; Dutton, P.L. Nature of Biological Electron Transfer. Nature 1992, 355, 796–802. [Google Scholar] [CrossRef]
- Marcus, R.A.; Sutin, N. Electron Transfers in Chemistry and Biology. Biochim. Biophys. Acta 1985, 811, 265–322. [Google Scholar] [CrossRef]
- Mayo, S.L.; Ellis, W.R.; Crutchley, R.J.; Gray, H.B. Long-Range Electron Transfer in Heme Proteins. Science 1986, 233, 953–959. [Google Scholar] [CrossRef]
- Gulaboski, R.; Mirčeski, V.; Kappl, R.; Hoth, M.; Bozem, M. Review—Quantification of Hydrogen Peroxide by Electrochemical Methods and Electron Spin Resonance Spectroscopy. J. Electrochem. Soc. 2019, 166, G82–G101. [Google Scholar] [CrossRef]
- Chenault, H.K.; Whitesides, G.M. Regeneration of Nicotinamide Cofactors for Use in Organic Synthesis. Appl. Biochem. Biotechnol. 1987, 14, 147–197. [Google Scholar] [CrossRef]
- Clark, W.M. Oxidation–Reduction Potentials of Organic Systems. J. Chem. Educ. 1960, 38, 487. [Google Scholar]
- Saleh, F.S.; Rahman, M.R.; Okajima, T.; Mao, L.; Ohsaka, T. Determination of Formal Potential of NADH/NAD+ Redox Couple and Catalytic Oxidation of NADH Using Poly(Phenosafranin)-Modified Carbon Electrodes. Bioelectrochemistry 2011, 80, 121–127. [Google Scholar] [CrossRef]
- Emahi, I.; Mitchell, M.P.; Baum, D.A. Electrochemistry of Pyrroloquinoline Quinone (PQQ) on Multi-Walled Carbon Nanotube-Modified Glassy Carbon Electrodes in Biological Buffers. J. Electrochem. Soc. 2017, 164, H3097–H3102. [Google Scholar] [CrossRef] [Green Version]
- Luong, J.H.T.; Glennon, J.D.; Gedanken, A.; Vashist, S.K. Achievement and Assessment of Direct Electron Transfer of Glucose Oxidase in Electrochemical Biosensing Using Carbon Nanotubes, Graphene, and Their Nanocomposites. Microchim. Acta 2017, 184, 369–388. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, M.; Zhao, F.; Xu, Z.; Dong, S. The Direct Electron Transfer of Glucose Oxidase and Glucose Biosensor Based on Carbon Nanotubes/Chitosan Matrix. Biosens. Bioelectron. 2005, 21, 984–988. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Lee, J.; Loew, N.; Takahashi-Inose, Y.; Okuda-Shimazaki, J.; Kojima, K.; Mori, K.; Tsugawa, W.; Sode, K. Engineered Glucose Oxidase Capable of Quasi-Direct Electron Transfer after a Quick-and-Easy Modification with a Mediator. Int. J. Mol. Sci. 2020, 21, 1137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamazaki, T.; Okuda-Shimazaki, J.; Sakata, C.; Tsuya, T.; Sode, K. Construction and Characterization of Direct Electron Transfer-Type Continuous Glucose Monitoring System Employing Thermostable Glucose Dehydrogenase Complex. Anal. Lett. 2008, 41, 2363–2373. [Google Scholar] [CrossRef]
- Kimmel, D.; LeBlanc, G.; Meschievitz, M.; Cliffel, D. Electrochemical Sensors and Biosensors. Anal. Chem. 2012, 24, 685–707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, A.P.F. Biosensors: Sense and Sensibility. Chem. Soc. Rev. 2013, 42, 3184–3196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, A.P.F. Biosensors-Sense and Sensitivity. Science 2000, 290, 1315–1317. [Google Scholar] [CrossRef]
- Ronkainen, N.J.; Halsall, H.B.; Heineman, W.R. Electrochemical Biosensors. Chem. Soc. Rev. 2010, 39, 1747–1763. [Google Scholar] [CrossRef]
- Grieshaber, D.; MacKenzie, R.; Vörös, J.; Reimhult, E. Electrochemical Biosensors—Sensor Principles and Architectures. Sensors 2008, 8, 1400–1458. [Google Scholar] [CrossRef]
- Harburry, H.A. Oxidation-Reduction Potentials of Horseradish Peroxidase. J. Biol. Chem. 1957, 225, 1009–1024. [Google Scholar] [CrossRef]
- Battistuzzi, G.; Borsari, M.; Ranieri, A.; Sola, M. Redox Thermodynamics of the Fe3+/Fe2+ Couple in Horseradish Peroxidase and Its Cyanide Complex. J. Am. Chem. Soc. 2002, 124, 26–27. [Google Scholar] [CrossRef]
- Battistuzzi, G.; Bellei, M.; Borsari, M.; Di Rocco, G.; Ranieri, A.; Sola, M. Axial Ligation and Polypeptide Matrix Effects on the Reduction Potential of Heme Proteins Probed on Their Cyanide Adducts. J. Biol. Inorg. Chem. 2005, 10, 643–651. [Google Scholar] [CrossRef]
- Lindgren, A.; Ruzgas, T.; Gorton, L.; Csöregi, E.; Bautista Ardila, G.; Sakharov, I.Y.; Gazaryan, I.G. Biosensors Based on Novel Peroxidases with Improved Properties in Direct and Mediated Electron Transfer. Biosens. Bioelectron. 2000, 15, 491–497. [Google Scholar] [CrossRef]
- Carolan, N. The Use of Soybean Peroxidase in Amperometric Biosensors. Ph.D. Thesis, Dublin City University, Dublin, Ireland, 2004. [Google Scholar]
- Xia, H.Q.; Kitazumi, Y.; Shirai, O.; Kano, K. Direct Electron Transfer-Type Bioelectrocatalysis of Peroxidase at Mesoporous Carbon Electrodes and Its Application for Glucose Determination Based on Bienzyme System. Anal. Sci. 2017, 33, 839–844. [Google Scholar] [CrossRef] [Green Version]
- Fang, Y.; Umasankar, Y.; Ramasamy, R.P. A Novel Bi-Enzyme Electrochemical Biosensor for Selective and Sensitive Determination of Methyl Salicylate. Biosens. Bioelectron. 2016, 81, 39–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, S.C.; Rawson, K.; McNeil, C.J. Disposable Tyrosinase-Peroxidase Bi-Enzyme Sensor for Amperometric Detection of Phenols. Biosens. Bioelectron. 2002, 17, 1015–1023. [Google Scholar] [CrossRef]
- Dai, M.; Huang, T.; Chao, L.; Xie, Q.; Tan, Y.; Chen, C.; Meng, W. Horseradish Peroxidase-Catalyzed Polymerization of l-DOPA for Mono-/Bi-Enzyme Immobilization and Amperometric Biosensing of H2O2 and Uric Acid. Talanta 2016, 149, 117–123. [Google Scholar] [CrossRef] [PubMed]
- López Marzo, A.M.; Mayorga-Martinez, C.C.; Pumera, M. 3D-Printed Graphene Direct Electron Transfer Enzyme Biosensors. Biosens. Bioelectron. 2020, 151, 111980. [Google Scholar] [CrossRef]
- Ferapontova, E.E.; Grigorenko, V.G.; Egorov, A.M.; Börchers, T.; Ruzgas, T.; Gorton, L. Mediatorless Biosensor for H2O2 Based on Recombinant Forms of Horseradish Peroxidase Directly Adsorbed on Polycrystalline Gold. Biosens. Bioelectron. 2001, 16, 147–157. [Google Scholar] [CrossRef]
- Shi, L.; Liu, X.; Niu, W.; Li, H.; Han, S.; Chen, J.; Xu, G. Hydrogen Peroxide Biosensor Based on Direct Electrochemistry of Soybean Peroxidase Immobilized on Single-Walled Carbon Nanohorn Modified Electrode. Biosens. Bioelectron. 2009, 24, 1159–1163. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, S.; Zimmermann, H.; Gazaryan, I.; Csöregi, E.; Schuhmann, W. Hydrogen Peroxide Biosensors Based on Direct Electron Transfer from Plant Peroxidases Immobilized on Self-Assembled Thiol-Monolayer Modified Gold Electrodes. Electroanalysis 2001, 13, 284–288. [Google Scholar] [CrossRef]
- Gazaryan, I.G.; Gorton, L.; Ruzgas, T.; Csoregi, E.; Schuhmann, W.; Lagrimini, L.M.; Khushpul’yan, D.M.; Tishkov, V.I. Tobacco Peroxidase as a New Reagent for Amperometric Biosensors. J. Anal. Chem. 2005, 60, 629–638. [Google Scholar] [CrossRef] [Green Version]
- Christenson, A.; Dock, E.; Gorton, L.; Ruzgas, T. Direct Heterogeneous Electron Transfer of Theophylline Oxidase. Biosens. Bioelectron. 2004, 20, 176–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferapontova, E.E.; Gorton, L. Direct Electrochemistry of Heme Multicofactor-Containing Enzymes on Alkanethiol-Modified Gold Electrodes. Bioelectrochemistry 2005, 66, 55–63. [Google Scholar] [CrossRef]
- Shipovskov, S.; Ferapontova, E.E. Biocatalysis of Theophylline Oxidation by Microbial Theophylline Oxidase in the Presence of Non-Physiological Electron Acceptors. Biocatal. Biotransform. 2008, 26, 455–465. [Google Scholar] [CrossRef]
- Ferapontova, E.E.; Shipovskov, S.; Gorton, L. Bioelectrocatalytic Detection of Theophylline at Theophylline Oxidase Electrodes. Biosens. Bioelectron. 2007, 22, 2508–2515. [Google Scholar] [CrossRef] [PubMed]
- Rodbard, D. Continuous Glucose Monitoring: A Review of Successes, Challenges, and Opportunities. Diabetes Technol. Ther. 2016, 18, S23–S213. [Google Scholar] [CrossRef] [Green Version]
- Bartlett, P.N.; Al-Lolage, F.A. There Is No Evidence to Support Literature Claims of Direct Electron Transfer (DET) for Native Glucose Oxidase (GOx) at Carbon Nanotubes or Graphene. J. Electroanal. Chem. 2018, 819, 26–37. [Google Scholar] [CrossRef] [Green Version]
- Sützl, L.; Foley, G.; Gillam, E.M.J.; Bodén, M.; Haltrich, D. The GMC Superfamily of Oxidoreductases Revisited: Analysis and Evolution of Fungal GMC Oxidoreductases. Biotechnol. Biofuels 2019, 12, 1–18. [Google Scholar] [CrossRef]
- Adachi, T.; Fujii, T.; Honda, M.; Kitazumi, Y.; Shirai, O.; Kano, K. Direct Electron Transfer-Type Bioelectrocatalysis of FAD-Dependent Glucose Dehydrogenase Using Porous Gold Electrodes and Enzymatically Implanted Platinum Nanoclusters. Bioelectrochemistry 2020, 133, 107457. [Google Scholar] [CrossRef] [PubMed]
- Schulz, C.; Kittl, R.; Ludwig, R.; Gorton, L. Direct Electron Transfer from the FAD Cofactor of Cellobiose Dehydrogenase to Electrodes. ACS Catal. 2016, 6, 555–563. [Google Scholar] [CrossRef]
- Kielb, P.; Sezer, M.; Katz, S.; Lopez, F.; Schulz, C.; Gorton, L.; Ludwig, R.; Wollenberger, U.; Zebger, I.; Weidinger, I.M. Spectroscopic Observation of Calcium-Induced Reorientation of Cellobiose Dehydrogenase Immobilized on Electrodes and Its Effect on Electrocatalytic Activity. ChemPhysChem 2015, 16, 1960–1968. [Google Scholar] [CrossRef]
- Yakovleva, M.E.; Killyéni, A.; Ortiz, R.; Schulz, C.; MacAodha, D.; Conghaile, P.Ó.; Leech, D.; Popescu, I.C.; Gonaus, C.; Peterbauer, C.K.; et al. Recombinant Pyranose Dehydrogenase—A Versatile Enzyme Possessing Both Mediated and Direct Electron Transfer. Electrochem. Commun. 2012, 24, 120–122. [Google Scholar] [CrossRef]
- Tasca, F.; Gorton, L.; Kujawa, M.; Patel, I.; Harreither, W.; Peterbauer, C.K.; Ludwig, R.; Nöll, G. Increasing the Coulombic Efficiency of Glucose Biofuel Cell Anodes by Combination of Redox Enzymes. Biosens. Bioelectron. 2010, 25, 1710–1716. [Google Scholar] [CrossRef]
- Oubrie, A.; Rozeboom, H.J.; Kalk, K.H.; Olsthoorn, A.J.J.; Duine, J.A.; Dijkstra, B.W. Structure and Mechanism of Soluble Quinoprotein Glucose Dehydrogenase. EMBO J. 1999, 18, 5187–5194. [Google Scholar] [CrossRef] [Green Version]
- Elias, M.; Tanaka, M.; Sakai, M.; Toyama, H.; Matsushita, K.; Adachi, O.; Yamada, M. C-Terminal Periplasmic Domain of Escherichia Coli Quinoprotein Glucose Dehydrogenase Transfers Electrons to Ubiquinone. J. Biol. Chem. 2001, 276, 48356–48361. [Google Scholar] [CrossRef] [Green Version]
- Flexer, V.; Durand, F.; Tsujimura, S.; Mano, N. Efficient Direct Electron Transfer of PQQ-Glucose Dehydrogenase on Carbon Cryogel Electrodes at Neutral PH. Anal. Chem. 2011, 83, 5721–5727. [Google Scholar] [CrossRef] [PubMed]
- Göbel, G.; Schubart, I.W.; Scherbahn, V.; Lisdat, F. Direct Electron Transfer of PQQ-Glucose Dehydrogenase at Modified Carbon Nanotubes Electrodes. Electrochem. Commun. 2011, 13, 1240–1243. [Google Scholar] [CrossRef]
- Milton, R.D.; Minteer, S.D. Direct Enzymatic Bioelectrocatalysis: Differentiating between Myth and Reality. J. R. Soc. Interface 2017, 14, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeda, K.; Igarashi, K.; Yoshida, M.; Nakamura, N. Discovery of a Novel Quinohemoprotein from a Eukaryote and Its Application in Electrochemical Devices. Bioelectrochemistry 2020, 131, 107372. [Google Scholar] [CrossRef]
- Takeda, K.; Kusuoka, R.; Inukai, M.; Igarashi, K.; Ohno, H.; Nakamura, N. An Amperometric Biosensor of L-Fucose in Urine for the First Screening Test of Cancer. Biosens. Bioelectron. 2021, 174, 112831. [Google Scholar] [CrossRef]
- Takeda, K.; Kusuoka, R.; Birrell, J.A.; Yoshida, M.; Igarashi, K.; Nakamura, N. Bioelectrocatalysis Based on Direct Electron Transfer of Fungal Pyrroloquinoline Quinone-Dependent Dehydrogenase Lacking the Cytochrome Domain. Electrochim. Acta 2020, 359, 136982. [Google Scholar] [CrossRef]
- Bogdanovskaya, V.A.; Arkad’eva, I.N.; Osina, M.A. Bioelectrocatalytic Oxygen Reduction by Laccase Immobilized on Various Carbon Carriers. Russ. J. Electrochem. 2017, 53, 1323–1333. [Google Scholar] [CrossRef]
- Thuesen, M.H.; Farver, O.; Reinhammar, B.; Ulstrup, J. Cyclic Voltammetry and Electrocatalysis of the Blue Copper Oxidase Polyporus Versicolor Laccase. Acta Chem. Scand. 1998, 52, 555–562. [Google Scholar] [CrossRef] [Green Version]
- Shleev, S.; Jarosz-Wilkolazka, A.; Khalunina, A.; Morozova, O.; Yaropolov, A.; Ruzgas, T.; Gorton, L. Direct Electron Transfer Reactions of Laccases from Different Origins on Carbon Electrodes. Bioelectrochemistry 2005, 67, 115–124. [Google Scholar] [CrossRef]
- Christenson, A.; Shleev, S.; Mano, N.; Heller, A.; Gorton, L. Redox Potentials of the Blue Copper Sites of Bilirubin Oxidases. Biochim. Biophys. Acta Bioenerg. 2006, 1757, 1634–1641. [Google Scholar] [CrossRef] [Green Version]
- Wayu, M.B.; Pannell, M.J.; Labban, N.; Case, W.S.; Pollock, J.A.; Leopold, M.C. Functionalized Carbon Nanotube Adsorption Interfaces for Electron Transfer Studies of Galactose Oxidase. Bioelectrochemistry 2019, 125, 116–126. [Google Scholar] [CrossRef]
- Santucci, R.; Ferri, T.; Morpurgo, L.; Savini, I.; Avigliano, L. Unmediated Heterogeneous Electron Transfer Reaction of Ascorbate Oxidase and Laccase at a Gold Electrode. Biochem. J. 1998, 615, 611–615. [Google Scholar] [CrossRef] [Green Version]
- Yaropolov, A.I.; Kharybin, A.N.; Emneus, J.; Marko-Varga, G.; Gorton, L. Electrochemical Properties of Some Copper-Containing Oxidases. Bioelectrochem. Bioener. 1996, 40, 49–57. [Google Scholar] [CrossRef]
- Haberska, K.; Vaz-Domínguez, C.; De Lacey, A.L.; Dagys, M.; Reimann, C.T.; Shleev, S. Direct Electron Transfer Reactions between Human Ceruloplasmin and Electrodes. Bioelectrochemistry 2009, 76, 34–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeshi, S.; Fumitaka, N.; Takayuki, F.; Shinnichiro, S. Reduction and Oxidation Processes of Blue Copper Proteins, Azurin, Pseudoazurin, Umecyanin, Stellacyanin, Plantacyanin, and Plastocyanin Approached by Cyclic and Potential Step Voltammetries. Bull. Chem. Soc. Jpn. 1996, 69, 2855–2862. [Google Scholar]
- Studnickova, M.; Pitrincova, J.; Kovar, J. The Electrochemical Behaviour of Copper Proteins Using Differential Pulse Polarography. Bioelectrochem. Bioener. 1991, 25, 109–120. [Google Scholar] [CrossRef]
- Reinhammar, B.R.M. Oxidation-Reduction Potentials of the Electron Acceptors in Laccases and Stellacyanin. Biochim. Biophys. Acta Bioenerg. 1972, 275, 245–259. [Google Scholar] [CrossRef]
- Shleev, S.V.; Morozova, O.V.; Nikitina, O.V.; Gorshina, E.S.; Rusinova, T.V.; Serezhenkov, V.A.; Burbaev, D.S.; Gazaryan, I.G.; Yaropolov, A.I. Comparison of Physico-Chemical Characteristics of Four Laccases from Different Basidiomycetes. Biochimie 2004, 86, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Saysell, C.G.; Borman, C.D.; Baron, A.J.; McPherson, M.J.; Sykes, A.G. Kinetic Studies on the Redox Interconversion of GOasesemi and GOaseox Forms of Galactose Oxidase with Inorganic Complexes as Redox Partners. Inorg. Chem. 1997, 36, 4520–4525. [Google Scholar] [CrossRef]
- Wright, C.; Sykes, A.G. Interconversion of CuI and CuII Forms of Galactose Oxidase: Comparison of Reduction Potentials. J. Inorg. Biochem. 2001, 85, 237–243. [Google Scholar] [CrossRef]
- Scheiblbrandner, S.; Ludwig, R. Cellobiose Dehydrogenase: Bioelectrochemical Insights and Applications. Bioelectrochemistry 2020, 131, 107345. [Google Scholar] [CrossRef]
- Larsson, T.; Elmgren, M.; Lindquist, S.E.; Tessema, M.; Gorton, L.; Henriksson, G. Electron Transfer between Cellobiose Dehydrogenase and Graphite Electrodes. Anal. Chim. Acta 1996, 331, 207–215. [Google Scholar] [CrossRef]
- Lindgren, A.; Larsson, T.; Ruzgas, T.; Gorton, L. Direct Electron Transfer between the Heme of Cellobiose Dehydrogenase and Thiol Modified Gold Electrodes. J. Electroanal. Chem. 2000, 494, 105–113. [Google Scholar] [CrossRef]
- Stoica, L.; Ludwig, R.; Haltrich, D.; Gorton, L. Third-Generation Biosensor for Lactose Based on Newly Discovered Cellobiose Dehydrogenase. Anal. Chem. 2006, 78, 393–398. [Google Scholar] [CrossRef]
- Bollella, P.; Gorton, L.; Antiochia, R. Direct Electron Transfer of Dehydrogenases for Development of 3rd Generation Biosensors and Enzymatic Fuel Cells. Sensors 2018, 18, 1319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lactose Biosensor Assay Kit System. Available online: https://www.directsens.com/Lactosens/ (accessed on 27 July 2021).
- Yoshida, H.; Kojima, K.; Shiota, M.; Yoshimatsu, K.; Yamazaki, T.; Ferri, S.; Tsugawa, W.; Kamitori, S.; Sode, K. X-Ray Structure of the Direct Electron Transfer-Type FAD Glucose Dehydrogenase Catalytic Subunit Complexed with a Hitchhiker Protein. Acta Crystallogr. Sect. D Struct. Biol. 2019, 75, 841–851. [Google Scholar] [CrossRef] [Green Version]
- Yamashita, Y.; Ferri, S.; Huynh, M.L.; Shimizu, H.; Yamaoka, H.; Sode, K. Direct Electron Transfer Type Disposable Sensor Strip for Glucose Sensing Employing an Engineered FAD Glucose Dehydrogenase. Enzyme Microb. Technol. 2013, 52, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Gineitytė, J.; Meškys, R.; Dagys, M.; Ratautas, D. Highly Efficient Direct Electron Transfer Bioanode Containing Glucose Dehydrogenase Operating in Human Blood. J. Power Sources 2019, 441, 1–6. [Google Scholar] [CrossRef]
- Ratautas, D.; Laurynenas, A.; Dagys, M.; Marcinkevičiene, L.; Meškys, R.; Kulys, J. High Current, Low Redox Potential Mediatorless Bioanode Based on Gold Nanoparticles and Glucose Dehydrogenase from Ewingella Americana. Electrochim. Acta 2016, 199, 254–260. [Google Scholar] [CrossRef]
- Ikeda, T.; Matsushita, F.; Senda, M. Amperometric Fructose Sensor Based on Direct Bioelectrocatalysis. Biosens. Bioelectron. 1991, 6, 299–304. [Google Scholar] [CrossRef]
- Kawai, S.; Yakushi, T.; Matsushita, K.; Kitazumi, Y.; Shirai, O.; Kano, K. The Electron Transfer Pathway in Direct Electrochemical Communication of Fructose Dehydrogenase with Electrodes. Electrochem. Commun. 2014, 38, 28–31. [Google Scholar] [CrossRef]
- Komori, K.; Huang, J.; Mizushima, N.; Ko, S.; Tatsuma, T.; Sakai, Y. Controlled Direct Electron Transfer Kinetics of Fructose Dehydrogenase at Cup-Stacked Carbon Nanofibers. Phys. Chem. Chem. Phys. 2017, 19, 27795–27800. [Google Scholar] [CrossRef]
- Bollella, P.; Hibino, Y.; Conejo-Valverde, P.; Soto-Cruz, J.; Bergueiro, J.; Calderón, M.; Rojas-Carrillo, O.; Kano, K.; Gorton, L. The Influence of the Shape of Au Nanoparticles on the Catalytic Current of Fructose Dehydrogenase. Anal. Bioanal. Chem. 2019, 411, 7645–7657. [Google Scholar] [CrossRef] [Green Version]
- Bollella, P.; Hibino, Y.; Kano, K.; Gorton, L.; Antiochia, R. The Influence of PH and Divalent/Monovalent Cations on the Internal Electron Transfer (IET), Enzymatic Activity, and Structure of Fructose Dehydrogenase. Anal. Bioanal. Chem. 2018, 410, 3253–3264. [Google Scholar] [CrossRef] [Green Version]
- Bollella, P.; Hibino, Y.; Kano, K.; Gorton, L.; Antiochia, R. Highly Sensitive Membraneless Fructose Biosensor Based on Fructose Dehydrogenase Immobilized onto Aryl Thiol Modified Highly Porous Gold Electrode: Characterization and Application in Food Samples. Anal. Chem. 2018, 90, 12131–12136. [Google Scholar] [CrossRef] [PubMed]
- Labeyrie, F.; Baudras, A. Differences in Quaternary Structure and Constitutive Chains between Two Homologous Forms of Cytochrome B2 (L-Lactate: Cytochrome c Oxidoreductase). Eur. J. Biochem. 1972, 25, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Jacq, C.; Lederer, F. Cytochrome b2 from Bakers Yeast (L-Lactate Dehydrogenase): A Double-Headed Enzyme. Eur. J. Biochem. 1974, 41, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Kay, C.J.; Lippay, E.W. Mutation of the Heme-Binding Crevice of Flavocytochrome b2 from Saccharomyces cerevisiae: Altered Heme Potential and Absence of Redox Cooperativity between Heme and FMN Centers. Am. Chem. Soc. 1992, 31, 11376–11382. [Google Scholar] [CrossRef]
- Staškevičien, S.L.; Čnas, N.K.; Kulys, J.J. Reagentless Lactate Electrodes Based on Electrocatalytic Oxidation of Flavocytochrome B2. Anal. Chim. Acta 1991, 243, 167–171. [Google Scholar] [CrossRef]
- Kulys, J.J.; Švirmickas, G.J.S. Reagentless Lactate Sensor Based on Cytochrome b2. Anal. Chim. Acta 1980, 117, 115–120. [Google Scholar] [CrossRef]
- Smutok, O.; Gayda, G.; Gonchar, M.; Schuhmann, W. A Novel L-Lactate-Selective Biosensor Based on Flavocytochrome b 2 from Methylotrophic Yeast Hansenula Polymorpha. Biosens. Bioelectron. 2005, 20, 1285–1290. [Google Scholar] [CrossRef]
- Smutok, O.; Karkovska, M.; Serkiz, R.; Vus, В.; Čenas, N.; Gonchar, M. A Novel Mediatorless Biosensor Based on Flavocytochrome b2 Immobilized onto Gold Nanoclusters for Non-Invasive L-Lactate Analysis of Human Liquids. Sens. Actuators B Chem. 2017, 250, 469–475. [Google Scholar] [CrossRef]
- Enroth, C.; Eger, B.T.; Okamoto, K.; Nishino, T.; Nishino, T.; Pai, E.F. Crystal Structures of Bovinemilk Xanthine Dehydrogenase and Xanthine Oxidase: Structure-Based Mechanism of Conversion. Proc. Natl. Acad. Sci. USA 2000, 97, 10723–10728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shan, D.; Wang, Y.N.; Xue, H.G.; Cosnier, S.; Ding, S.N. Xanthine Oxidase/Laponite Nanoparticles Immobilized on Glassy Carbon Electrode: Direct Electron Transfer and Multielectrocatalysis. Biosens. Bioelectron. 2009, 24, 3556–3561. [Google Scholar] [CrossRef]
- Smith, V.J. Determination of Sulfite Using a Sulfite Oxidase Enzyme Electrode. Anal. Chem. 1987, 59, 2256–2259. [Google Scholar] [CrossRef]
- Elliott, S.J.; McElhaney, A.E.; Feng, C.; Enemark, J.H.; Armstrong, F.A. A Voltammetric Study of Interdomain Electron Transfer within Sulfite Oxidase. J. Am. Chem. Soc. 2002, 124, 11612–11613. [Google Scholar] [CrossRef] [PubMed]
- Ferapontova, E.E.; Ruzgas, T.; Gorton, L. Direct Electron Transfer of Heme- and Molybdopterin Cofactor-Containing Chicken Liver Sulfite Oxidase on Alkanethiol-Modified Gold Electrodes. Anal. Chem. 2003, 75, 4841–4850. [Google Scholar] [CrossRef]
- Spricigo, R.; Richter, C.; Leimkühler, S.; Gorton, L.; Scheller, F.W.; Wollenberger, U. Sulfite Biosensor Based on Osmium Redox Polymer Wired Sulfite Oxidase. Colloids Surf. A Physicochem. Eng. Asp. 2010, 354, 314–319. [Google Scholar] [CrossRef]
- Saengdee, P.; Promptmas, C.; Zeng, T.; Wollenberger, U. Third-Generation Sulfite Biosensor Based on Sulfite Oxidase Immobilized on Aminopropyltriethoxysilane Modified Indium Tin Oxide. Electroanalysis 2017, 29, 110–115. [Google Scholar] [CrossRef] [Green Version]
- Karppi, J.; Zhao, H.; Chong, S.L.; Koistinen, A.E.; Tenkanen, M.; Master, E. Quantitative Comparison of Pyranose Dehydrogenase Action on Diverse Xylooligosaccharides. Front. Chem. 2020, 8, 1–15. [Google Scholar] [CrossRef]
- Takeda, K.; Matsumura, H.; Ishida, T.; Yoshida, M.; Igarashi, K.; Samejima, M.; Ohno, H.; Nakamura, N. pH-Dependent Electron Transfer Reaction and Direct Bioelectrocatalysis of the Quinohemoprotein Pyranose Dehydrogenase. Biochem. Biophys. Res. Commun. 2016, 477, 369–373. [Google Scholar] [CrossRef]
- Takeda, K.; Matsumura, H.; Ishida, T.; Samejima, M.; Ohno, H.; Yoshida, M.; Igarashi, K.; Nakamura, N. Characterization of a Novel PQQ-Dependent Quinohemoprotein Pyranose Dehydrogenase from Coprinopsis Cinerea Classified into Auxiliary Activities Family 12 in Carbohydrate-Active Enzymes. PLoS ONE 2015, 10, e0115722. [Google Scholar] [CrossRef]
- Ikeda, T.; Kobayashi, D.; Matsushita, F.; Sagara, T.; Niki, K. Bioelectrocatalysis at Electrodes Coated with Alcohol Dehydrogenase, a Quinohemoprotein with Heme c Serving as a Built-in Mediator. J. Electroanal. Chem. 1993, 361, 221–228. [Google Scholar] [CrossRef]
- Ramanavicius, A.; Habermuller, K.; Csöregi, E.; Laurinavicius, V.; Schuhmann, W. Polypyrrole-Entrapped Quinohemoprotein Alcohol Dehydrogenase. Evidence for Direct Electron Transfer via Conducting-Polymer Chains. Anal. Chem. 1999, 71, 3581–3586. [Google Scholar] [CrossRef] [PubMed]
- Treu, B.L.; Minteer, S.D. Isolation and Purification of PQQ-Dependent Lactate Dehydrogenase from Gluconobacter and Use for Direct Electron Transfer at Carbon and Gold Electrodes. Bioelectrochemistry 2008, 74, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Ludwig, R. Direct Electron Transfer of Enzymes Facilitated by Cytochromes. ChemElectroChem 2019, 6, 958–975. [Google Scholar] [CrossRef] [PubMed]
- Okuda, J.; Sode, K. PQQ Glucose Dehydrogenase with Novel Electron Transfer Ability. Biochem. Biophys. Res. Commun. 2004, 314, 793–797. [Google Scholar] [CrossRef]
- Algov, I.; Grushka, J.; Zarivach, R.; Alfonta, L. Highly Efficient Flavin–Adenine Dinucleotide Glucose Dehydrogenase Fused to a Minimal Cytochrome C Domain. ACS Catal. 2017, 139, 17217–17220. [Google Scholar] [CrossRef]
- Jones, S.; Wilson, T.; Brown, M.; Rahn-Lee, L.; Yu, Y.; Frederiksen, L.; Ozyamak, E.; Komeili, A.; Chang, M. Genetic and Biochemical Investigations of the Role of MamP in Redox Control of Iron Biomineralization in Magnetospirillum Magneticum. Proc. Natl. Acad. Sci. USA 2015, 112, 3904–3909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, K.; Okuda-Shimazaki, J.; Mori, K.; Kojima, K.; Tsugawa, W.; Ikebukuro, K.; Lin, C.E.; La Belle, J.; Yoshida, H.; Sode, K. Designer Fungus FAD Glucose Dehydrogenase Capable of Direct Electron Transfer. Biosens. Bioelectron. 2019, 123, 114–123. [Google Scholar] [CrossRef]
- Igarashi, K.; Verhagen, M.F.J.M.; Samejima, M.; Schülein, M.; Eriksson, K.E.L.; Nishino, T. Cellobiose Dehydrogenase from the Fungi Phanerochaete Chrysosporium and Humicola Insolens. A Flavohemoprotein from Humicola Insolens Contains 6- Hydroxy-Fad as the Dominant Active Cofactor. J. Biol. Chem. 1999, 274, 3338–3344. [Google Scholar] [CrossRef] [Green Version]
- Yanase, T.; Okuda-Shimazaki, J.; Mori, K.; Kojima, K.; Tsugawa, W.; Sode, K. Creation of a Novel DET Type FAD Glucose Dehydrogenase Harboring Escherichia Coli Derived Cytochrome b562 as an Electron Transfer Domain. Biochem. Biophys. Res. Commun. 2020, 530, 82–86. [Google Scholar] [CrossRef]
- Ferapontova, E.E.; Grigorenko, V.G.; Egorov, A.M.; Börchers, T.; Ruzgas, T.; Gorton, L. Direct Electron Transfer in the System Gold Electrode-Recombinant Horseradish Peroxidases. J. Electroanal. Chem. 2001, 509, 19–26. [Google Scholar] [CrossRef]
- Tasca, F.; Harreither, W.; Ludwig, R.; Gooding, J.J.; Gorton, L. Cellobiose Dehydrogenase Aryl Diazonium Modified Single Walled Carbon Nanotubes: Enhanced Direct Electron Transfer through a Positively Charged Surface. Anal. Chem. 2011, 83, 3042–3049. [Google Scholar] [CrossRef]
- Hitaishi, V.P.; Clement, R.; Bourassin, N.; Baaden, M.; de Poulpiquet, A.; Sacquin-Mora, S.; Ciaccafava, A.; Lojou, E. Controlling Redox Enzyme Orientation at Planar Electrodes. Catalysts 2018, 8, 192. [Google Scholar] [CrossRef] [Green Version]
- Ghindilis, A.L.; Atanasov, P.; Wilkins, E. Enzyme-Catalyzed Direct Electron Transfer: Fundamentals and Analytical Applications. Electroanalysis 1997, 9, 661–674. [Google Scholar] [CrossRef]
- Schuhmann, W.; Zimmermann, H.; Habermüller, K.; Laurinavicius, V. Electron-Transfer Pathways between Redox Enzymes and Electrode Surfaces: Reagentless Biosensors Based on Thiol-Monolayer-Bound and Polypyrrole-Entrapped Enzymes. Faraday Discuss. 2000, 116, 245–255. [Google Scholar] [CrossRef]
- Yan, X.; Tang, J.; Tanner, D.; Ulstrup, J.; Xiao, X. Direct Electrochemical Enzyme Electron Transfer on Electrodes Modified by Self-Assembled Molecular Monolayers. Catalysts 2020, 10, 1458. [Google Scholar] [CrossRef]
- Bollella, P.; Katz, E. Enzyme-Based Biosensors: Tackling Electron Transfer Issues. Sensors 2020, 20, 3517. [Google Scholar] [CrossRef]
- Adachi, T.; Kaida, Y.; Kitazumi, Y.; Shirai, O.; Kano, K. Bioelectrocatalytic Performance of D-Fructose Dehydrogenase. Bioelectrochemistry 2019, 129, 1–9. [Google Scholar] [CrossRef]
- Ikeda, T.; Fushimi, F.; Miki, K.; Senda, M. Direct Bioelectrocatalysis at Electrodes Modified with D—Gluconate Dehydrogenase. Agric. Biol. Chem. 1988, 52, 2655–2658. [Google Scholar]
- Ikeda, T.; Miyaoka, S.; Matsushita, F.; Kobayashi, D.; Senda, M. Direct Bioelectrocatalysis at Metal and Carbon Electrodes Modified with Adsorbed D-Gluconate Dehydrogenase or Adsorbed Alcohol Dehydrogenase from Bacterial Mebranes. Chem. Lett. 1992, 21, 847–850. [Google Scholar] [CrossRef]
- Tsujimura, S.; Abo, T.; Ano, Y.; Matsushita, K.; Kano, K. Electrochemistry of D-Gluconate 2-Dehydrogenase from Gluconobacter Frateurii on Indium Tin Oxide Electrode Surface. Chem. Lett. 2007, 36, 1164–1165. [Google Scholar] [CrossRef]
- Ikeda, T.; Miyaoka, S.; Miki, K. Enzyme-Catalysed Electrochemical Oxidation of D-Gluconate at Electrodes Coated with d-Gluconate Dehydrogenase, a Membrane-Bound Flavohemoprotein. J. Electroanal. Chem. 1993, 352, 267–278. [Google Scholar] [CrossRef]
- Razumiene, J.; Niculescu, M.; Ramanavicius, A.; Laurinavicius, V.; Csöregi, E. Direct Bioelectrocatalysis at Carbon Electrodes Modified with Quinohemoprotein Alcohol Dehydrogenase from Gluconobacter Sp. 33. Electroanalysis 2002, 14, 43–49. [Google Scholar] [CrossRef]
- Lindgren, A.; Gorton, L.; Ruzgas, T.; Baminger, U.; Haltrich, D.; Schülein, M. Direct Electron Transfer of Cellobiose Dehydrogenase from Various Biological Origins at Gold and Graphite Electrodes. J. Electroanal. Chem. 2001, 496, 76–81. [Google Scholar] [CrossRef]
- Matsumura, H.; Ortiz, R.; Ludwig, R.; Igarashi, K.; Samejima, M.; Gorton, L. Direct Electrochemistry of Phanerochaete Chrysosporium Cellobiose Dehydrogenase Covalently Attached onto Gold Nanoparticle Modified Solid Gold Electrodes. Langmuir 2012, 28, 10925–10933. [Google Scholar] [CrossRef]
- Tanne, J.; Kracher, D.; Dietzel, B.; Schulz, B.; Ludwig, R.; Lisdat, F.; Scheller, F.W.; Bier, F.F. Carboxylated or Aminated Polyaniline-Multiwalled Carbon Nanotubes Nanohybrids for Immobilization of Cellobiose Dehydrogenase on Gold Electrodes. Biosensors 2014, 4, 370–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeda, K.; Umezawa, K.; Várnai, A.; Eijsink, V.G.; Igarashi, K.; Yoshida, M.; Nakamura, N. Fungal PQQ-Dependent Dehydrogenases and Their Potential in Biocatalysis. Curr. Opin. Chem. Biol. 2019, 49, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Loew, N.; Tsugawa, W.; Lin, C.E.; Probst, D.; La Belle, J.T.; Sode, K. The Electrochemical Behavior of a FAD Dependent Glucose Dehydrogenase with Direct Electron Transfer Subunit by Immobilization on Self-Assembled Monolayers. Bioelectrochemistry 2018, 121, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Yates, N.D.J.; Fascione, M.A.; Parkin, A. Methodologies for “Wiring” Redox Proteins/Enzymes to Electrode Surfaces. Chem. A Eur. J. 2018, 24, 12164–12182. [Google Scholar] [CrossRef] [Green Version]
- Stoica, L.; Dimcheva, N.; Haltrich, D.; Ruzgas, T.; Gorton, L. Electrochemical Investigation of Cellobiose Dehydrogenase from New Fungal Sources on Au Electrodes. Biosens. Bioelectron. 2005, 20, 2010–2018. [Google Scholar] [CrossRef]
- Sugimoto, Y.; Kitazumi, Y.; Shirai, O.; Yamamoto, M.; Kano, K. Role of 2-Mercaptoethanol in Direct Electron Transfer-Type Bioelectrocatalysis of Fructose Dehydrogenase at Au Electrodes. Electrochim. Acta 2015, 170, 242–247. [Google Scholar] [CrossRef] [Green Version]
- Kawai, S.; Yakushi, T.; Matsushita, K.; Kitazumi, Y.; Shirai, O.; Kano, K. Role of a Non-Ionic Surfactant in Direct Electron Transfer-Type Bioelectrocatalysis by Fructose Dehydrogenase. Electrochim. Acta 2015, 152, 19–24. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.; McNeil, C.J.; Cooper, J.M. Direct Electron Transfer Reactions of Glucose Oxidase Immobilised at a Self-Assembled Monolayer. J. Chem. Soc. Chem. Commun. 1995, 1293–1295. [Google Scholar] [CrossRef]
- Adachi, T.; Kitazumi, Y.; Shirai, O.; Kano, K. Direct Electron Transfer-Type Bioelectrocatalysis of Redox Enzymes at Nanostructured Electrodes. Catalysts 2020, 10, 236. [Google Scholar] [CrossRef] [Green Version]
- Bollella, P.; Ludwig, R.; Gorton, L. Cellobiose Dehydrogenase: Insights on the Nanostructuration of Electrodes for Improved Development of Biosensors and Biofuel Cells. Appl. Mater. Today 2017, 9, 319–332. [Google Scholar] [CrossRef] [Green Version]
- Blanford, C.F.; Armstrong, F.A. The Pyrolytic Graphite Surface as an Enzyme Substrate: Microscopic and Spectroscopic Studies. J. Solid State Electrochem. 2006, 10, 826–832. [Google Scholar] [CrossRef]
- Tominaga, M.; Shirakihara, C.; Taniguchi, I. Direct Heterogeneous Electron Transfer Reactions and Molecular Orientation of Fructose Dehydrogenase Adsorbed onto Pyrolytic Graphite Electrodes. J. Electroanal. Chem. 2007, 610, 1–8. [Google Scholar] [CrossRef]
- Sakai, K.; Kitazumi, Y.; Shirai, O.; Kano, K. Nanostructured Porous Electrodes by the Anodization of Gold for an Application as Scaffolds in Direct-Electron-Transfer-Type Bioelectrocatalysis. Anal. Sci. 2018, 34, 1317–1322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, Y.; Wanibuchi, M.; Kitazumi, Y.; Shirai, O.; Kano, K. Improved Direct Electron Transfer-Type Bioelectrocatalysis of Bilirubin Oxidase Using Porous Gold Electrodes. J. Electroanal. Chem. 2019, 843, 47–53. [Google Scholar] [CrossRef]
- Qiu, H.; Xu, C.; Huang, X.; Ding, Y.; Qu, Y.; Gao, P. Adsorption of Laccase on the Surface of Nanoporous Gold and the Direct Electron Transfer between Them. J. Phys. Chem. C 2008, 112, 14781–14785. [Google Scholar] [CrossRef]
- Iannlello, R.M.; Lindsay, T.J.; Yacynych, A.M. Differential Pulse Voltammetric Study of Direct Electron Transfer in Glucose Oxidase Chemically Modified Graphite Electrodes. Anal. Chem. 1982, 54, 1098–1101. [Google Scholar] [CrossRef]
- Muguruma, H.; Iwasa, H.; Hidaka, H.; Hiratsuka, A.; Uzawa, H. Mediatorless Direct Electron Transfer between Flavin Adenine Dinucleotide-Dependent Glucose Dehydrogenase and Single-Walled Carbon Nanotubes. ACS Catal. 2017, 7, 725–734. [Google Scholar] [CrossRef]
- Shimizu, H.; Tsugawa, W. Glucose Monitoring by Direct Electron Transfer Needle-Type Miniaturized Electrode. Electrochemistry 2012, 80, 375–378. [Google Scholar] [CrossRef] [Green Version]
- Tashima, D.; Kishita, T.; Maeno, S.; Nagasawa, Y. Mesoporous Graphitized Ketjenblack as Conductive Nanofiller for Supercapacitors. Mater. Lett. 2013, 110, 105–107. [Google Scholar] [CrossRef]
- Filipiak, M.S.; Vetter, D.; Thodkar, K.; Gutiérrez-Sanz, O.; Jönsson-Niedziółka, M.; Tarasov, A. Electron Transfer from FAD-Dependent Glucose Dehydrogenase to Single-Sheet Graphene Electrodes. Electrochim. Acta 2020, 330, 134998. [Google Scholar] [CrossRef]
- Ito, K.; Okuda-Shimazaki, J.; Kojima, K.; Mori, K.; Tsugawa, W.; Asano, R.; Ikebukuro, K.; Sode, K. Strategic Design and Improvement of the Internal Electron Transfer of Heme b Domain-Fused Glucose Dehydrogenase for Use in Direct Electron Transfer-Type Glucose Sensors. Biosens. Bioelectron. 2021, 176, 112911. [Google Scholar] [CrossRef] [PubMed]
- Grosse, W.; Champavert, J.; Gambhir, S.; Wallace, G.G.; Moulton, S.E. Aqueous Dispersions of Reduced Graphene Oxide and Multi Wall Carbon Nanotubes for Enhanced Glucose Oxidase Bioelectrode Performance. Carbon N. Y. 2013, 61, 467–475. [Google Scholar] [CrossRef] [Green Version]
- Komori, K.; Tatsuma, T.; Sakai, Y. Direct Electron Transfer Kinetics of Peroxidase at Edge Plane Sites of Cup-Stacked Carbon Nanofibers and Their Comparison with Single-Walled Carbon Nanotubes. Langmuir 2016, 32, 9163–9170. [Google Scholar] [CrossRef]
- Mao, X.; Guo, F.; Yan, E.H.; Rutledge, G.C.; Alan Hatton, T. Remarkably High Heterogeneous Electron Transfer Activity of Carbon-Nanotube-Supported Reduced Graphene Oxide. Chem. Mater. 2016, 28, 7422–7432. [Google Scholar] [CrossRef]
- Tsujimura, S.; Nishina, A.; Kamitaka, Y.; Kano, K. Coulometric D-Fructose Biosensor Based on Direct Electron Transfer Using D-Fructose Dehydrogenase. Anal. Chem. 2009, 81, 9383–9387. [Google Scholar] [CrossRef] [PubMed]
- Tavahodi, M.; Ortiz, R.; Schulz, C.; Ekhtiari, A.; Ludwig, R.; Haghighi, B.; Gorton, L. Direct Electron Transfer of Cellobiose Dehydrogenase on Positively Charged Polyethyleneimine Gold Nanoparticles. ChemPlusChem 2017, 82, 546–552. [Google Scholar] [CrossRef] [PubMed]
- Bozorgzadeh, S.; Hamidi, H.; Ortiz, R.; Ludwig, R.; Gorton, L. Direct Electron Transfer of Phanerochaete Chrysosporium Cellobiose Dehydrogenase at Platinum and Palladium Nanoparticles Decorated Carbon Nanotubes Modified Electrodes. Phys. Chem. Chem. Phys. 2015, 17, 24157–24165. [Google Scholar] [CrossRef] [PubMed]
- Tasca, F.; Zafar, M.N.; Harreither, W.; Nöll, G.; Ludwig, R.; Gorton, L. A Third Generation Glucose Biosensor Based on Cellobiose Dehydrogenase from Corynascus Thermophilus and Single-Walled Carbon Nanotubes. Analyst 2011, 136, 2033–2036. [Google Scholar] [CrossRef]
- Ratautas, D.; Tetianec, L.; Marcinkevičienė, L.; Meškys, R.; Kulys, J. Bioanode with Alcohol Dehydrogenase Undergoing a Direct Electron Transfer on Functionalized Gold Nanoparticles for an Application in Biofuel Cells for Glycerol Conversion. Biosens. Bioelectron. 2017, 98, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Rafighi, P.; Tavahodi, M.; Haghighi, B. Fabrication of a Third-Generation Glucose Biosensor Using Graphene-Polyethyleneimine-Gold Nanoparticles Hybrid. Sens. Actuators B Chem. 2016, 232, 454–461. [Google Scholar] [CrossRef]
- Kovács, G.; Ortiz, R.; Coman, V.; Harreither, W.; Popescu, C.; Ludwig, R.; Gorton, L. Influence of SAM Structure on Direct Electron Transfer at Au Electrodes Modified with Cellobiose Dehydrogenase. Rev. Roum. Chim 2012, 57, 361–368. [Google Scholar]
- Meneghello, D.M.; Al-Lolage, D.F.A.; Ma, D.S.; Ludwig, P.R.; Bartlett, P.P.N. Studying Direct Electron Transfer by Site-Directed Immobilization of Cellobiose Dehydrogenase. ChemElectroChem 2019, 6, 700–713. [Google Scholar] [CrossRef]
















| Analyte | Prosthetic Group | Enzyme | Electrode Base | Electrode Modification | Sensitivity [µA mM−1 cm−2] | Ref. |
|---|---|---|---|---|---|---|
| Theophylline | heme | Cytochrome P450 | SPGE | +/− DDAB * | 0.05 | [39] |
| Theophylline | heme | Cytochrome P450 | Au | 52.1 | ||
| H2O2 | heme | Horseradish peroxidase | Au | 1400–1500 | [113] | |
| H2O2 | heme | Soybean peroxidase | GC | SWCNT | 16.625 µA mM−1 | [33] |
| H2O2 | heme | Tobacco peroxidase | Au | SAM | ND | [34] |
| PhenolsDiamines | heme | Tobacco peroxidase | Au, SPGE | SAM | ND | [35] |
| L-Fucose | PQQ | Pyranose dehydrogenase | Au | Au-NPs | 3.12 | [54] |
| Lactose | FAD, heme b | Cellobiose dehydrogenase | GC | SWCNT/aryldiazonium | 500 µA cm−2 | [114] |
| Fructose, food sample | FAD, 3 heme c | Fructose dehydrogenase | Au | porous Au | 175 | [84] |
| L-Lactate (saliva, sweat) | FMN, heme b | Lactate dehydrogenase | Au | Au-NPs | ND | [92] |
| Glucose | PQQ, heme c | Glucose dehydrogenase/cytochrome c | graphite/paraffin paste | / | 131.8 | [107] |
| Glucose | FAD, heme c | Glucose dehydrogenase/cytochrome c | GC | / | ND | [108] |
| Glucose | FAD, heme b | Glucose dehydrogenase, cytochrome b | SPGE | MWCNT | ND | [110] |
| Glucose | FAD, heme b | Glucose dehydrogenase, cytochrome b562 | Au | mesoporous carbon particles | 2.14–8.57 µA cm−2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schachinger, F.; Chang, H.; Scheiblbrandner, S.; Ludwig, R. Amperometric Biosensors Based on Direct Electron Transfer Enzymes. Molecules 2021, 26, 4525. https://doi.org/10.3390/molecules26154525
Schachinger F, Chang H, Scheiblbrandner S, Ludwig R. Amperometric Biosensors Based on Direct Electron Transfer Enzymes. Molecules. 2021; 26(15):4525. https://doi.org/10.3390/molecules26154525
Chicago/Turabian StyleSchachinger, Franziska, Hucheng Chang, Stefan Scheiblbrandner, and Roland Ludwig. 2021. "Amperometric Biosensors Based on Direct Electron Transfer Enzymes" Molecules 26, no. 15: 4525. https://doi.org/10.3390/molecules26154525