Deconstruction of Lignin: From Enzymes to Microorganisms
Abstract
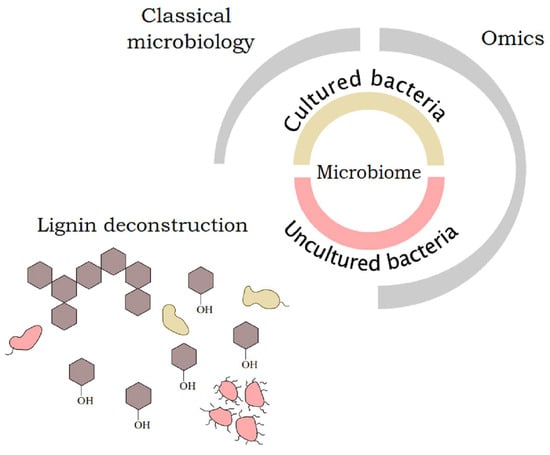
:1. Introduction
2. Lignin
2.1. Structure and Composition of Natural Lignin
2.2. Technical Lignins
3. Bacterial Enzymes for Lignin Deconstruction
4. Approaches to Access Bacterial Community Structure and Functionality
4.1. Microbiomes Characterized Using Metagenomic Approaches
4.1.1. Soil
4.1.2. Invertebrate Digestive Tract
4.1.3. Vertebrate Digestive Tract
4.2. Culture Enrichment
4.3. Metatranscriptomics and Metaproteomics Approaches
4.3.1. Metatranscriptomics
4.3.2. Metaproteomics
5. Application of Lignin-Active Enzymes
6. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- De Bhowmick, G.; Sarmah, A.K.; Sen, R. Lignocellulosic biorefifinery as a model for sustainable development of biofuels and value added products. Bioresour. Technol. 2018, 247, 1144–1154. [Google Scholar] [CrossRef] [PubMed]
- Tursi, A. A review on biomass: Importance, chemistry, classification, and conversion. Biofuel Res. J. 2019, 6, 962–979. [Google Scholar] [CrossRef]
- Barapatre, A.; Jha, H. Degradation of alkali lignin by two ascomycetes and free radical scavenging activity of the products. Biocatal. Biotransform. 2017, 35, 269–286. [Google Scholar] [CrossRef]
- Bugg, T.D.H.; Ahmad, M.; Hardiman, E.M.; Rahmanpour, R. Pathways for degradation of lignin in bacteria and fungi. Nat. Prod. Rep. 2011, 28, 1883–1896. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kang, M.; Bae, J.H.; Sohn, J.H.; Sung, B.H. Bacterial Valorization of Lignin: Strains, Enzymes, Conversion Pathways, Biosensors, and Perspectives. Front. Bioeng. Biotechnol. 2019, 7, 209. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.H.; Kim, C.S. Recent efforts to prevent undesirable reactions from fractionation to depolymerization of lignin: Toward maximizing the value from lignin. Front. Energy Res. 2018, 6, 92. [Google Scholar] [CrossRef] [Green Version]
- Pu, Y.; Hu, F.; Huang, F.; Davison, B.H.; Ragauskas, A.J. Assessing the molecular structure basis for biomass recalcitrance during dilute acid and hydrothermal pretreatments. Biotechnol. Biofuels 2013, 6, 15. [Google Scholar] [CrossRef] [Green Version]
- Schoenherr, S.; Ebrahimi, M.; Czermak, P. Lignin Degradation Processes and the Purification of Valuable Products. In Lignin: Trends and Applications; Books on Demand: Norderstedt, Germany, 2018. [Google Scholar]
- Ravi, K.; García-Hidalgo, J.; Gorwa-Grauslund, M.F.; Lidén, G. Conversion of lignin model compounds by Pseudomonas putida KT2440 and isolates from compost. Appl. Microbiol. Biotechnol. 2017, 101, 5059–5070. [Google Scholar] [CrossRef] [Green Version]
- Dessbesell, L.; Paleologou, M.; Leitch, M.; Pulkki, R.; Xu, C. (Charles) Global lignin supply overview and kraft lignin potential as an alternative for petroleum-based polymers. Renew. Sustain. Energy Rev. 2020, 123, 109768. [Google Scholar] [CrossRef]
- Dashtban, M.; Schraft, H.; Syed, T.A.; Qin, W. Fungal biodegradation and enzymatic modification of lignin. Int. J. Biochem. Mol. Biol. 2010, 1, 36–50. [Google Scholar]
- Zhu, D.; Zhang, P.; Xie, C.; Zhang, W.; Sun, J.; Qian, W.J.; Yang, B. Biodegradation of alkaline lignin by Bacillus ligniniphilus L1. Biotechnol. Biofuels 2017, 10, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, Z.B.; Luo, Q.; Wang, H.R.; Chen, Y.; Liao, X.R. Bacterial laccases: Promising biological green tools for industrial applications. Cell. Mol. Life Sci. 2018, 75, 3569–3592. [Google Scholar] [CrossRef] [PubMed]
- Riyadi, F.A.; Tahir, A.A.; Yusof, N.; Sabri, N.S.A.; Noor, M.J.M.M.; Akhir, F.N.M.D.; Othman, N.; Zakaria, Z.; Hara, H. Enzymatic and genetic characterization of lignin depolymerization by Streptomyces sp. S6 isolated from a tropical environment. Sci. Rep. 2020, 10, 7813–7819. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Lei, P.; Zhai, R.; Wen, Z.; Jin, M. Recent advances in lignin valorization with bacterial cultures: Microorganisms, metabolic pathways, and bio-products. Biotechnol. Biofuels 2019, 12, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.; Chai, L.; Tang, C.; Yang, Z.; Zheng, Y.; Chen, Y.; Jing, Q. Biochemical investigation of kraft lignin degradation by pandoraea sp. B-6 isolated from bamboo slips. Bioprocess Biosyst. Eng. 2013, 36, 1957–1965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scully, E.D.; Geib, S.M.; Hoover, K.; Tien, M.; Tringe, S.G.; Barry, K.W.; Glavina del Rio, T.; Chovatia, M.; Herr, J.R.; Carlson, J.E. Metagenomic Profiling Reveals Lignocellulose Degrading System in a Microbial Community Associated with a Wood-Feeding Beetle. PLoS ONE 2013, 8, e73827. [Google Scholar] [CrossRef] [Green Version]
- Oh, H.N.; Park, D.; Seong, H.J.; Kim, D.; Sul, W.J. Antarctic tundra soil metagenome as useful natural resources of cold-active lignocelluolytic enzymes. J. Microbiol. 2019, 57, 865–873. [Google Scholar] [CrossRef]
- Ralph, J. Hydroxycinnamates in lignification. Phytochem. Rev. 2010, 9, 65–83. [Google Scholar] [CrossRef]
- Ferreira, S.S.; Simões, M.S.; Carvalho, G.G.; de Lima, L.G.A.; de Almeida Svartman, R.M.; Cesarino, I. The lignin toolbox of the model grass Setaria viridis. Plant Mol. Biol. 2019, 101, 235–255. [Google Scholar] [CrossRef]
- Cesarino, I.; Araújo, P.; Domingues, A.P.; Mazzafera, P. An overview of lignin metabolism and its effect on biomass recalcitrance. Rev. Bras. Bot. 2012, 35, 303–311. [Google Scholar] [CrossRef]
- Ragauskas, A.J.; Beckham, G.T.; Biddy, M.J.; Chandra, R.; Chen, F.; Davis, M.F.; Davison, B.H.; Dixon, R.A.; Gilna, P.; Keller, M.; et al. Lignin valorization: Improving lignin processing in the biorefinery. Science 2014, 344, 1246843. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhao, X.; Wang, A.; Huber, G.W.; Zhang, T. Catalytic Transformation of Lignin for the Production of Chemicals and Fuels. Chem. Rev. 2015, 115, 11559–11624. [Google Scholar] [CrossRef] [PubMed]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin Biosynthesis. Annu. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef]
- Lourenço, A.; Pereira, H. Compositional Variability of Lignin in Biomass. In Lignin: Trends and Applications; Books on Demand: Norderstedt, Germany, 2018. [Google Scholar]
- Molinari, H.B.C.; Pellny, T.K.; Freeman, J.; Shewry, P.R.; Mitchell, R.A.C. Grass cell wall feruloylation: Distribution of bound ferulate and candidate gene expression in Brachypodium distachyon. Front. Plant Sci. 2013, 4, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ponnusamy, V.K.; Nguyen, D.D.; Dharmaraja, J.; Shobana, S.; Banu, J.R.; Saratale, R.G.; Chang, S.W.; Kumar, G. A review on lignin structure, pretreatments, fermentation reactions and biorefinery potential. Bioresour. Technol. 2019, 271, 462–472. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Lu, Y.C.; Hu, H.Q.; Xie, F.J.; Wei, X.Y.; Fan, X. Structural characterization of lignin and its degradation products with spectroscopic methods. J. Spectrosc. 2017, 2017, 8951658. [Google Scholar] [CrossRef] [Green Version]
- Giummarella, N.; Pu, Y.; Ragauskas, A.J.; Lawoko, M. A critical review on the analysis of lignin carbohydrate bonds. Green Chem. 2019, 21, 1573–1595. [Google Scholar] [CrossRef]
- Yoo, C.G.; Meng, X.; Pu, Y.; Ragauskas, A.J. The critical role of lignin in lignocellulosic biomass conversion and recent pretreatment strategies: A comprehensive review. Bioresour. Technol. 2020, 301, 122784. [Google Scholar] [CrossRef]
- Vogel, J. Unique aspects of the grass cell wall. Curr. Opin. Plant Biol. 2008, 11, 301–307. [Google Scholar] [CrossRef]
- Amin, F.R.; Khalid, H.; Zhang, H.; Rahman, S.; Zhang, R.; Liu, G.; Chen, C. Pretreatment methods of lignocellulosic biomass for anaerobic digestion. AMB Express 2017, 7, 72. [Google Scholar] [CrossRef] [Green Version]
- Jedrzejczyk, M.; Soszka, E.; Czapnik, M.; Ruppert, A.M.; Grams, J. Physical and chemical pretreatment of lignocellulosic biomass. In Second and Third Generation of Feedstocks; Basile, A., Dalena, F., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 143–196. ISBN 9780128151624. [Google Scholar]
- Jönsson, L.J.; Martín, C. Pretreatment of lignocellulose: Formation of inhibitory by-products and strategies for minimizing their effects. Bioresour. Technol. 2015, 199, 103–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gil-Chávez, J.; Gurikov, P.; Hu, X.; Meyer, R.; Reynolds, W.; Smirnova, I. Application of novel and technical lignins in food and pharmaceutical industries: Structure-function relationship and current challenges. Biomass Convers. Biorefinery 2019. [Google Scholar] [CrossRef]
- Li, T.; Takkellapati, S. The current and emerging sources of technical lignins and their applications. Biofuels Bioprod. Biorefining 2018, 12, 756–787. [Google Scholar] [CrossRef]
- Eraghi Kazzaz, A.; Fatehi, P. Technical lignin and its potential modification routes: A mini-review. Ind. Crop. Prod. 2020, 154, 112732. [Google Scholar] [CrossRef]
- Kumar, A.K.; Sharma, S. Recent updates on different methods of pretreatment of lignocellulosic feedstocks: A review. Bioresour. Bioprocess. 2017, 4, 7. [Google Scholar] [CrossRef] [Green Version]
- Baruah, J.; Nath, B.K.; Sharma, R.; Kumar, S.; Deka, R.C.; Baruah, D.C.; Kalita, E. Recent Trends in the Pretreatment of Lignocellulosic Biomass for Value-Added Products. Front. Chem. 2018, 6, 1–19. [Google Scholar] [CrossRef]
- Li, M.; Pu, Y.; Ragauskas, A.J. Current understanding of the correlation of lignin structure with biomass recalcitrance. Front. Chem. 2016, 4, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angeler, D.G.; Fried-Petersen, H.; Allen, C.R.; Garmestani, A.; Chuang, W.; Donovan, V.M.; Eason, T.; Roberts, C.P.; Wonkka, C.L.; Survey, G.; et al. The current and emerging sources of technical lignins and their applications. Adv. Ecol. Res. 2019, 60, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Ramulu, H.G.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, 490–495. [Google Scholar] [CrossRef] [Green Version]
- Xu, R.; Zhang, K.; Liu, P.; Han, H.; Zhao, S.; Kakade, A.; Khan, A.; Du, D.; Li, X. Lignin depolymerization and utilization by bacteria. Bioresour. Technol. 2018, 269, 557–566. [Google Scholar] [CrossRef]
- Crawford, D.L.; Crawford, R.L. Microbial degradation of lignocellulose: The lignin component. Appl. Environ. Microbiol. 1976, 31, 714–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Claus, H. Laccases: Structure, reactions, distribution. Micron 2004, 35, 93–96. [Google Scholar] [CrossRef] [PubMed]
- de Gonzalo, G.; Colpa, D.I.; Habib, M.H.M.; Fraaije, M.W. Bacterial enzymes involved in lignin degradation. J. Biotechnol. 2016, 236, 110–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eugenio, M.E.; HErnández, M.; Moya, R.; Sampedro, R.M.; Villar, J.C.; Arias, E. Evaluation of a new laccase produced by Streptomyces ipo- moea on biobleaching and ageing of kraft pilps. Bioresources 2011, 6, 3231–3241. [Google Scholar]
- Givaudan, A.; Effosse, A.; Faure, D.; Potier, P.; Bouillant, M.L.; Bally, R. Polyphenol oxidase in Azospirillum lipoferum isolated from rice rhizosphere: Evidence for laccase activity in non-motile strains of Azospirillum lipoferum. FEMS Microbiol. Lett. 1993, 108, 205–210. [Google Scholar] [CrossRef]
- Rezaei, S.; Shahverdi, A.R.; Faramarzi, M.A. Isolation, one-step affinity purification, and characterization of a polyextremotolerant laccase from the halophilic bacterium Aquisalibacillus elongatus and its application in the delignification of sugar beet pulp. Bioresour. Technol. 2017, 230, 67–75. [Google Scholar] [CrossRef]
- Chen, C.; Huang, Y.; Wei, C.; Meng, M.; Liu, W.; Yang, C. Properties of the newly isolated extracellular thermo-alkali-stable laccase from thermophilic actinomycetes, Thermobifida fusca and its application in dye intermediates oxidation. AMB Express 2013, 3, 49. [Google Scholar] [CrossRef] [Green Version]
- de Oliveira, P.L.; Duarte, M.C.T.; Ponezi, A.N.; Durrant, L.R. Purification and partial characterization of manganese peroxidase from Bacillus pumilus and Paenibacillus sp. Braz. J. Microbiol. 2009, 40, 818–826. [Google Scholar] [CrossRef] [Green Version]
- Anderson, C.R.; Johnson, H.A.; Caputo, N.; Davis, R.E.; Torpey, J.W.; Tebo, B.M. Mn(II) oxidation is catalyzed by heme peroxidases in “Aurantimonas manganoxydans” strain SI85-9A1 and Erythrobacter sp. strain SD-21. Appl. Environ. Microbiol. 2009, 75, 4130–4138. [Google Scholar] [CrossRef] [Green Version]
- Nayanashree, G.; Thippeswamy, B. Biodegradation of Natural Rubber by Laccase and Manganese Peroxidase Enzyme of Bacillus subtilis. Environ. Process. 2015, 2, 761–772. [Google Scholar] [CrossRef] [Green Version]
- Kharayat, Y.; Thakur, I.S. Isolation of bacterial strain from sediment core of Pulp and Paper Mill industries for production and purification of lignin peroxidase (LiP) enzyme. Bioremediat. J. 2012, 16, 125–130. [Google Scholar] [CrossRef]
- Gomare, S.S.; Jadhav, J.P.; Govindwar, S.P. Degradation of sulfonated azo dyes by the purified lignin peroxidase from Brevibacillus laterosporus MTCC 2298. Biotechnol. Bioprocess Eng. 2008, 13, 136–143. [Google Scholar] [CrossRef]
- Yee, D.C.; Wood, T.K. 2,4-Dichlorophenol Degradation Using Streptomyces visidosporus T7A Lignin Peroxidase. Biotechnol. Prog. 1997, 13, 53–59. [Google Scholar] [CrossRef]
- Jeon, J.H.; Han, Y.J.; Kang, T.G.; Kim, E.S.; Hong, S.K.; Jeong, B.C. Purification and characterization of 2,4-dichlorophenol oxidizing peroxidase from Streptomyces sp. AD001. J. Microbiol. Biotechnol. 2002, 12, 972–978. [Google Scholar]
- Sonoki, T.; Iimura, Y.; Masai, E.; Kajita, S.; Katayama, Y. Specific degradation of β-aryl ether linkage in synthetic lignin (dehydrogenative polymerizate) by bacterial enzymes of Sphingomonas paucimobilis SYK-6 produced in recombinant Escherichia coli. J. Wood Sci. 2002, 48, 429–433. [Google Scholar] [CrossRef]
- Pereira, J.H.; Heins, R.A.; Gall, D.L.; McAndrew, R.P.; Deng, K.; Holland, K.C.; Donohue, T.J.; Noguera, D.R.; Simmons, B.A.; Sale, K.L.; et al. Structural and biochemical characterization of the early and late enzymes in the lignin β-aryl ether cleavage pathway from sphingobium sp. SYK-6. J. Biol. Chem. 2016, 291, 10228–10238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmeisser, C.; Steele, H.; Streit, W.R. Metagenomics, biotechnology with non-culturable microbes. Appl. Microbiol. Biotechnol. 2007, 75, 955–962. [Google Scholar] [CrossRef] [PubMed]
- Batista-García, R.A.; del Rayo Sánchez-Carbente, M.; Talia, P.; Jackson, S.A.; O’Leary, N.D.; Dobson, A.D.W.; Folch-Mallol, J.L. From lignocellulosic metagenomes to lignocellulolytic genes: Trends, challenges and future prospects. Biofuels, Bioprod. Biorefining 2016, 10, 864–882. [Google Scholar] [CrossRef]
- Quince, C.; Walker, A.W.; Simpson, J.T.; Loman, N.J.; Segata, N. Supplement-study design Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 2017, 35, 833–844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breitwieser, F.P.; Lu, J.; Salzberg, S.L. A review of methods and databases for metagenomic classification and assembly. Brief. Bioinform. 2018, 20, 1125–1139. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Lu, S.; Yang, J.; Jin, D.; Pu, J.; Díaz Moyá, S.; Xiong, Y.; Rossello-Mora, R.; Xu, J. Precise Fecal Microbiome of the Herbivorous Tibetan Antelope Inhabiting High-Altitude Alpine Plateau. Front. Microbiol. 2018, 9, 2321. [Google Scholar] [CrossRef] [Green Version]
- Simon, C.; Daniel, R. Metagenomic analyses: Past and future trends. Appl. Environ. Microbiol. 2011, 77, 1153–1161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, K.N.; Cheng, J.; Engel, K.; Neufeld, J.D.; Charles, T.C. Current and future resources for functional metagenomics. Front. Microbiol. 2015, 6, 1196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ngara, T.R.; Zhang, H. Recent Advances in Function-based Metagenomic Screening. Genom. Proteom. Bioinform. 2018, 16, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Lacerda Júnior, G.V.; Noronha, M.F.; de Sousa, S.T.P.; Cabral, L.; Domingos, D.F.; Sáber, M.L.; de Melo, I.S.; Oliveira, V.M. Potential of semiarid soil from Caatinga biome as a novel source for mining lignocellulose-degrading enzymes. FEMS Microbiol. Ecol. 2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Žifčáková, L.; Větrovský, T.; Lombard, V.; Henrissat, B.; Howe, A.; Baldrian, P. Feed in summer, rest in winter: Microbial carbon utilization in forest topsoil. Microbiome 2017, 5, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilhelm, R.C.; Singh, R.; Eltis, L.D.; Mohn, W.W. Bacterial contributions to delignification and lignocellulose degradation in forest soils with metagenomic and quantitative stable isotope probing. ISME J. 2019, 13, 413–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salam, L.B. Detection of carbohydrate-active enzymes and genes in a spent engine oil-perturbed agricultural soil. Bull. Natl. Res. Cent. 2018, 42, 10. [Google Scholar] [CrossRef]
- Grieco, M.B.; Lopes, F.A.C.; Oliveira, L.S.; Tschoeke, D.A.; Popov, C.C.; De Souza, W.; Thompson, F.L. Metagenomic Analysis of the Whole Gut Microbiota in Brazilian Termitidae Termites Cornitermes cumulans, Cyrilliotermes strictinasus. Curr. Microbiol. 2019, 76, 687–697. [Google Scholar] [CrossRef]
- Joynson, R.; Pritchard, L.; Osemwekha, E.; Ferry, N. Metagenomic analysis of the gut microbiome of the common black slug arion ater in search of novel lignocellulose degrading enzymes. Front. Microbiol. 2017, 8, 2181. [Google Scholar] [CrossRef] [Green Version]
- Agamennone, V.; Le, N.G.; van Straalen, N.M.; Brouwer, A.; Roelofs, D. Antimicrobial activity and carbohydrate metabolism in the bacterial metagenome of the soil-living invertebrate Folsomia candida. Sci. Rep. 2019, 9, 7308. [Google Scholar] [CrossRef] [PubMed]
- Jose, V.L.; Appoothy, T.; More, R.P.; Arun, A.S. Metagenomic insights into the rumen microbial fibrolytic enzymes in Indian crossbred cattle fed finger millet straw. AMB Express 2017, 7, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gharechahi, J.; Salekdeh, G.H. A metagenomic analysis of the camel rumen’s microbiome identifies the major microbes responsible for lignocellulose degradation and fermentation. Biotechnol. Biofuels 2018, 11, 216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Xu, B.; Lu, T.; Huang, Z. Metagenomic Analysis of the Fecal Microbiomes of Wild Asian Elephants Reveals Microflora and Enzymes that Mainly Digest Hemicellulose. J. Microbiol. Biotechnol. 2019, 29, 1255–1265. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Li, Y.; Chen, Y.; Fu, C.; Long, W.; Xiao, X.; Liao, H.; Yang, Y. Bamboo lignocellulose degradation by gut symbiotic microbiota of the bamboo snout beetle Cyrtotrachelus buqueti. Biotechnol. Biofuels 2019, 12, 70. [Google Scholar] [CrossRef] [PubMed]
- Tokuda, G. Plant Cell Wall Degradation in Insects: Recent Progress on Endogenous Enzymes Revealed by Multi-Omics Technologies, 1st ed.; Elsevier Ltd.: Amsterdam, The Netherlands, 2019; Volume 57, ISBN 9780128186602. [Google Scholar]
- Do, T.H.; Nguyen, T.T.; Nguyen, T.N.; Le, Q.G.; Nguyen, C.; Kimura, K.; Truong, N.H. Mining biomass-degrading genes through Illumina-based de novo sequencing and metagenomic analysis of free-living bacteria in the gut of the lower termite Coptotermes gestroi harvested in Vietnam. J. Biosci. Bioeng. 2014, 118, 665–671. [Google Scholar] [CrossRef]
- Brune, A. Symbiotic digestion of lignocellulose in termite guts. Nat. Rev. Microbiol. 2014, 12, 168–180. [Google Scholar] [CrossRef]
- Zhou, J.; Duan, J.; Gao, M.; Wang, Y.; Wang, X.; Zhao, K. Diversity, Roles, and Biotechnological Applications of Symbiotic Microorganisms in the Gut of Termite. Curr. Microbiol. 2018, 76, 755–761. [Google Scholar] [CrossRef]
- Suman, S.K.; Dhawaria, M.; Tripathi, D.; Raturi, V.; Adhikari, D.K.; Kanaujia, P.K. Investigation of lignin biodegradation by Trabulsiella sp. isolated from termite gut. Int. Biodeterior. Biodegrad. 2016, 112, 12–17. [Google Scholar] [CrossRef]
- Chung, S.Y.; Maeda, M.; Song, E.; Horikoshi, K.; Kudo, T. A Gram-positive Polychlorinated Biphenyl-degrading Bacterium, Rhodococcus erythropolis Strain TA421, Isolated from a Termite Ecosystem. Biosci. Biotechnol. Biochem. 1994, 58, 2111–2113. [Google Scholar] [CrossRef]
- Le Roes-Hill, M.; Rohland, J.; Burton, S. Actinobacteria isolated from termite guts as a source of novel oxidative enzymes. Antonie Van Leeuwenhoek 2011, 100, 589–605. [Google Scholar] [CrossRef] [PubMed]
- Terrapon, N.; Lombard, V.; Drula, E.; Coutinho, P.M.; Henrissat, B. The CAZy Database/the Carbohydrate-Active Enzyme (CAZy) Database: Principles and Usage Guidelines. In A Practical Guide to Using Glycomics Databases; Springer: Tokyo, Japan, 2017; pp. 117–131. [Google Scholar]
- Su, L.; Yang, L.; Huang, S.; Su, X.; Li, Y.; Wang, F.; Wang, E.; Kang, N.; Xu, J.; Song, A. Comparative Gut Microbiomes of Four Species Representing the Higher and the Lower Termites. J. Insect Sci. 2016, 16, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Brune, A.; Ohkuma, M. Role of the termite gut microbiota in symbiotic digestion. In Biology of Termites: A Modern Synthesis; Springer: Dordrecht, The Netherlands, 2011; pp. 439–475. ISBN 9789048139767. [Google Scholar]
- Bateman, A.; Coin, L.; Durbin, R.; Finn, R.D.; Hollich, V.; Griffiths-Jones, S.; Khanna, A.; Marshall, M.; Moxon, S.; Sonnhammer, E.L.L.; et al. The Pfam protein families database. Nucleic Acids Res. 2004, 32, D138–D141. [Google Scholar] [CrossRef] [PubMed]
- Nagaraja, T.G. Microbiology of the Rumen. In Rumenology; Springer International Publishing: Cham, Switzerland, 2016; pp. 39–61. ISBN 9783319305332. [Google Scholar]
- De Nardi, R.; Marchesini, G.; Li, S.; Khafipour, E.; Plaizier, K.J.C.; Gianesella, M. Metagenomic analysis of rumen microbial population in dairy heifers fed a high grain diet supplemented with dicarboxylic acids or polyphenols. BMC Vet. Res. 2016, 12, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Hatem, A.; Catalyurek, U.V.; Morrison, M.; Yu, Z. Metagenomic insights into the carbohydrate-active enzymes carried by the microorganisms adhering to solid digesta in the rumen of cows. PLoS ONE 2013, 8, e78507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kajikawa, H.; Kudo, H.; Kondo, T.; Jodai, K.; Honda, Y.; Kuwahara, M.; Watanabe, T. Degradation of benzyl ether bonds of lignin by ruminal microbes. FEMS Microbiol. Lett. 2000, 187, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Beloqui, A.; Pita, M.; Polaina, J.; Martínez-Arias, A.; Golyshina, O.V.; Zumárraga, M.; Yakimov, M.M.; García-Arellano, H.; Alcalde, M.; Fernández, V.M.; et al. Novel polyphenol oxidase mined from a metagenome expression library of bovine rumen: Biochemical properties, structural analysis, and phylogenetic relationships. J. Biol. Chem. 2006, 281, 22933–22942. [Google Scholar] [CrossRef] [Green Version]
- Ufarté, L.; Potocki-Veronese, G.; Cecchini, D.; Tauzin, A.S.; Rizzo, A.; Morgavi, D.P.; Cathala, B.; Moreau, C.; Cleret, M.; Robe, P.; et al. Highly promiscuous oxidases discovered in the bovine rumen microbiome. Front. Microbiol. 2018, 9, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Carlos, C.; Fan, H.; Currie, C.R. Substrate shift reveals roles for members of bacterial consortia in degradation of plant cell wall polymers. Front. Microbiol. 2018, 9, 364. [Google Scholar] [CrossRef] [Green Version]
- Moraes, E.C.; Alvarez, T.M.; Persinoti, G.F.; Tomazetto, G.; Brenelli, L.B.; Paixão, D.A.A.; Ematsu, G.C.; Aricetti, J.A.; Caldana, C.; Dixon, N.; et al. Lignolytic-consortium omics analyses reveal novel genomes and pathways involved in lignin modification and valorization. Biotechnol. Biofuels 2018, 11, 75. [Google Scholar] [CrossRef]
- Jiménez, D.J.; de Lima Brossi, M.J.; Schückel, J.; Kračun, S.K.; Willats, W.G.T.; van Elsas, J.D. Characterization of three plant biomass-degrading microbial consortia by metagenomics- and metasecretomics-based approaches. Appl. Microbiol. Biotechnol. 2016, 100, 10463–10477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, N.; Yang, J.; Ji, L.; Liu, J.; Yang, Y.; Yuan, H. Metagenomic and metaproteomic analyses of a corn stover-adapted microbial consortium EMSD5 reveal its taxonomic and enzymatic basis for degrading lignocellulose. Biotechnol. Biofuels 2016, 9, 243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Dong, D.; Wang, H.; Müller, K.; Qin, Y.; Wang, H.; Wu, W. Metagenomic analysis of microbial consortia enriched from compost: New insights into the role of Actinobacteria in lignocellulose decomposition. Biotechnol. Biofuels 2016, 9, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’haeseleer, P.; Gladden, J.M.; Allgaier, M.; Chain, P.S.G.; Tringe, S.G.; Malfatti, S.A.; Aldrich, J.T.; Nicora, C.D.; Robinson, E.W.; Paša-Tolić, L.; et al. Proteogenomic Analysis of a Thermophilic Bacterial Consortium Adapted to Deconstruct Switchgrass. PLoS ONE 2013, 8, e68465. [Google Scholar] [CrossRef] [PubMed]
- Lemos, L.N.; Pereira, R.V.; Quaggio, R.B.; Martins, L.F.; Moura, L.M.S.; da Silva, A.R.; Antunes, L.P.; da Silva, A.M.; Setubal, J.C. Genome-centric analysis of a thermophilic and cellulolytic bacterial consortium derived from composting. Front. Microbiol. 2017, 8, 644. [Google Scholar] [CrossRef] [PubMed]
- Aguiar-Pulido, V.; Huang, W.; Suarez-Ulloa, V.; Cickovski, T.; Mathee, K.; Narasimhan, G. Metagenomics, metatranscriptomics, and metabolomics approaches for microbiome analysis. Evol. Bioinforma. 2016, 12, 5–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassa, J.; Maus, I.; Off, S.; Pühler, A.; Scherer, P.; Klocke, M.; Schlüter, A. Metagenome, metatranscriptome, and metaproteome approaches unraveled compositions and functional relationships of microbial communities residing in biogas plants. Appl. Microbiol. Biotechnol. 2018, 102, 5045–5063. [Google Scholar] [CrossRef] [Green Version]
- Antunes, L.P.; Martins, L.F.; Pereira, R.V.; Thomas, A.M.; Barbosa, D.; Lemos, L.N.; Silva, G.M.M.; Moura, L.M.S.; Epamino, G.W.C.; Digiampietri, L.A.; et al. Microbial community structure and dynamics in thermophilic composting viewed through metagenomics and metatranscriptomics. Sci. Rep. 2016, 6, 38915. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, D.J.; De Mares, M.C.; Salles, J.F. Temporal expression dynamics of plant biomass-degrading enzymes by a synthetic bacterial consortium growing on sugarcane bagasse. Front. Microbiol. 2018, 9, 299. [Google Scholar] [CrossRef]
- Tokuda, G.; Mikaelyan, A.; Fukui, C.; Matsuura, Y.; Watanabe, H.; Fujishima, M. Fiber-associated spirochetes are major agents of hemicellulose degradation in the hindgut of wood-feeding higher termites. Proc. Natl. Acad. Sci. USA 2018, 115, E11996–E12004. [Google Scholar] [CrossRef] [Green Version]
- Marynowska, M.; Goux, X.; Sillam-Dussès, D.; Rouland-Lefèvre, C.; Roisin, Y.; Delfosse, P.; Calusinska, M. Optimization of a metatranscriptomic approach to study the lignocellulolytic potential of the higher termite gut microbiome. BMC Genom. 2017, 18, 681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, R.; Bennett, J.P.; Gupta, M.; Sharma, M.; Eqbal, D.; Alessi, A.M.; Dowle, A.A.; McQueen-Mason, S.J.; Bruce, N.C.; Yazdani, S.S. Mining the biomass deconstructing capabilities of rice yellow stem borer symbionts. Biotechnol. Biofuels 2019, 12, 265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleiner, M. Metaproteomics: Much More than Measuring Gene Expression in Microbial Communities. mSystems 2019, 4, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Zhou, Y.; Xiao, X.; Zheng, J.; Zhou, H. Metaproteomics: A strategy to study the taxonomy and functionality of the gut microbiota. J. Proteom. 2020, 219, 103737. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Guo, W.; Xu, B.; Teng, Z.; Tao, D.; Lou, Y.; Gao, Y. Screening and identification of lignin-degrading bacteria in termite gut and the construction of LiP-expressing recombinant Lactococcus lactis. Microb. Pathog. 2017, 112, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Han, X.; Qian, Y.; Liu, G.; Yao, G.; Zhong, Y.; Qu, Y.; Han, X.; Qu, Y.; Zhong, Y.; et al. Proteomic analysis of the biomass hydrolytic potentials of Penicillium oxalicum lignocellulolytic enzyme system. Biotechnol. Biofuels 2016, 9, 68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gómez-Mendoza, D.P.; Junqueira, M.; Do Vale, L.H.F.; Domont, G.B.; Ferreira Filho, E.X.; De Sousa, M.V.; Ricart, C.A.O. Secretomic survey of trichoderma harzianum grown on plant biomass substrates. J. Proteome Res. 2014, 13, 1810–1822. [Google Scholar] [CrossRef] [PubMed]
- Rocha-Martín, J.; Martínez-Bernal, C.; Zamorano, L.S.; Reyes-Sosa, F.M.; Díez, G.B. Inhibition of enzymatic hydrolysis of pretreated corn stover and sugar cane straw by laccases. Process Biochem. 2018, 67, 88–91. [Google Scholar] [CrossRef]
- Arias, M.E.; Rodrı, J.; Soliveri, J.; Ball, A.S.; Herna, M. Kraft Pulp Biobleaching and Mediated Oxidation of a Nonphenolic Substrate by Laccase from Streptomyces cyaneus CECT 3335. Appl. Environ. Microbiol. 2003, 69, 1953–1958. [Google Scholar] [CrossRef] [Green Version]






| DNA Source | Bacteria Associated with Lignin Degradation | AAs Families | References |
|---|---|---|---|
| Brazilian Caatinga soil | Proteobacteria, Actinobacteria, Acidobacteria | AA1, AA3, AA7 | [68] |
| Forest soil | Proteobacteria, Acidobacteria, and Actinobacteria | AA3 | [69] |
| Soil | Caulobacteraceae, Acidobacteria, Solirubrobacterales, Elusimicrobia, Nevskiales, and Cystobacteraceae | AA1, AA3, AA4, AA5, AA6, and AA7 | [70] |
| Antarctic soil | Geodermatophilus, Thermobispora, and Amycolatopsis | AA3, AA4, and AA7 | [18] |
| Agricultural soil | Proteobacteria | AA3, and AA6 | [71] |
| Termite gut microbiome | Legionella, Acinetobacter, and Pseudomonas, Myxococcus, Streptomyces, and Actinoplanes | AA1, AA3, AA4, AA5, and AA6 | [72] |
| Arion gut microbiome | n.s. | AA2, AA3, AA4, and AA6 | [73] |
| Folsomia gut microbiome | Proteobacteria and Actinobacteria | AA3, AA6, and AA7 | [74] |
| Bovine rumen | Prevotella, Bacteroides, Clostridium, Fibrobacter, and Ruminococcus | AA6, AA5, AA4, AA7, and AA3 | [75] |
| Camel rumen | Firmicutes, Bacteroidetes, Spirochaetaes, Fibrobacteres, and Proteobacteria | AA3, AA4, AA6, and AA7 | [76] |
| Elephant feces | n.s. | AA4 and AA6 | [77] |
| Consortium Source and Bacteria with Ligninolytic Potential | Substrate | AA Families | Pathways of Consuming Aromatic Compounds | References |
|---|---|---|---|---|
| Enrichment from chicken feces and soil (Pseudomonas, Klebsiella, Variovorax, Leclercia, and Enterobacter) | Alkali lignin | - | Catechol ortho- cleavage and benzoate degradation pathways | [96] |
| Enrichment from sugarcane plantation soil (Proteobacteria, Actinobacteria, Firmicutes, Alcaligenaceae, and Micrococcaceae) | Low-molecular- weight (LW) lignin | AA2, AA3, AA4, AA6, and AA7 | Benzoate degradation to catechol, catechol ortho-cleavage, catechol meta- cleavage, and phthalate degradation | [97] |
| Enrichment from soil (Brevundimonas, Caulobacter, Pseudomonas, Citrobacter, and Aeromonas) | Wheat straw switchgrass and corn stover | AA2, AA4, AA6, and AA7 | - | [98] |
| Enrichment from compost ecosystems (Proteobacteria and Firmicutes) | Corn stover | AA2, AA3, AA4, AA6 and AA7 | - | [99] |
| Enrichment from compost ecosystems (Symbiobacterium thermophilum, T. curvata, Mycobacterium xenopi, Amycolicicoccus subflavus and Mycobacterium thermoresistibile) | Rice straw | AA2 | - | [100] |
| Enrichment from compost ecosystems (Sphaerobacter, Hyphomicrobium, Thermus thermophilus, Sphaerobacter, Gemmatimonadetes, Paenibacillus) | Switchgrass | AA2 | Ortho-cleavage of protocatechuate and 4- hydroxyphenylacetate degradation | [101] |
| Enriched from compost ecosystems (Thermobacillus species, Bacillus) | CMC | AA1, AA2, AA4, AA6, and AA7 | - | [102] |
| Sample | Omics Techniques Applied | AAs Families | References |
|---|---|---|---|
| Compost ecosystems | Metatranscriptomics | AA2, AA3, AA4, AA6, and AA7 | [105] |
| Soil microbiota | Metatranscriptomics | AA2 and AA6 | [106] |
| Forest soil | Metatranscriptomics | AA1 and AA3 | [69] |
| Termite gut | Metatranscriptomics | AA2, AA4, and AA6 | [107] |
| Termite gut | Metatranscriptomics | AA4 and AA6 | [108] |
| Bacterial consortium | Metaproteomics | AA2 and AA7 | [109] |
| Bacterial consortium | Metaproteomics | AA2 | [99] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, J.P.; Ticona, A.R.P.; Hamann, P.R.V.; Quirino, B.F.; Noronha, E.F. Deconstruction of Lignin: From Enzymes to Microorganisms. Molecules 2021, 26, 2299. https://doi.org/10.3390/molecules26082299
Silva JP, Ticona ARP, Hamann PRV, Quirino BF, Noronha EF. Deconstruction of Lignin: From Enzymes to Microorganisms. Molecules. 2021; 26(8):2299. https://doi.org/10.3390/molecules26082299
Chicago/Turabian StyleSilva, Jéssica P., Alonso R. P. Ticona, Pedro R. V. Hamann, Betania F. Quirino, and Eliane F. Noronha. 2021. "Deconstruction of Lignin: From Enzymes to Microorganisms" Molecules 26, no. 8: 2299. https://doi.org/10.3390/molecules26082299