Heme Binding to HupZ with a C-Terminal Tag from Group A Streptococcus
Abstract
:1. Introduction
2. Results
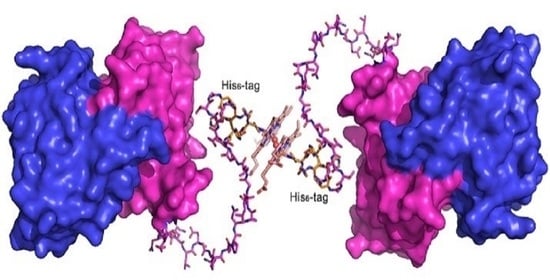
2.1. The C-Terminally Tagged HupZ Presents an Unusual Heme-Protein Interaction Mode
2.2. Probing the Oxidation State of the HupZ-Heme Complex
2.3. Resonance Raman Spectroscopy Suggests a Six-Coordinate Low-Spin Heme with Histidine Axial Ligand(s)
2.4. The Role of Dioxygen in HupZ-Heme Interactions
2.5. Side-Directed Mutagenesis Analysis for Identification of the Axial Ligand
2.6. Heme-Induced Higher-Order Oligomerization in Protein Quaternary Structure
2.7. The Heme-Degradation Activity of HupZ and H111A
3. Discussion
4. Materials and Methods
4.1. Cloning, Expression, Purification of HupZ and H111A Variant
4.2. Preparation of HupZ-Heme Complex
4.3. Anaerobic Sample Preparation
4.4. UV-Visible Absorbance Spectroscopy
4.5. EPR Spectroscopy
4.6. Resonance Raman Spectroscopy
4.7. Hemin Titration
4.8. Size-Exclusion Chromatography
4.9. Crystallization, X-Ray Diffraction Data Collection, and Refinement
4.10. Heme Degradation Activity Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Cunningham, M.W. Pathogenesis of group A streptococcal infections and their sequelae. Adv. Exp. Med. Biol. 2008, 609, 29–42. [Google Scholar] [PubMed]
- The Centers for Disease Control and Prevention, Antibiotic Resistance Threats in the United States. 2019. Available online: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf (accessed on 19 January 2021).
- Hammer, N.D.; Skaar, E.P. Molecular mechanisms of Staphylococcus aureus iron acquisition. Annu. Rev. Microbiol. 2011, 65, 129–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrade, M.A.; Ciccarelli, F.D.; Perez-Iratxeta, C.; Bork, P. NEAT: A domain duplicated in genes near the components of a putative Fe3+ siderophore transporter from Gram-positive pathogenic bacteria. Genome Biol. 2002, 3, 1–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grigg, J.C.; Vermeiren, C.L.; Heinrichs, D.E.; Murphy, M.E. Haem recognition by a Staphylococcus aureus NEAT domain. Mol. Microbiol. 2007, 63, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Mazmanian, S.K.; Skaar, E.P.; Gaspar, A.H.; Humayun, M.; Gornicki, P.; Jelenska, J.; Joachmiak, A.; Missiakas, D.M.; Schneewind, O. Passage of heme-iron across the envelope of Staphylococcus aureus. Science 2003, 299, 906–909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bates, C.S.; Montanez, G.E.; Woods, C.R.; Vincent, R.M.; Eichenbaum, Z. Identification and characterization of a Streptococcus pyogenes operon involved in binding of hemoproteins and acquisition of iron. Infect. Immun. 2003, 71, 1042–1055. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Liu, M.; Lei, B. The surface protein Shr of Streptococcus pyogenes binds heme and transfers it to the streptococcal heme-binding protein Shp. BMC Microbiol. 2008, 8, 15. [Google Scholar] [CrossRef] [Green Version]
- Nygaard, T.K.; Blouin, G.C.; Liu, M.; Fukumura, M.; Olson, J.S.; Fabian, M.; Dooley, D.M.; Lei, B. The mechanism of direct heme transfer from the streptococcal cell surface protein Shp to HtsA of the HtsABC transporter. J. Biol. Chem. 2006, 281, 20761–20771. [Google Scholar] [CrossRef] [Green Version]
- Lei, B.; Liu, M.; Voyich, J.M.; Prater, C.I.; Kala, S.V.; DeLeo, F.R.; Musser, J.M. Identification and characterization of HtsA, a second heme-binding protein made by Streptococcus pyogenes. Infect. Immun. 2003, 71, 5962–5969. [Google Scholar] [CrossRef] [Green Version]
- Tenhunen, R.; Marver, H.S.; Schmid, R. The enzymatic conversion of heme to bilirubin by microsomal heme oxygenase. Proc. Natl. Acad. Sci. USA 1968, 61, 748–755. [Google Scholar] [CrossRef] [Green Version]
- Schmitt, M.P. Transcription of the Corynebacterium diphtheriae hmuO gene is regulated by iron and heme. Infect. Immun. 1997, 65, 4634–4641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ratliff, M.; Zhu, W.; Deshmukh, R.; Wilks, A.; Stojiljkovic, I. Homologues of neisserial heme oxygenase in gram-negative bacteria: Degradation of heme by the product of the pigA gene of Pseudomonas aeruginosa. J. Bacteriol. 2001, 183, 6394–6403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, W.; Hunt, D.J.; Richardson, A.R.; Stojiljkovic, I. Use of heme compounds as iron sources by pathogenic neisseriae requires the product of the hemO gene. J. Bacteriol. 2000, 182, 439–447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skaar, E.P.; Gaspar, A.H.; Schneewind, O. IsdG and IsdI, heme-degrading enzymes in the cytoplasm of Staphylococcus aureus. J. Biol. Chem. 2004, 279, 436–443. [Google Scholar] [CrossRef] [Green Version]
- Chim, N.; Iniguez, A.; Nguyen, T.Q.; Goulding, C.W. Unusual diheme conformation of the heme-degrading protein from Mycobacterium tuberculosis. J. Mol. Biol. 2010, 395, 595–608. [Google Scholar] [CrossRef] [Green Version]
- Lyles, K.V.; Eichenbaum, Z. From host heme to iron: The expanding spectrum of heme degrading enzymes used by pathogenic bacteria. Front. Cell. Infect. Microbiol. 2018, 8, 198. [Google Scholar] [CrossRef]
- Eichenbaum, Z.; Muller, E.; Morse, S.A.; Scott, J.R. Acquisition of iron from host proteins by the group A streptococcus. Infect. Immun. 1996, 64, 5428–5429. [Google Scholar] [CrossRef] [Green Version]
- Aranda, R., 4th; Worley, C.E.; Liu, M.; Bitto, E.; Cates, M.S.; Olson, J.S.; Lei, B.; Phillips, G.N., Jr. Bis-methionyl coordination in the crystal structure of the heme-binding domain of the streptococcal cell surface protein Shp. J. Mol. Biol. 2007, 374, 374–383. [Google Scholar] [CrossRef] [Green Version]
- Ouattara, M.; Cunha, E.B.; Li, X.; Huang, Y.S.; Dixon, D.; Eichenbaum, Z. Shr of group A streptococcus is a new type of composite NEAT protein involved in sequestering haem from methaemoglobin. Mol. Microbiol. 2010, 78, 739–756. [Google Scholar] [CrossRef] [Green Version]
- Sook, B.R.; Block, D.R.; Sumithran, S.; Montanez, G.E.; Rodgers, K.R.; Dawson, J.H.; Eichenbaum, Z.; Dixon, D.W. Characterization of SiaA, a streptococcal heme-binding protein associated with a heme ABC transport system. Biochemistry 2008, 47, 2678–2688. [Google Scholar] [CrossRef]
- Toukoki, C.; Gold, K.M.; McIver, K.S.; Eichenbaum, Z. MtsR is a dual regulator that controls virulence genes and metabolic functions in addition to metal homeostasis in the group A streptococcus. Mol. Microbiol. 2010, 76, 971–989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sachla, A.J.; Ouattara, M.; Romero, E.; Agniswamy, J.; Weber, I.T.; Gadda, G.; Eichenbaum, Z. In vitro heme biotransformation by the HupZ enzyme from Group A streptococcus. Biometals 2016, 29, 593–609. [Google Scholar] [CrossRef] [PubMed]
- Peisach, J.; Blumberg, W.E.; Ogawa, S.; Rachmilewitz, E.A.; Oltzik, R. The effects of protein conformation on the heme symmetry in high spin ferric heme proteins as studied by electron paramagnetic resonance. J. Biol. Chem. 1971, 246, 3342–3355. [Google Scholar] [CrossRef]
- Fu, R.; Gupta, R.; Geng, J.F.; Dornevil, K.; Wang, S.M.; Zhang, Y.; Hendrich, M.P.; Liu, A. The reactivation mechanism of tryptophan 2,3-dioxygenase by hydrogen peroxide. J. Biol. Chem. 2011, 286, 26541–26554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spiro, T.G.; Soldatova, A.V.; Balakrishnan, G. CO, NO, and O2 as vibrational probes of heme protein interactions. Coord. Chem. Rev. 2013, 257, 511–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engh, R.A.; Huber, R. Accurate bond and angle parameters for X-ray protein-structure refinement. Acta Crystallogr. A 1991, 47, 392–400. [Google Scholar] [CrossRef]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 2010, 66, 12–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Naik, S.G.; Krzystek, J.; Shin, S.; Nelson, W.H.; Xue, S.; Yang, J.J.; Davidson, V.L.; Liu, A. Role of calcium in metalloenzymes: Effects of calcium removal on the axial ligation geometry and magnetic properties of the catalytic diheme center in MauG. Biochemistry 2012, 51, 1586–1597. [Google Scholar] [CrossRef] [Green Version]
- Zoppellaro, G.; Bren, K.L.; Ensign, A.A.; Harbitz, E.; Kaur, R.; Hersleth, H.-P.; Ryde, U.; Hederstedt, L.; Andersson, K.K. Review: Studies of ferric heme proteins with highly anisotropic/highly axial low spin (S = 1/2) electron paramagnetic resonance signals with bis-histidine and histidine-methionine axial iron coordination. Biopolymers 2009, 91, 1064–1082. [Google Scholar] [CrossRef] [Green Version]
- Blumberg, W.E.; Peisach, J. Bioinorganic chemistry. Adv. Chem. Ser. 1971, 100, 271–291. [Google Scholar]
- Peisach, J.; Blumberg, W.E.; Adler, A. Electron paramagnetic resonance studies of iron porphin and chlorin systems. Ann. N. Y. Acad. Sci. 1973, 206, 310–327. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Tanaka, Y.; Suenaga, A.; Kuroda, M.; Yao, M.; Watanabe, N.; Arisaka, F.; Ohta, T.; Tanaka, I.; Tsumoto, K. Structural basis for multimeric heme complexation through a specific protein-heme interaction: The case of the third neat domain of IsdH from Staphylococcus aureus. J. Biol. Chem. 2008, 283, 28649–28659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pokkuluri, P.R.; Londer, Y.Y.; Duke, N.E.C.; Pessanha, M.; Yang, X.; Orshonsky, V.; Orshonsky, L.; Erickson, J.; Zagyanskiy, Y.; Salgueiro, C.A.; et al. Structure of a novel dodecaheme cytochrome c from Geobacter sulfurreducens reveals an extended 12 nm protein with interacting hemes. J. Struct. Biol. 2011, 174, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Einsle, O.; Stach, P.; Messerschmidt, A.; Simon, J.; Kröger, A.; Huber, R.; Kroneck, P.M. Cytochrome c nitrite reductase from Wolinella succinogenes. Structure at 1.6 Å resolution, inhibitor binding, and heme-packing motifs. J. Biol. Chem. 2000, 275, 39608–39616. [Google Scholar] [CrossRef] [Green Version]
- Ferrand, L.; Soorkia, S.; Grégoire, G.; Broquier, M.; Soep, B.; Shafizadeh, N. Bonding of heme FeIII with dioxygen: Observation and characterization of an incipient bond. Phys. Chem. Chem. Phys. 2015, 17, 25693–25699. [Google Scholar] [CrossRef]
- Paul, G.C.; Banerjee, S.; Mukherjee, C. Dioxygen reactivity of an iron complex of 2-aminophenol-appended ligand: Crystallographic evidence of the aromatic ring cleavage product of the 2-aminophenol Unit. Inorg. Chem. 2017, 56, 729–736. [Google Scholar] [CrossRef]
- Boffi, A.; Das, T.K.; della Longa, S.; Spagnuolo, C.; Rousseau, D.L. Pentacoordinate hemin derivatives in sodium dodecyl sulfate micelles: Model systems for the assignment of the fifth ligand in ferric heme proteins. Biophys. J. 1999, 77, 1143–1149. [Google Scholar] [CrossRef] [Green Version]
- Lansky, I.B.; Lukat-Rodgers, G.S.; Block, D.; Rodgers, K.R.; Ratliff, M.; Wilks, A. The cytoplasmic heme-binding protein (PhuS) from the heme uptake system of Pseudomonas aeruginosa is an intracellular heme-trafficking protein to the delta-regioselective heme oxygenase. J. Biol. Chem. 2006, 281, 13652–13662. [Google Scholar] [CrossRef] [Green Version]
- O’Neill, M.J.; Bhakta, M.N.; Fleming, K.G.; Wilks, A. Induced fit on heme binding to the Pseudomonas aeruginosa cytoplasmic protein (PhuS) drives interaction with heme oxygenase (HemO). Proc. Natl. Acad. Sci. USA 2012, 109, 5639–5644. [Google Scholar] [CrossRef] [Green Version]
- Mathew, L.G.; Beattie, N.R.; Pritchett, C.; Lanzilotta, W.N. New insight into the mechanism of anaerobic heme degradation. Biochemistry 2019, 58, 4641–4654. [Google Scholar] [CrossRef]
- Edgar, R.J.; Chen, J.; Kant, S.; Rechkina, E.; Rush, J.S.; Forsberg, L.S.; Jaehrig, B.; Azadi, P.; Tchesnokova, V.; Sokurenko, E.V.; et al. SpyB, a small heme-binding protein, affects the composition of the cell wall in Streptococcus pyogenes. Front. Cell. Infect. Microbiol. 2016, 6, 126. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, T.A.; Park, J.; Dang, T.L.; Choi, Y.G.; Kim, V.N. Microprocessor depends on hemin to recognize the apical loop of primary microRNA. Nucleic Acids Res. 2018, 46, 5726–5736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Partin, A.C.; Ngo, T.D.; Herrell, E.; Jeong, B.C.; Hon, G.; Nam, Y. Heme enables proper positioning of Drosha and DGCR8 on primary microRNAs. Nat. Commun. 2017, 8, 1737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Owens, C.P.; Du, J.; Dawson, J.H.; Goulding, C.W. Characterization of heme ligation properties of Rv0203, a secreted heme binding protein involved in Mycobacterium tuberculosis heme uptake. Biochemistry 2012, 51, 1518–1531. [Google Scholar] [CrossRef] [Green Version]
- Thakuri, B.; Graves, A.B.; Chao, A.; Johansen, S.L.; Goulding, C.W.; Liptak, M.D. The affinity of MhuD for heme is consistent with a heme degrading function in vivo. Metallomics 2018, 10, 1560–1563. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, R.C.; Yang, Y.; Wang, Y.; Davis, I.; Liu, A. Substrate-assisted hydroxylation and O-demethylation in the peroxidase-like cytochrome P450 enzyme CYP121. ACS Catal. 2020, 10, 1628–1639. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Davis, I.; Chan, Y.; Naik, S.G.; Griffith, W.P.; Liu, A. Characterization of the nonheme iron center of cysteamine dioxygenase and its interaction with substrates. J. Biol. Chem. 2020, 295, 11789–11802. [Google Scholar] [CrossRef]
- Fielding, A.J.; Dornevil, K.; Ma, L.; Davis, I.; Liu, A. Probing ligand exchange in the P450 enzyme CYP121 from Mycobacterium tuberculosis: Dynamic equilibrium of the distal heme ligand as a function of pH and temperature. J. Am. Chem. Soc. 2017, 139, 17481–17499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar]










| Identity | Soret Band (nm) | α Band (nm) | β Band (nm) | Charge Transfer (nm) |
|---|---|---|---|---|
| Free ferric heme (hemin) | 385 | 548 | 515 | 649 |
| Ferric heme-HupZ complex | 414 | 564 | 536 | - |
| Ferrous heme-HupZ complex | 424 | 559 | 530 | - |
| Ferrous CO-heme-HupZ complex | 421 | 567 | 537 | 623 |
| Ferric CN-heme-HupZ complex | 422 | 543 | - | - |
| Ferric heme-HupZ H111A complex | 414 | 568 | 531 | - |
| HupZ-V5-His6 | H111A HupZ-V5-His6 | |
|---|---|---|
| Data Collection | ||
| Wavelength (Å) | 0.97913 | 0.97946 |
| Space group | P212121 | P6522 |
| Cell dimensions | ||
| a, b, c (Å) | 40.0, 62.4, 94.9 | 54.9, 54.9, 333.3 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 120 |
| Resolution (Å) | 50.00–1.70 | 50–1.97 |
| (1.73–1.70) a | (2.00–1.97) | |
| Total reflection | 258352 | 661695 |
| Unique reflection | 26686 | 22490 |
| Redundancy | 9.7 (9.9) | 29.4 (32.0) |
| Rsym or Rmerge b (%) | 5.9 (18.3) | 17.1 (97.2) |
| CC1/2 e | 99.8(98.6) | 98.8 (90.6) |
| I/σI | 40.9 (9.7) | 25.4 (3.6) |
| Completeness (%) | 99.7 (99.9) | 99.9 (100) |
| Refinement | ||
| Resolution (Å) | 37.8–1.70 | 38.71–1.98 |
| No. reflections | 26636 | 21371 |
| Rworkc/Rfreed (%) | 18.36/21.87 | 19.54/22.65 |
| No. Atoms/B-factors (Å2) | ||
| Protein (Chain A) | 985/27.0 | 945/34.4 |
| Protein (Chain B) | 994/27.9 | 959/34.3 |
| Water (Chain S) | 208/34.7 | 161/40.9 |
| r.m.s. deviations e | ||
| Bond lengths (Å) | 0.007 | 0.009 |
| Bond angles (°) | 1.056 | 1.012 |
| Ramachandran Statistics f | ||
| Favored (%) | 96.8 | 98.3 |
| Allowed (%) | 3.2 | 1.7 |
| Outlier (%) | 0.00 | 0.00 |
| PDB Code | 7KPZ | 7KQ2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Traore, E.S.; Li, J.; Chiura, T.; Geng, J.; Sachla, A.J.; Yoshimoto, F.; Eichenbaum, Z.; Davis, I.; Mak, P.J.; Liu, A. Heme Binding to HupZ with a C-Terminal Tag from Group A Streptococcus. Molecules 2021, 26, 549. https://doi.org/10.3390/molecules26030549
Traore ES, Li J, Chiura T, Geng J, Sachla AJ, Yoshimoto F, Eichenbaum Z, Davis I, Mak PJ, Liu A. Heme Binding to HupZ with a C-Terminal Tag from Group A Streptococcus. Molecules. 2021; 26(3):549. https://doi.org/10.3390/molecules26030549
Chicago/Turabian StyleTraore, Ephrahime S., Jiasong Li, Tapiwa Chiura, Jiafeng Geng, Ankita J. Sachla, Francis Yoshimoto, Zehava Eichenbaum, Ian Davis, Piotr J. Mak, and Aimin Liu. 2021. "Heme Binding to HupZ with a C-Terminal Tag from Group A Streptococcus" Molecules 26, no. 3: 549. https://doi.org/10.3390/molecules26030549