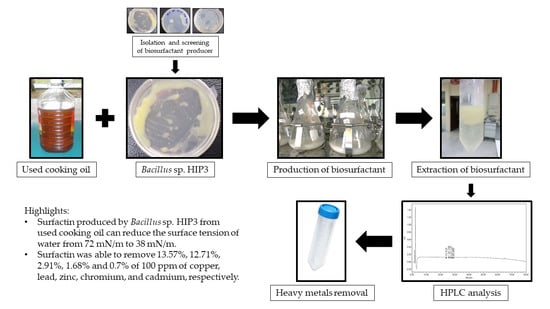
Production of Biosurfactant Produced from Used Cooking Oil by Bacillus sp. HIP3 for Heavy Metals Removal
Abstract
:1. Introduction
2. Results and Discussion
2.1. Characterization of Used Cooking Oil
2.2. Isolation and Screening of Biosurfactant-Producing Bacteria
2.3. Identification and Characterization of Biosurfactant-Producing Bacteria
2.4. Production of Biosurfactant from Used Cooking Oil
2.5. Analysis of Biosurfactant
2.6. Heavy Metals Removal
3. Materials and Methods
3.1. Characterization and Preparation of Raw Material/Substrate
3.2. Isolation and Screening of Biosurfactant-Producing Bacteria
3.3. Preparation of Culture Medium
3.4. Characterization and Identification of Biosurfactant-Producing Bacteria
3.5. Production of Biosurfactant from Used Cooking Oil as Carbon Source
3.6. Analytical Methods
3.6.1. Cell Biomass Determination
3.6.2. Surface Activity Measurements
3.7. Extraction of Biosurfactant
3.8. Analysis of Biosurfactant
3.9. Heavy Metals Remediation Efficiency
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yaakob, Z.; Mohammad, M.; Alherbawi, M.; Alam, Z.; Sopian, K. Overview of the production of biodiesel from waste cooking oil. Renew. Sustain. Energy Rev. 2013, 18, 184–193. [Google Scholar] [CrossRef]
- Kheang, L.S.; May, C.Y.; Foon, C.S.; Ngan, M.A. Recovery and conversion of plam olein-derived used frying oil to methyl esters for biodiesel. J. Oil Palm Res. 2006, 18, 247–252. [Google Scholar]
- Sanli, H.; Canakci, M.; Alptekin, E. Characterization of waste frying oils obtained from different facilities. In World Renewable Energy Congress-Sweden; Linköping University Electronic Press: Linköping, Sweden, 2011; pp. 479–485. [Google Scholar]
- Banat, I.M. Biosurfactants production and possible uses in microbial enhanced oil recovery and oil pollution remediation: A review. Bioresour. Technol. 1995, 51, 1–12. [Google Scholar] [CrossRef]
- Dos Santos, S.C.; Fernandez, L.G.; Rossi-Alva, J.C.; de Abreu Roque, M.R. Evaluation of substrates from renewable-resources in biosurfactants production by Pseudomonas strains. Afr. J. Biotechnol. 2010, 9, 5704–5711. [Google Scholar]
- Jamal, P.; Alam, M.Z.; Mohd Yusof, U.S.M.; Wan Nawawi, W.M.F. Optimization of process conditions for extraction of biosurfactant. Aust. J. Basic Appl. Sci. 2011, 7, 2156–2161. [Google Scholar]
- De Oliveira, J.G.; Garcia-Cruz, C.H. Properties of a biosurfactant produced by Bacillus pumilus using vinasse and waste frying oil as alternative carbon sources. Braz. Arch. Biol. Technol. 2013, 56, 155–160. [Google Scholar] [CrossRef]
- Haba, E.; Espuny, M.J.; Busquets, M.; Manresa, A. Screening and production of rhamnolipids by Pseudomonas aeruginosa 47T2 NCIB 40044 from waste frying oils. J. Appl. Microbiol. 2000, 88, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Makkar, R.S.; Cameotra, S.S. Biosurfactant Production by Microorganisms on Unconventional Carbon Sources. J. Surfactants Deterg. 1999, 2, 237–241. [Google Scholar] [CrossRef]
- Pacwa-Płociniczak, M.; Płaza, G.A.; Piotrowska-Seget, Z.; Cameotra, S.S. Environmental applications of biosurfactants: Recent advances. Int. J. Mol. Sci. 2011, 12, 633–654. [Google Scholar] [CrossRef]
- Argun, M.E.; Dursun, S.; Ozdemir, C.; Karatas, M. Heavy metal adsorption by modified oak sawdust: Thermodynamics and kinetics. J. Hazard. Mater. 2007, 141, 77–85. [Google Scholar] [CrossRef]
- Mulligan, C.N. Environmental applications for biosurfactants. Environ. Pollut. 2005, 133, 183–198. [Google Scholar] [CrossRef] [PubMed]
- Maikudi Usman, M.; Dadrasnia, A.; Tzin Lim, K.; Fahim Mahmud, A.; Ismail, S. Application of biosurfactants in environmental biotechnology; remediation of oil and heavy metal. AIMS Bioeng. 2016, 3, 289–304. [Google Scholar] [CrossRef]
- Olaniran, A.O.; Balgobind, A.; Pillay, B. Bioavailability of heavy metals in soil: Impact on microbial biodegradation of organic compounds and possible improvement strategies. Int. J. Mol. Sci. 2013, 14, 10197–10228. [Google Scholar] [CrossRef] [PubMed]
- Arima, K.; Kakinuma, A.; Tamura, G. Surfactin, a crystalline peptidelipid surfactant produced by Bacillus subtilis: Isolation, characterization and its inhibition of fibrin clot formation. Biochem. Biophys. Res. Commun. 1968, 31, 488–494. [Google Scholar] [CrossRef]
- Thimon, L.; Peyoux, F.; Maget-Dana, R.; Michel, G. Surface-active properties of antifungal lipopeptides produced by Bacillus subtilis. J. Am. Oil Chem. Soc. 1992, 69, 92–93. [Google Scholar] [CrossRef]
- Esteban, B.; Riba, J.-R.; Baquero, G.; Puig, R.; Rius, A. Characterization of the surface tension of vegetable oils to be used as fuel in diesel engines. Fuel 2012, 102, 231–238. [Google Scholar] [CrossRef]
- Gertz, C. Chemical and physical parameters as quality deep-frying process—Changes at elevated temperature. Eur. J. Lipid Sci. Technol. 2000, 102, 566–572. [Google Scholar] [CrossRef]
- Tan, J.; Zuniga, C.; Zengler, K. Unraveling interactions in microbial communities—From co-cultures to microbiomes. J. Microbiol. 2015, 53, 295–305. [Google Scholar] [CrossRef]
- Prakash, B.; Irfan, M. Pseudomonas aeruginosa is present in crude oil contaminated sites of barmer region (India). J. Bioremediat. Biodegrad. 2011, 2, 5. [Google Scholar] [CrossRef]
- Morikawa, M.; Hirata, Y.; Imanaka, T. A study on the structure-function relationship of lipopeptides biosurfactants. Biochim. Biophys. Acta 2000, 1488, 211–218. [Google Scholar] [CrossRef]
- Pandey, A.; Anis, R. Optimization and characterization of biosurfactant producing microbes isolated from oil contaminated soil and expression of biosurfactant genes in E.coli. Int. J. Pharm. Res. Bio-Sci. 2013, 2, 46–67. [Google Scholar]
- Beveridge, T.J. Use of the gram stain in microbiology. Biotechnol. Histochem. 2001, 76, 111–118. [Google Scholar] [CrossRef]
- Varadavenkatesan, T.; Murty, V.R. Production of a lipopeptide biosurfactant by a novel Bacillus sp. and its applicability to enhanced oil recovery. ISRN Microbiol. 2013, 2013, 621519. [Google Scholar] [CrossRef] [PubMed]
- Kavitha, V.; Mandal, A.B.; Gnanamani, A. Microbial biosurfactant mediated removal and/or solubilization of crude oil contamination from soil and aqueous phase: An approach with Bacillus licheniformis MTCC 5514. Int. Biodeterior. Biodegrad. 2014, 94, 24–30. [Google Scholar] [CrossRef]
- Zhou, H.; Chen, J.; Yang, Z.; Qin, B.; Li, Y.; Kong, X. Biosurfactant production and characterization of Bacillus sp. ZG0427 isolated from oil-contaminated soil. Ann. Microbiol. 2015, 65, 2255–2264. [Google Scholar] [CrossRef]
- Tarntip, R.; Sirichom, T. Isolation of proteolytic, lipolytic, and bioemulsifying bacteria for improvement of the aerobic treatment of poultry processing wastewater. Afr. J. Micriobiol. Res. 2011, 5, 5493–5497. [Google Scholar]
- Wu, S.J.; Hu, Z.H.; Zhang, L.L.; Yu, X.; Chen, J.M. A novel dichloromethane-degrading Lysinibacillus sphaericus strain Wh22 and its degradative plasmid. Appl. Microbiol. Biotechnol. 2009, 82, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. Path-O-Gen v1.4. Available online: http://tree.bio.ed.ac.uk/software/pathogen/ (accessed on 7 May 2016).
- Li, J.; Yang, G.; Wu, M.; Zhao, Y.; Zhou, S. Bacillus huizhouensis sp. nov., isolated from a paddy field soil. Antonie Van Leeuwenhoek 2014, 106, 357–363. [Google Scholar] [CrossRef]
- Dong, K.; Lee, S. Bacillus kyonggiensis sp. nov., isolated from soil of a lettuce field. J. Microbiol. 2011, 49, 776–781. [Google Scholar] [CrossRef]
- Madhaiyan, M.; Poonguzhali, S.; Lee, J.S.; Lee, K.C.; Hari, K. Bacillus rhizosphaerae sp. nov., an novel diazotrophic bacterium isolated from sugarcane rhizosphere soil. Antonie Van Leeuwenhoek 2011, 100, 437–444. [Google Scholar] [CrossRef]
- Peng, L.; Huihui, L.; Rong, L.; Shunpeng, L.; Xing, H. Biodegradation of fomesafan by Lysinibacillus sp.ZB-1 isolated from soil. Chemosphere 2009, 77, 1614–1619. [Google Scholar]
- Al-Bahry, S.N.; Al-Wahaibi, Y.M.; Elshafie, A.E.; Al-Bemani, A.S.; Joshi, S.J.; Al-Makhmari, H.S.; Al-Sulaimani, H.S. Biosurfactant production by Bacillus subtilis B20 using date molasses and its possible application in enhanced oil recovery. Int. Biodeterior. Biodegrad. 2013, 81, 141–146. [Google Scholar] [CrossRef]
- Neves, L.C.M.; Oliveira, K.S.; Kobayashi, M.J.; Penna, T.C.V.; Converti, A. Biosurfactant produdion by cultivation of Bacillus atrophaeus ATCC 9372 in semidefined Glucose/casein-based media. Appl. Biochem. Biotechnol. 2007, 137, 539–554. [Google Scholar] [PubMed]
- Chandankere, R.; Yao, J.; Choi, M.M.F.; Masakorala, K.; Chan, Y. An efficient biosurfactant-producing and crude-oil emulsifying bacterium Bacillus methylotrophicus USTBa isolated from petroleum reservoir. Biochem. Eng. J. 2013, 74, 46–53. [Google Scholar] [CrossRef]
- Elazzazy, A.M.; Abdelmoneim, T.S.; Almaghrabi, O.A. Isolation and characterization of biosurfactant production under extreme environmental conditions by alkali-halo-thermophilic bacteria from Saudi Arabia. Saudi J. Biol. Sci. 2015, 22, 466–475. [Google Scholar] [CrossRef]
- Satpute, S.K.; Bhuyan, S.S.; Pardesi, K.R.; Mujumdar, S.S.; Dhakephalkar, P.K.; Shete, A.M.; Chopade, B.A. Molecular Genetics of Biosurfactant Synthesis in Microorganisms, 1st ed.; Sen, R., Ed.; Landes Bioscience: Austin, TX, USA, 2010; ISBN 9781441959782. [Google Scholar]
- Helmy, Q.; Kardena, E.; Funamizu, N. Wisjnuprapto Strategies toward commercial scale of biosurfactant production as potential substitute for it’s chemically counterparts. Int. J. Biotechnol. 2011, 12, 66. [Google Scholar]
- Joshi, S.J.; Desai, A.J. Biosurfactant’s role in bioremediation of NAPL and fermentative production. Adv. Exp. Med. Biol. 2010, 672, 222–235. [Google Scholar]
- Shaligram, N.S.; Singhal, R.S. Surfactin—A review on biosynthesis, fermentation, purification and applications. Food Technol. Biotechnol. 2010, 48, 119–134. [Google Scholar]
- Baker, S.C.; Chen, C.Y. Enrichment and purification of lipopeptide biosurfactants. In Advances in Experimental Medicine and Biology; Sen, R., Ed.; Landes Bioscience: Austin, TX, USA; Springer Science + Business Media: New York, NY, USA, 2010; Volume 672, pp. 281–288. ISBN 9781441959782. [Google Scholar]
- Chen, H.R.; Chen, C.C.; Reddy, A.S.; Chen, C.Y.; Li, W.R.; Tseng, M.J.; Liu, H.T.; Pan, W.; Maity, J.P.; Atla, S.B. Removal of mercury by foam fractionation using surfactin, a biosurfactant. Int. J. Mol. Sci. 2011, 12, 8245–8258. [Google Scholar] [CrossRef]
- Makkar, R.S.; Cameotra, S.S.; Banat, I.M. Advances in utilization of renewable substrates for biosurfactant production. AMB Express 2011, 1, 5. [Google Scholar] [CrossRef]
- Smyth, T.J.P.; Perfumo, A.; McClean, S.; Marchant, R.; Banat, I.M. Isolation and analysis of lipopeptides and high molecular weight biosurfactants. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Northern Ireland, UK, 2010; Volume 78, pp. 3687–3704. ISBN 9783540775874. [Google Scholar]
- Swapna, T.H.; Papathoti, N.K.; Khan, M.Y.; Reddy, G.; Hameeda, B. Bioreduction of Cr (VI) by biosurfactant producing marine bacterium Bacillus subtilis SHB 13. J. Sci. Ind. Res. 2016, 75, 432–438. [Google Scholar]
- Thavasi, R.; Jayalakshmi, S.; Balasubramanian, T.; Banat, I.M. Production and characterization of a glycolipid biosurfactant from Bacillus megaterium using economically cheaper sources. World J. Microbiol. Biotechnol. 2008, 24, 917–925. [Google Scholar] [CrossRef]
- Pereira, J.F.B.; Gudiña, E.J.; Costa, R.; Vitorino, R.; Teixeira, J.A.; Coutinho, J.A.P.; Rodrigues, L.R. Optimization and characterization of biosurfactant production by Bacillus subtilis isolates towards microbial enhanced oil recovery applications. Fuel 2013, 111, 259–268. [Google Scholar] [CrossRef]
- Mnif, I.; Ghribi, D. Review lipopeptides biosurfactants: Mean classes and new insights for industrial, biomedical, and environmental applications. Biopolymers 2015, 104, 129–147. [Google Scholar] [CrossRef] [PubMed]
- Hathout, Y.; Ho, Y.P.; Ryzhov, V.; Demirev, P.; Fenselau, C. Kurstakins: A new class of lipopeptides isolated from Bacillus thuringiensis. J. Nat. Prod. 2000, 63, 1492–1496. [Google Scholar] [CrossRef] [PubMed]
- Sousa, M.; Dantas, I.T.; Feitosa, F.X.; Alencar, A.E.V.; Soares, S.A.; Melo, V.M.M. Performance of a biosurfactant produced by Bacillus subtilis LAMI005 on the formation of oil/biosurfactant/water emulsion: Study of the phase behaviour of emulsified systems. Braz. J. Chem. Eng. 2014, 31, 613–623. [Google Scholar] [CrossRef]
- Anburajan, L.; Meena, B.; Raghavan, R.V.; Shridhar, D.; Joseph, T.C.; Vinithkumar, N.V.; Dharani, G.; Dheenan, P.S.; Kirubagaran, R. Heterologous expression, purification, and phylogenetic analysis of oil-degrading biosurfactant biosynthesis genes from the marine sponge-associated Bacillus licheniformis NIOT-06. Bioprocess Biosyst. Eng. 2014, 38, 1009–1018. [Google Scholar] [CrossRef] [PubMed]
- Sarubbo, L.A.; Brasileiro, P.P.F.; Silveira, G.N.M.; Juliana, M. Application of a low cost biosurfactant in the removal of heavy metals in soil. Chem. Eng. Trans. 2018, 64, 433–438. [Google Scholar]
- Mohammed, A.S.; Kapri, A.; Goel, R. Use of Biosurfactants in the Removal of Heavy Metal Ions from Soils; Springer: Dordrecht, The Netherlands, 2011; Volume 20, ISBN 978-94-007-1913-2. [Google Scholar]
- Singh, A.K.; Cameotra, S.S. Efficiency of lipopeptide biosurfactants in removal of petroleum hydrocarbons and heavy metals from contaminated soil. Environ. Sci. Pollut. Res. 2013, 20, 7367–7376. [Google Scholar] [CrossRef]
- Das, P.; Mukherjee, S.; Sen, R. Biosurfactant of marine origin exhibiting heavy metal remediation properties. Bioresour. Technol. 2009, 100, 4887–4890. [Google Scholar] [CrossRef]
- Lima, T.M.S.; Procópio, L.C.; Brandão, F.D.; Carvalho, A.M.X.; Tótola, M.R.; Borges, A.C. Simultaneous phenanthrene and cadmium removal from contaminated soil by a ligand/biosurfactant solution. Biodegradation 2011, 22, 1007–1015. [Google Scholar] [CrossRef] [PubMed]
- Abidin, S.Z.; Patel, D.; Saha, B. Quantitative analysis of fatty acids composition in the used cooking oil (UCO) by gas chromatography-mass spectrometry (GC-MS). Can. J. Chem. Eng. 2013, 91, 1896–1903. [Google Scholar] [CrossRef]
- Batista, S.B.; Mounteer, A.H.; Amorim, F.R.; Tótola, M.R. Isolation and characterization of biosurfactant/bioemulsifier-producing bacteria from petroleum contaminated sites. Bioresour. Technol. 2006, 97, 868–875. [Google Scholar] [CrossRef] [PubMed]
- Rollins, D.M.; Joseph, S.W. The Gram Stain; University of Maryland, College Park: College Park, MD, USA, 2000. [Google Scholar]
- Xia, W.J.; Luo, Z.B.; Dong, H.P.; Yu, L.; Cui, Q.F.; Bi, Y.Q. Synthesis, characterization, and oil recovery application of biosurfactant produced by indigenous Pseudomonas aeruginosa WJ-1 using waste vegetable oils. Appl. Biochem. Biotechnol. 2012, 166, 1148–1166. [Google Scholar] [CrossRef] [PubMed]
- Bodour, A.A.; Miller-Maier, R.M. Application of a modified drop-collapse technique for surfactant quantitation and screening of biosurfactant-producing microorganisms. J. Microbiol. Methods 1998, 32, 273–280. [Google Scholar] [CrossRef]
- Vater, J.; Kablitz, B.; Wilde, C.; Franke, P.; Mehta, N.; Cameotra, S.S. Matrix-assisted laser desorption ionization—Time of flight mass spectrometry of lipopeptide biosurfactants in whole cells and culture filtrates of Bacillus subtilis C-1 isolated from petroleum sludge. Appl. Environ. Microbiol. 2002, 68, 6210–6219. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, M.-I. HPLC of Peptides and Proteins: Methods and Protocols; Humana Press: New York, NY, USA, 2004; ISBN 1064-3745. [Google Scholar]
Sample Availability: Samples of the compounds surfactin are not available from the authors. |





| Microorganisms | Isolate | Source |
|---|---|---|
| Bacteria | HIP1 | POME |
| HIP2 | POME | |
| HIP3 | POME | |
| HIO1 | UCO | |
| Yeast | HIP4 | POME |
| HIP5 | POME | |
| HIP6 | POME | |
| HIS1 | Cooking oil contaminated soil | |
| Fungi | HIS2 | Cooking oil contaminated soil |
| HIS3 | Cooking oil contaminated soil |
| Isolates | Oil Displacement Area (cm2) | Surface Tension (mN/m) |
|---|---|---|
| HIP1 | 33.18 | 43.5 |
| HIP2 | 29.9 | 43.33 |
| HIP3 | 92.12 | 38.15 |
| HIO1 | 26.7 | 50.17 |
| Isolate | Shape | Gram | Genus/Species | Similarity (%) |
|---|---|---|---|---|
| HIP1 | Rod | Positive | Lysinibacillus sphaericus (KJ576904.1) | 99 |
| HIP2 | Rod | Positive | Lysinibacillus xylanilyticus (LK391655.1) | 75 |
| HIP3 | Rod | Positive | Bacillus sp. (AY787805.1) | 83 |
| HIO1 | Rod | Positive | Bacillus sp. (KC160846.1) | 58 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Md Badrul Hisham, N.H.; Ibrahim, M.F.; Ramli, N.; Abd-Aziz, S. Production of Biosurfactant Produced from Used Cooking Oil by Bacillus sp. HIP3 for Heavy Metals Removal. Molecules 2019, 24, 2617. https://doi.org/10.3390/molecules24142617
Md Badrul Hisham NH, Ibrahim MF, Ramli N, Abd-Aziz S. Production of Biosurfactant Produced from Used Cooking Oil by Bacillus sp. HIP3 for Heavy Metals Removal. Molecules. 2019; 24(14):2617. https://doi.org/10.3390/molecules24142617
Chicago/Turabian StyleMd Badrul Hisham, Nurul Hanisah, Mohamad Faizal Ibrahim, Norhayati Ramli, and Suraini Abd-Aziz. 2019. "Production of Biosurfactant Produced from Used Cooking Oil by Bacillus sp. HIP3 for Heavy Metals Removal" Molecules 24, no. 14: 2617. https://doi.org/10.3390/molecules24142617