Reversible Immobilization of Lipases on Heterofunctional Octyl-Amino Agarose Beads Prevents Enzyme Desorption
Abstract
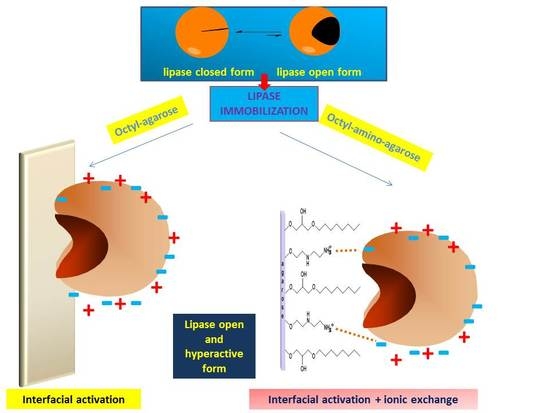
:1. Introduction
2. Results
2.1. Immobilization on EDA and HDA Supports
2.2. Immobilization of Enzymes in OC, OCEDA and OCHDA Support
2.3. Confirmation of the Mixed Adsorption of the Different Enzymes and Reversibility of the Immobilization
2.4. Thermal Stabilities of the Differently Immobilized Lipases
2.5. Solvent Stabilities of the Differently Immobilized Lipases
2.6. Reuse of OCEDA-CALB in Hydrolysis of Triacetin to Produce 1,2-Diacetin
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Standard Determination of Enzyme Activity
4.3. Preparation of Amino Supports
4.4. Immobilization of Enzymes
4.4.1. Immobilization of Enzymes on Octyl (OC) and Octyl-Amino (OCHDA and OCEDA) Supports
4.4.2. Immobilization of Enzymes on HDA and EDA Supports
4.5. Thermal Inactivation of Different Enzymatic Preparations
4.6. Inactivation of Different Enzymatic Preparations by Incubation in Organic Co-Solvents
4.7. SDS-PAGE Analysis
4.8. Hydrolysis of Triacetin
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| OC | OCTYL SUPPORT Multidisciplinary Digital Publishing Institute |
| EDA | DIETHYLENAMINE MODIFIED SUPPORT |
| HDA | DIHEXYENAMINE MODIFIED SUPPORT |
| OCEDA | OCTYL-DIETHYLENAMINE MODIFIED SUPPORT |
| OCHDA | OCTYL-DIHEXYENAMINE MODIFIED SUPPORT |
References
- Sheldon, R.A.; van Pelt, S. Enzyme immobilisation in biocatalysis: Why, what and how. Chem. Soc. Rev. 2013, 42, 6223–6235. [Google Scholar] [CrossRef] [PubMed]
- Adlercreutz, P. Immobilisation and application of lipases in organic media. Chem. Soc. Rev. 2013, 42, 6406. [Google Scholar] [CrossRef] [PubMed]
- DiCosimo, R.; McAuliffe, J.; Poulose, A.J.; Bohlmann, G. Industrial use of immobilized enzymes. Chem. Soc. Rev. 2013, 42, 6437. [Google Scholar] [CrossRef] [PubMed]
- Liese, A.; Hilterhaus, L. Evaluation of immobilized enzymes for industrial applications. Chem. Soc. Rev. 2013, 42, 6236. [Google Scholar] [CrossRef] [PubMed]
- Ansorge-Schumacher, M.B.; Thum, O. Immobilised lipases in the cosmetics industry. Chem. Soc. Rev. 2013, 42, 6475. [Google Scholar] [CrossRef] [PubMed]
- Min, K.; Yoo, Y.J. Recent progress in nanobiocatalysis for enzyme immobilization and its application, Biotechnol. Bioprocess Eng. 2014, 19, 553–567. [Google Scholar] [CrossRef]
- Homaei, A.A.; Sariri, R.; Vianello, F.; Stevanato, R. Enzyme immobilization: An update. J. Chem. Biol. 2013, 6, 185–205. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Tiwari, M.K.; Singh, R.; Lee, J.K. From protein engineering to immobilization: Promising strategies for the upgrade of industrial enzymes. Int. J. Mol. Sci. 2013, 14, 1232–1277. [Google Scholar] [CrossRef] [PubMed]
- Tran, D.N.; Balkus, K.J. Perspective of recent progress in immobilization of enzymes. ACS Catal. 2011, 1, 956–968. [Google Scholar] [CrossRef]
- Barbosa, O.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Rodrigues, R.C.; Fernandez-Lafuente, R. Strategies for the one-step immobilization-purification of enzymes as industrial biocatalysts. Biotechnol. Adv. 2015, 33, 435–456. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Lafuente, R.; Armisén, P.; Sabuquillo, P.; Fernández-Lorente, G.; Guisán, J.M. Immobilization of lipases by selective adsorption on hydrophobic supports. Chem. Phys. Lipids 1998, 93, 185–197. [Google Scholar] [CrossRef]
- Brzozowski, A.M.; Derewenda, U.; Derewenda, Z.S.; Dodson, G.G.; Lawson, D.M.; Turkenburg, J.P.; Bjorkling, F.; Huge-Jensen, B.; Patkar, S.A.; Thim, L. A model for interfacial activation in lipases from the structure of a fungal lipase-inhibitor complex. Nature 1991, 351, 491–494. [Google Scholar] [CrossRef] [PubMed]
- Manoel, E.A.; dos Santos, J.C.S.; Freire, D.M.G.; Rueda, N.; Fernandez-Lafuente, R. Immobilization of lipases on hydrophobic supports involves the open form of the enzyme. Enzyme Microb. Technol. 2015, 71, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Uppenberg, J.; Hansen, M.T.; Patkar, S.; Jones, T.A. The sequence, crystal structure determination and refinement of two crystal forms of lipase B from Candida antarctica. Structure 1994, 2, 293–308. [Google Scholar] [CrossRef]
- Carrasco-López, C.; Godoy, C.; de las Rivas, B.; Fernández-Lorente, G.; Palomo, J.M.; Guisán, J.M.; Fernández-Lafuente, R.; Martínez-Ripoll, M.; Hermoso, J.A. Activation of bacterial thermo alkalophilic lipases is spurred by dramatic structural rearrangements. J. Biol. Chem. 2009, 284, 4365–4372. [Google Scholar] [CrossRef] [PubMed]
- Verger, R. “Interfacial activation” of lipases: Facts and artifacts. Trends Biotechnol. 1997, 15, 32–38. [Google Scholar] [CrossRef]
- Brzozowski, A.M.; Savage, H.; Verma, C.S.; Turkenburg, J.P.; Lawson, D.M.; Svendsen, A.; Patkar, S. Structural origins of the interfacial activation in Thermomyces (Humicola) lanuginosa lipase. Biochemistry 2000, 39, 15071–15082. [Google Scholar] [CrossRef]
- Ericsson, D.J.; Kasrayan, A.; Johansson, P.; Bergfors, T.; Sandström, A.G.; Bäckvall, J.E.; Mowbray, S.L. X-ray structure of Candida antarctica lipase a shows a novel lid structure and a likely mode of interfacial activation. J. Mol. Biol. 2008, 376, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Palomo, J.M.; Peñas, M.M.; Fernández-Lorente, G.; Mateo, C.; Pisabarro, A.G.; Fernández-Lafuente, R.; Ramírez, L.; Guisán, J.M. Solid-phase handling of hydrophobins: Immobilized hydrophobins as a new tool to study lipases. Biomacromolecules 2003, 4, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Palomo, J.M.; Fuentes, M.; Fernández-Lorente, G.; Mateo, C.; Guisan, J.M.; Fernández-Lafuente, R. General trend of lipase to self-assemble giving bimolecular aggregates greatly modifies the enzyme functionality. Biomacromolecules 2003, 4, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Palomo, J.M.; Ortiz, C.; Fuentes, M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernandez-Lafuente, R. Use of immobilized lipases for lipase purification via specific lipase-lipase interactions. J. Chromatogr. A 2004, 1038, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Palomo, J.M.; Ortiz, C.; Fernández-Lorente, G.; Fuentes, M.; Guisán, J.M.; Fernández-Lafuente, R. Lipase-lipase interactions as a new tool to immobilize and modulate the lipase properties. Enzyme Microb. Technol. 2005, 36, 447–454. [Google Scholar] [CrossRef]
- Al-Duri, B.; Yong, Y.P. Lipase immobilisation: An equilibrium study of lipases immobilised on hydrophobic and hydrophilic/hydrophobic supports. Biochem. Eng. J. 2000, 4, 207–215. [Google Scholar] [CrossRef]
- Fernández-Lorente, G.; Palomo, J.M.; Cabrera, Z.; Guisán, J.M.; Fernández-Lafuente, R. Specificity enhancement towards hydrophobic substrates by immobilization of lipases by interfacial activation on hydrophobic supports. Enzyme Microb. Technol. 2007, 41, 565–569. [Google Scholar] [CrossRef]
- Boros, Z.; Weiser, D.; Márkus, M.; Abaháziová, E.; Magyar, Á.; Tomin, A.; Koczka, B.; Kovács, P.; Poppe, L. Hydrophobic adsorption and covalent immobilization of Candida antarctica lipase B on mixed-function-grafted silica gel supports for continuous-flow biotransformations. Process Biochem. 2013, 48, 1039–1047. [Google Scholar] [CrossRef] [Green Version]
- Jaeger, K.E.; Ransac, S.; Koch, H.B.; Ferrato, F.; Dijkstra, B.W. Topological characterization and modeling of the 3D structure of lipase from Pseudomonas aeruginosa. FEBS Lett. 1993, 332, 143–149. [Google Scholar] [CrossRef]
- Cygler, M.; Schrag, J.D. Structure and conformational flexibility of Candida rugosa lipase. Biochim. Biophys. Acta 1999, 1441, 205–214. [Google Scholar] [CrossRef]
- Palomo, J.M.; Muñoz, G.; Fernández-Lorente, G.; Mateo, C.; Fernández-Lafuente, R.; Guisán, J.M. Interfacial adsorption of lipases on very hydrophobic support (octadecyl–Sepabeads): Immobilization, hyperactivation and stabilization of the open form of lipases. J. Mol. Catal. B Enzym. 2002, 19–20, 279–286. [Google Scholar] [CrossRef]
- Dos Santos, J.C.S.; Rueda, N.; Gonçalves, L.R.B.; Fernandez-Lafuente, R. Tuning the catalytic properties of lipases immobilized on divinylsulfone activated agarose by altering its nanoenvironment. Enzyme Microb. Technol. 2015, 77, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, J.C.S.; Rueda, N.; Sanchez, A.; Villalonga, R.; Gonçalves, L.R.B.; Fernandez-Lafuente, R. Versatility of divinylsulfone supports permits the tuning of CALB properties during its immobilization. RSC Adv. 2015, 5, 35801–35810. [Google Scholar] [CrossRef]
- Pizarro, C.; Brañes, M.C.; Markovits, A.; Fernández-Lorente, G.; Guisán, J.M.; Chamy, R.; Wilson, L. Influence of different immobilization techniques for Candida cylindracea lipase on its stability and fish oil hydrolysis. J. Mol. Catal. B Enzym. 2012, 78, 111–118. [Google Scholar] [CrossRef]
- Fernandez-Lorente, G.; Filice, M.; Lopez-Vela, D.; Pizarro, C.; Wilson, L.; Betancor, L.; Avila, Y.; Guisan, J.M. Cross-linking of lipases adsorbed on hydrophobic supports: Highly selective hydrolysis of fish oil catalyzed by RML. J. Am. Oil Chem. Soc. 2010, 88, 801–807. [Google Scholar] [CrossRef]
- Suescun, A.; Rueda, N.; dos Santos, J.C.S.; Castillo, J.J.; Ortiz, C.; Torres, R.; Barbosa, O.; Fernandez-Lafuente, R. Immobilization of lipases on glyoxyl–octyl supports: Improved stability and reactivation strategies. Process Biochem. 2015, 50, 1211–1217. [Google Scholar] [CrossRef]
- Rueda, N.; dos Santos, J.C.S.; Ortiz, C.; Barbosa, O.; Fernandez-Lafuente, R.; Torres, R. Chemical amination of lipases improves their immobilization on octyl-glyoxyl agarose beads. Catal. Today 2016, 259, 107–118. [Google Scholar] [CrossRef]
- Rueda, N.; dos Santos, J.C.S.; Torres, R.; Barbosa, O.; Ortiz, C.; Fernandez-Lafuente, R. Reactivation of lipases by the unfolding and refolding of covalently immobilized biocatalysts. RSC Adv. 2015, 5, 55588–55594. [Google Scholar] [CrossRef]
- Rueda, N.; dos Santos, J.C.S.; Torres, R.; Ortiz, C.; Barbosa, O.; Fernandez-Lafuente, R. Improved performance of lipases immobilized on heterofunctional octyl-glyoxyl agarose beads. RSC Adv. 2015, 5, 11212–11222. [Google Scholar] [CrossRef]
- Bernal, C.; Illanes, A.; Wilson, L. Heterofunctional hydrophilic-hydrophobic porous silica as support for multipoint covalent immobilization of lipases: Application to lactulose palmitate synthesis. Langmuir 2014, 30, 3557–3566. [Google Scholar] [CrossRef] [PubMed]
- Bernal, C.; Illanes, A.; Wilson, L. Improvement of efficiency in the enzymatic synthesis of lactulose palmitate. J. Agric. Food Chem. 2015, 63, 3716–3724. [Google Scholar] [CrossRef] [PubMed]
- Guajardo, N.; Bernal, C.; Wilson, L.; Cabrera, Z. Selectivity of R-α-monobenzoate glycerol synthesis catalyzed by Candida antarctica lipase B immobilized on heterofunctional supports. Process Biochem. 2015, 50, 1870–1877. [Google Scholar] [CrossRef]
- Guajardo, N.; Bernal, C.; Wilson, L.; Cabrera, Z. Asymmetric hydrolysis of dimethyl-3-phenylglutarate in sequential batch reactor operation catalyzed by immobilized Geobacillus thermocatenulatus lipase. Catal. Today 2015, 255, 21–26. [Google Scholar] [CrossRef]
- Garcia-Galan, C.; Berenguer-Murcia, Á.; Fernandez-Lafuente, R.; Rodrigues, R.C. Potential of different enzyme immobilization strategies to improve enzyme performance. Adv. Synth. Catal. 2011, 353, 2885–2904. [Google Scholar] [CrossRef]
- Barbosa, O.; Torres, R.; Ortiz, C.; Berenguer-Murcia, A.; Rodrigues, R.C.; Fernandez-Lafuente, R. Heterofunctional supports in enzyme immobilization: From traditional immobilization protocols to opportunities in tuning enzyme properties. Biomacromolecules 2013, 14, 2433–2462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez-Lafuente, R.; Rosell, C.M.; Rodriguez, V.; Santana, C.; Soler, G.; Bastida, A.; Guisán, J.M. Preparation of activated supports containing low pK amino groups. A new tool for protein immobilization via the carboxyl coupling method. Enzyme Microb. Technol. 1993, 15, 546–550. [Google Scholar] [CrossRef]
- Bolivar, J.M.; Nidetzky, B. Positively charged mini-protein zbasic2 as a highly efficient silica binding module: Opportunities for enzyme immobilization on unmodified silica supports. Langmuir 2012, 28, 10040–10049. [Google Scholar] [CrossRef] [PubMed]
- Wiesbauer, J.; Bolivar, J.M.; Mueller, M.; Schiller, M.; Nidetzky, B. Oriented immobilization of enzymes made fit for applied biocatalysis: Non-covalent attachment to anionic supports using zbasic2 module. ChemCatChem 2011, 3, 1299–1303. [Google Scholar] [CrossRef]
- Kumar, A.; Galaev, I.Y.; Mattiasson, B. Polymer displacement/shielding in protein chromatography. J. Chromatogr. B Biomed. Sci. Appl. 2000, 741, 103–113. [Google Scholar] [CrossRef]
- Fuentes, M.; Mateo, C.; Pessela, B.C.C.; Batalla, P.; Fernandez-Lafuente, R.; Guisán, J.M. Solid phase proteomics: Dramatic reinforcement of very weak protein-protein interactions. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2007, 849, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Domínguez De María, P.; Carboni-Oerlemans, C.; Tuin, B.; Bargeman, G.; Van Der Meer, A.; Van Gemert, R. Biotechnological applications of Candida antarctica lipase A: State-of-the-art. J. Mol. Catal. B Enzym. 2005, 37, 36–46. [Google Scholar] [CrossRef]
- Kirk, O.; Christensen, M.W. Lipases from Candida antarctica: Unique biocatalysts from a unique origin. Org. Process Res. Dev. 2002, 6, 446–451. [Google Scholar] [CrossRef]
- Benjamin, S.; Pandey, A. Candida rugosa lipases: Molecular biology and versatility in biotechnology. Yeast 1998, 14, 1069–1087. [Google Scholar] [CrossRef]
- Fernandez-Lafuente, R. Lipase from Thermomyces lanuginosus: Uses and prospects as an industrial biocatalyst. J. Mol. Catal. B Enzym. 2010, 62, 197–212. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Fernandez-Lafuente, R. Lipase from Rhizomucor miehei as an industrial biocatalyst in chemical process. J. Mol. Catal. B Enzym. 2010, 64, 1–22. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Fernandez-Lafuente, R. Lipase from Rhizomucor miehei as a biocatalyst in fats and oils modification. J. Mol. Catal. B Enzym. 2010, 66, 15–32. [Google Scholar] [CrossRef]
- Dos Santos, J.C.S.; Garcia-Galan, C.; Rodrigues, R.C.; de Sant’Ana, H.B.; Gonçalves, L.R.B.; Fernandez-Lafuente, R. Stabilizing hyperactivated lecitase structures through physical treatment with ionic polymers. Process Biochem. 2014, 49, 1511–1515. [Google Scholar] [CrossRef]
- Garcia-Galan, C.; dos Santos, J.C.S.; Barbosa, O.; Torres, R.; Pereira, E.B.; Corberan, V.C.; Gonçalves, L.R.B.; Fernandez-Lafuente, R. Tuning of Lecitase features via solid-phase chemical modification: Effect of the immobilization protocol. Process Biochem. 2014, 49, 604–616. [Google Scholar] [CrossRef]
- De Maria, L.; Vind, J.; Oxenbøll, K.M.; Svendsen, A.; Patkar, S. Phospholipases and their industrial applications. Appl. Microbiol. Biotechnol. 2007, 74, 290–300. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, J.C.S.; Barbosa, O.; Ortiz, C.; Berenguer-Murcia, A.; Rodrigues, R.C.; Fernandez-Lafuente, R. Importance of the support properties for immobilization or purification of enzymes. ChemCatChem 2015, 7, 2413–2432. [Google Scholar] [CrossRef]
- Sanchez, A.; Cruz, J.; Rueda, N.; Dos Santos, J.C.S.; Torres, R.; Ortiz, C.; Villalonga, R.; Fernandez-Lafuente, R. Inactivation of immobilized trypsin under dissimilar conditions produces trypsin molecules with different structures. RSC Adv. 2016, 6, 27329–27334. [Google Scholar] [CrossRef]
- Hernandez, K.; Fernandez-Lafuente, R. Control of protein immobilization: Coupling immobilization and site-directed mutagenesis to improve biocatalyst or biosensor performance. Enzyme Microb. Technol. 2011, 48, 107–122. [Google Scholar] [CrossRef] [PubMed]
- Rueda, N.; Dos Santos, C.S.; Rodriguez, M.D.; Albuquerque, T.L.; Barbosa, O.; Torres, R.; Ortiz, C.; Fernandez-Lafuente, R. Reversible immobilization of lipases on octyl-glutamic agarose beads: A mixed adsorption that reinforces enzyme immobilization. J. Mol. Catal. B Enzym. 2016, 128, 10–18. [Google Scholar] [CrossRef]
- Fuentes, M.; Batalla, P.; Grazu, V.; Pessela, B.C.C.; Mateo, C.; Montes, T.; Hermoso, J.A.; Guisan, J.M.; Fernandez-Lafuente, R. Mixed ion exchange supports as useful ion exchangers for protein purification: Purification of penicillin G acylase from Escherichia coli. Biomacromolecules 2007, 8, 703–707. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Dong, X.; Sun, Y. Enhanced protein adsorption and facilitated refolding of like-charged protein with highly charged silica nanoparticles fabricated by sequential double modifications. Langmuir 2015, 31, 655–658. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.-Z.; Dong, X.-Y.; Sun, Y. Ion-exchange resins greatly facilitate refolding of like-charged proteins at high concentrations. Biotechnol.Bioeng. 2011, 108, 1068–1077. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Du, W.-J.; Dong, X.-Y.; Sun, Y. Integrative refolding and purification of histidine-tagged protein by like-charge facilitated refolding and metal-chelate affinity adsorption. J. Chromat. A 2014, 1344, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.-L.; Dong, X.-Y.; Sun, Y.J. Ion-exchange resins facilitate like-charged protein refolding: Effects of porous solid phase properties. J. Chromat. A 2012, 122, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.-Y.; Yu, L.-L.; Dong, X.-Y.; Sun, Y. Mono-sized microspheres modified with poly(ethylenimine) facilitate the refolding of like-charged lysozyme. Reac. Funct. Polym. 2012, 72, 889–896. [Google Scholar] [CrossRef]
- Barbosa, O.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Rodrigues, R.C.; Fernandez-Lafuente, R. Glutaraldehyde in bio-catalysts design: A useful crosslinker and a versatile tool in enzyme immobilization. RSC Adv. 2014, 4, 1583–1600. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Fernández-Lafuente, R. Modifying enzyme activity and selectivity by immobilization. Chem. Soc. Rev. 2013, 42, 6290–6307. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M. Rapid and sensitive method for quantification of microgram quantities of protein utilizing principle of protein-dye-binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Pessela, B.C.C.; Munilla, R.; Betancor, L.; Fuentes, M.; Carrascosa, A.V.; Vian, A.; Fernandez-Lafuente, R.; Guisán, J.M. Ion exchange using poorly activated supports, an easy way for purification of large proteins. J. Chromat. A 2004, 1034, 155–159. [Google Scholar] [CrossRef]
- Fuentes, M.; Mateo, C.; Pessela, B.C.C.; Guisán, J.M.; Fernandez-Lafuente, R. Purification, stabilization, and concentration of very weak protein-protein complexes: Shifting the association equilibrium via complex selective adsorption on lowly activated support. Proteomics 2005, 5, 4062–4069. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, M.; Pessela, B.C.C.; Mateo, C.; Munilla, R.; Guisán, J.M.; Fernandez-Lafuente, R. Detection and purification of two antibody-antigen complexes via selective adsorption on lowly activated anion exchangers. J. Chromat. A 2004, 1059, 89–94. [Google Scholar] [CrossRef]
- Fuentes, M.; Pessela, B.C.C.; Mateo, C.; Palomo, J.M.; Batalla, P.; Fernández-Lafuente, R.; Guisán, J.M. Adsorption behavior of bovine serum albumin on lowly activated anionic exchangers suggests a new strategy for solid-phase proteomics. Biomacromolecules 2006, 7, 1357–1361. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, K.; Garcia-Galan, C.; Fernandez-Lafuente, R. Simple and efficient immobilization of lipase B from Candida antarctica on porous styrene-divinylbenzene beads. Enzyme Microb. Technol. 2011, 49, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, K.; Garcia-Verdugo, E.; Porcar, R.; Fernandez-Lafuente, R. Hydrolysis of triacetin catalyzed by immobilized lipases: Effect of the immobilization protocol and experimental conditions on diacetin yield. Enzyme Microb. Technol. 2011, 48, 510–517. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Not Available.









| Biocatalyst | Experimental Conditions | ||
|---|---|---|---|
| pH 5.0 | pH 7.0 | pH 9.0 | |
| OCCALA | 360 ± 10 | 60 ± 5 | 42 ± 4 |
| OCEDACALA | 240 ± 10 | 18 ± 2 | 90 ± 9 |
| OCHDACALA | 120 ± 11 | 15 ± 1 | 24 ± 3 |
| EDACALA | 120 ± 8 | 12 ± 1 | 15 ± 2 |
| HDACALA | 120 ± 9 | 9 ± 1 | 12 ± 1 |
| OCCALB | 120 ± 10 | 300 ± 18 | 60 ± 5 |
| OCEDACALB | 18 ± 2 | 300 ± 20 | 300 ± 31 |
| OCHDACALB | 12 ± 1 | 108 ± 9 | 180 ± 19 |
| OCCRL | 180 ± 11 | 60 ± 4 | 48 ± 5 |
| OCEDACRL | 12 ± 1 | 18 ± 3 | 12 ± 2 |
| OCHDACRL | 12 ± 1 | 18 ± 2 | 360 ± 14 |
| EDACRL | 240 (100%) * | 240 (100%) * | 420 ± 21 |
| HDACRL | 240 (100%) * | 240 (100%) * | 420 ± 30 |
| OCRML | 360 ± 20 | 330 ± 32 | 720 ± 75 |
| OCEDARML | 210 ± 15 | 108 ± 12 | 29 ± 1 |
| OCHDARML | 60 ± 7 | 48 ± 4 | 45 ± 5 |
| EDARML | 360 (100%) * | 360 (100%) * | 120 ± 4 |
| HDARML | 360 (100%) * | 360 (70%) * | 18 ± 3 |
| OCTLL | 300 ± 25 | 480 (80%) * | 72 ± 6 |
| OCEDATLL | 1440 (80%) * | 10 ± 1 | 180 ± 11 |
| OCHDATLL | 1440 (80%) * | 228 ± 15 | 90 ± 10 |
| EDATLL | 1440 (80%) * | 60 ± 6 | 30 ± 2 |
| HDATLL | 1440 (80%) * | 60 ± 5 | 24 ± 3 |
| OCLU | 18 ± 1 | 24 ± 5 | 60 ± 11 |
| OCEDALU | 240 ± 13 | 180 ± 12 | 600 ± 90 |
| OCHDALU | 240 ± 12 | 180 ± 21 | 600 ± 80 |
| EDALU | 180 (80%) * | 180 (100%) * | 1440 (100%) * |
| HDALU | 180 (100%) * | 180 (100%) * | 1440 (100%) * |
| Biocatalyst | Experimental Conditions | |||
|---|---|---|---|---|
| ACN 25% | DMSO 80% | ACN 40% | Dioxane 60% | |
| OCCALA | - | 210 ± 18 | - | - |
| OCEDACALA | - | 48 ± 3 | - | - |
| OCHDACALA | - | 24 ± 3 | - | - |
| EDACALA | - | 15 ± 1 | - | - |
| HDACALA | - | 15 ± 2 | - | - |
| OCCALB | - | 150 ± 11 | - | - |
| OCEDACALB | - | 162 ± 10 | - | - |
| OCHDACALB | - | 150 ± 10 | - | - |
| OCCRL | - | - | 30 ± 2 | - |
| OCEDACRL | - | - | 18 ± 1 | - |
| OCHDACRL | - | - | 18 ± 2 | - |
| EDACRL | - | - | 28 ± 3 | - |
| HDACRL | - | - | 42 ± 3 | - |
| OCRML | 24 ± 2 | - | - | - |
| OCEDARML | 12 ± 1 | - | - | - |
| OCHDARML | 12 ± 2 | - | - | - |
| EDARML | 12 ± 1 | - | - | - |
| HDARML | 18 ± 2 | - | - | - |
| OCTLL | - | - | - | 348 ± 33 |
| OCEDATLL | - | - | - | 1440 (80%) * |
| OCHDATLL | - | - | - | 1440 ± 100 |
| EDATLL | - | - | - | 1440 (100%) * |
| HDATLL | - | - | - | 1440 (100%) * |
| OCLU | 15 ± 2 | - | - | - |
| OCEDALU | 102 ± 9 | - | - | - |
| OCHDALU | 84 ± 5 | - | - | - |
| EDALU | 18 ± 1 | - | - | - |
| HDALU | 30 ± 2 | - | - | - |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rueda, N.; Albuquerque, T.L.; Bartolome-Cabrero, R.; Fernandez-Lopez, L.; Torres, R.; Ortiz, C.; Dos Santos, J.C.S.; Barbosa, O.; Fernandez-Lafuente, R. Reversible Immobilization of Lipases on Heterofunctional Octyl-Amino Agarose Beads Prevents Enzyme Desorption. Molecules 2016, 21, 646. https://doi.org/10.3390/molecules21050646
Rueda N, Albuquerque TL, Bartolome-Cabrero R, Fernandez-Lopez L, Torres R, Ortiz C, Dos Santos JCS, Barbosa O, Fernandez-Lafuente R. Reversible Immobilization of Lipases on Heterofunctional Octyl-Amino Agarose Beads Prevents Enzyme Desorption. Molecules. 2016; 21(5):646. https://doi.org/10.3390/molecules21050646
Chicago/Turabian StyleRueda, Nazzoly, Tiago L. Albuquerque, Rocio Bartolome-Cabrero, Laura Fernandez-Lopez, Rodrigo Torres, Claudia Ortiz, Jose C. S. Dos Santos, Oveimar Barbosa, and Roberto Fernandez-Lafuente. 2016. "Reversible Immobilization of Lipases on Heterofunctional Octyl-Amino Agarose Beads Prevents Enzyme Desorption" Molecules 21, no. 5: 646. https://doi.org/10.3390/molecules21050646