An Electrostatically Self-Assembled Ternary Nanocomplex as a Non-Viral Vector for the Delivery of Plasmid DNA into Human Adipose-Derived Stem Cells
Abstract
:1. Introduction
2. Results and Discussion
2.1. Characterization of pDNA/PEI Binary and pDNA/PEI/HA Ternary Nanocomplexes
2.2. Viability of hASCs Treated with pDNA/PEI Binary and pDNA/PEI/HA Ternary Nanocomplexes
2.3. Transfection Efficiency of pDNA/PEI Binary and pDNA/PEI/HA Ternary Nanocomplexes
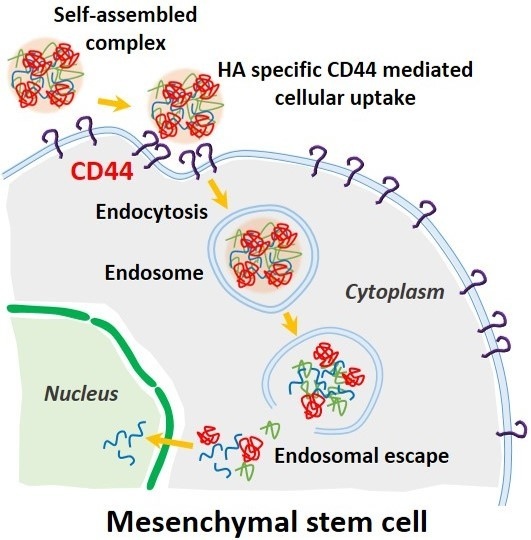
2.4. CD44 Targeting Effect of pDNA/PEI/HA Ternary Nanocomplexes
3. Materials and Methods
3.1. Materials
3.2. hASCs Culture
3.3. Preparation and Characterization of Ternary Nanocomplexes
3.4. Gel Retardation Assay
3.5. Cell Viability Assay
3.6. Determining Transfection Efficiency by Flow Cytometry
3.7. Fluorescence Microscopy Imaging
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Pittenger, M.F.; Mackay, A.M.; Beck, S.C.; Jaiswal, R.K.; Douglas, R.; Mosca, J.D.; Moorman, M.A.; Simonetti, D.W.; Craig, S.; Marshak, D.R. Multilineage potential of adult human mesenchymal stem cells. Science 1999, 284, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Bianco, P.; Robey, P.G.; Simmons, P.J. Mesenchymal stem cells: Revisiting history, concepts, and assays. Cell Stem Cell 2008, 2, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Baksh, D.; Song, L.; Tuan, R.S. Adult mesenchymal stem cells: Characterization, differentiation, and application in cell and gene therapy. J. Cell. Mol. Med. 2004, 8, 301–316. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.H.; Tuan, R.S. Mesenchymal stem cells in arthritic diseases. Arthritis Res. Ther. 2008, 10. [Google Scholar] [CrossRef] [PubMed]
- Marolt, D.; Knezevic, M.; Novakovic, G.V. Bone tissue engineering with human stem cells. Stem Cell Res. Ther. 2010, 1. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.L.; Pandita, D.; Rodrigues, J.; Pego, A.P.; Granja, P.L.; Tomas, H. Non-viral gene delivery to mesenchymal stem cells: Methods, strategies and application in bone tissue engineering and regeneration. Curr. Gene Ther. 2011, 11, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.; O’Donoghue, K.; de la Fuente, J.; Roberts, I.A.; Kumar, S.; Morgan, J.E.; Fisk, N.M. Human fetal mesenchymal stem cells as vehicles for gene delivery. Stem Cells 2005, 23, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Godbey, W.T. Viral vectors for gene delivery in tissue engineering. Adv. Drug Deliv. Rev. 2006, 58, 515–534. [Google Scholar] [CrossRef] [PubMed]
- Nayerossadat, N.; Maedeh, T.; Ali, P.A. Viral and nonviral delivery systems for gene delivery. Adv. Biomed. Res. 2012, 1. [Google Scholar] [CrossRef] [PubMed]
- Collins, M.N.; Birkinshaw, C. Hyaluronic acid based scaffolds for tissue engineering—A review. Carbohydr. Polym. 2013, 92, 1262–1279. [Google Scholar] [CrossRef] [PubMed]
- Li, S.D.; Huang, L. Non-viral is superior to viral gene delivery. J. Control. Release 2007, 123, 181–183. [Google Scholar] [CrossRef] [PubMed]
- Babister, J.C.; Tare, R.S.; Green, D.W.; Inglis, S.; Mann, S.; Oreffo, R.O.C. Genetic manipulation of human mesenchymal progenitors to promote chondrogenesis using “bead-in-bead” polysaccharide capsules. Biomaterials 2008, 29, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Jeon, O.; Yang, H.S.; Lee, T.-J.; Kim, B.-S. Heparin-conjugated polyethylenimine for gene delivery. J. Control. Release 2008, 132, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.F.; Tseng, M.T.; Ho, Y.C.; Wei, M.C.; Liao, Z.X.; Sung, H.W. Mechanisms of cellular uptake and intracellular trafficking with chitosan/DNA/poly(gamma-glutamic acid) complexes as a gene delivery vector. Biomaterials 2011, 32, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Via, A.G.; Frizziero, A.; Oliva, F. Biological properties of mesenchymal Stem Cells from different sources. Muscles Ligaments Tendons J. 2012, 2, 154–162. [Google Scholar] [PubMed]
- Hornof, M.; de la Fuente, M.; Hallikainen, M.; Tammi, R.H.; Urtti, A. Low molecular weight hyaluronan shielding of DNA/PEI polyplexes facilitates CD44 receptor mediated uptake in human corneal epithelial cells. J. Gene Med. 2008, 10, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Kircheis, R.; Wightman, L.; Wagner, E. Design and gene delivery activity of modified polyethylenimines. Adv. Drug Deliv. Rev. 2001, 53, 341–358. [Google Scholar] [CrossRef]
- Kichler, A.; Leborgne, C.; Coeytaux, E.; Danos, O. Polyethylenimine-mediated gene delivery: A mechanistic study. J. Gene Med. 2001, 3, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Ahn, H.H.; Lee, J.H.; Kim, K.S.; Lee, J.Y.; Kim, M.S.; Khang, G.; Lee, I.W.; Lee, H.B. Polyethyleneimine-mediated gene delivery into human adipose derived stem cells. Biomaterials 2008, 29, 2415–2422. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Luo, X.; Zhao, Y.; Han, L.; Zeng, X.; Feng, M.; Pan, S.; Peng, H.; Wu, C. Influence of the polyanion on the physico-chemical properties and biological activities of polyanion/DNA/polycation ternary polyplexes. Acta Biomater. 2012, 8, 3014–3026. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds are available from the authors.








| Formulation | Size (nm) | PDI | Zeta Potential (mV) |
|---|---|---|---|
| pDNA/PEI (1:8) | 626 ± 106 | 0.260 | +12.24 ± 0.27 |
| pDNA/PEI/HA (1:8:5) | 806 ± 170 | 0.232 | −7.49 ± 0.44 |
| pDNA/PEI/HA (1:8:10) | 649 ± 150 | 0.297 | −12.40 ± 0.55 |
| pDNA/PEI/HA (1:8:20) | 683 ± 152 | 0.177 | −16.05 ± 0.25 |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, S.-H.; Noh, Y.-W.; Cho, M.Y.; Lim, Y.T. An Electrostatically Self-Assembled Ternary Nanocomplex as a Non-Viral Vector for the Delivery of Plasmid DNA into Human Adipose-Derived Stem Cells. Molecules 2016, 21, 572. https://doi.org/10.3390/molecules21050572
Cho S-H, Noh Y-W, Cho MY, Lim YT. An Electrostatically Self-Assembled Ternary Nanocomplex as a Non-Viral Vector for the Delivery of Plasmid DNA into Human Adipose-Derived Stem Cells. Molecules. 2016; 21(5):572. https://doi.org/10.3390/molecules21050572
Chicago/Turabian StyleCho, Sun-Hee, Young-Woock Noh, Mi Young Cho, and Yong Taik Lim. 2016. "An Electrostatically Self-Assembled Ternary Nanocomplex as a Non-Viral Vector for the Delivery of Plasmid DNA into Human Adipose-Derived Stem Cells" Molecules 21, no. 5: 572. https://doi.org/10.3390/molecules21050572